Figure 1.
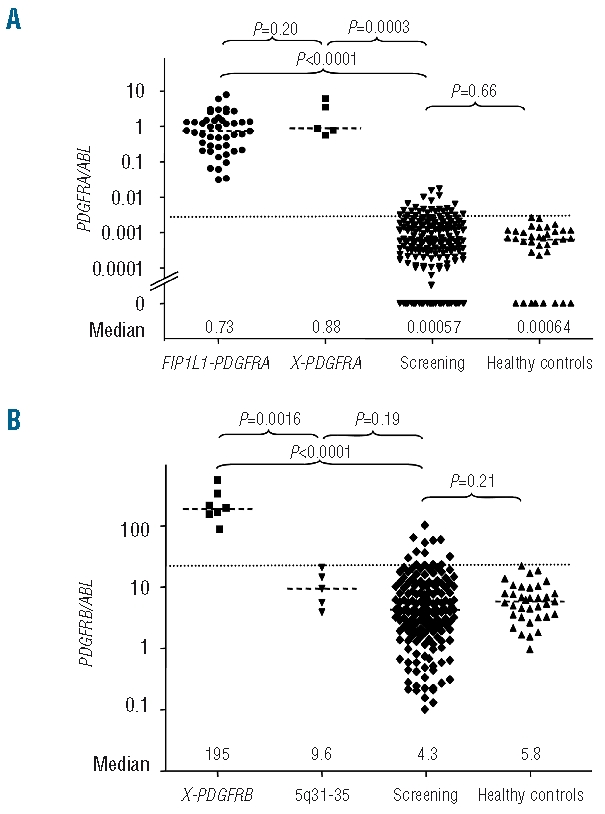
(A) Patients with PDGFRA fusion genes showed significantly increased PDGFRA expression levels (PDGFRA/ABL) as compared to patients with non-reactive eosinophilia without known molecular aberrations (n=191) and healthy controls (n=35). The expression level was not different between patients with FIP1L1-PDGFRA (n=45) and alternative PDGFRA fusion genes (X-PDGFRA: BCR-PDGFRA, CDK5RAP2-PDGFRA, ETV6-PDGFRA, KIF5B-PDGFRA). The cut-off point for overexpression of PDGFRA was determined for PDGFRA at a ratio of 0.030 PDGFRA/ABL (35 healthy controls, mean + 3 SD, dotted line). PDGFRA expression levels were not different between patients with non-reactive eosinophilia and healthy controls. (B) Patients with different PDGFRB fusion genes (X-PDGFRB [n=7]: ETV6-PDGFRB, CCDC6-PDGFRB, GIT2-PDGFRB, GPIAP1-PDGFRB and MYO18A-PDGFRB showed significantly increased PDGFRB expression levels compared to patients with non-reactive eosinophilia without known molecular aberrations and healthy controls. In the screening group, 13 patients showed significant overexpression of PDGFRB. In one of these patients, with uninformative cytogenetics and an excellent response to imatinib a new SART3-PDGFRB fusion gene was identified by 5′-RACE-PCR. No increased PDGFRB expression was found in five patients with chromosomal aberrations and involvement of chromosome bands 5q31–32. In four of these cases, alternative fusion genes with involvement of ETV6, NPM1, MLF1 and ACSL6 could be confirmed by RT-PCR. The cut-off point for overexpression was determined at a ratio of 23 PDGFRB/ABL (35 healthy controls, mean+ 3 SD, dotted line). PDGFRB expression levels were not different between patients with non-reactive eosinophilia and healthy controls.
