Abstract
Background
Multiple myeloma is a life-threatening disease and despite the introduction of stem cell transplantation and novel agents such as thalidomide, lenalidomide, and bortezomib most patients will relapse and develop chemoresistant disease. Therefore, alternative therapeutic modes for myeloma are needed and cancer-testis antigens such as MAGE-C1/CT7 and MAGE-A3 have been suggested to represent a class of tumor-specific proteins particularly suited for targeted immunotherapies. Surprisingly, the biological role of cancer-testis genes in myeloma remains poorly understood.
Design and Methods
We performed the first investigation of the function of two cancer-testis antigens most commonly expressed in myeloma, MAGE-C1/CT7 and MAGE-A3, using an RNA interference-based gene silencing model in myeloma cell lines. Functional assays were used to determine changes in proliferation, cell adhesion, chemosensitivity, colony formation, and apoptosis resulting from gene-specific silencing.
Results
We show that the investigated genes are not involved in regulating cell proliferation or adhesion; however, they play an important role in promoting the survival of myeloma cells. Accordingly, knock-down of MAGE-C1/CT7 and MAGE-A3 led to the induction of apoptosis in the malignant plasma cells and, importantly, both genes were also essential for the survival of clonogenic myeloma precursors. Finally, silencing of cancer-testis genes further improved the response of myeloma cells to conventional therapies.
Conclusions
Cancer-testis antigens such as MAGE-C1/CT7 and MAGE-A3 play an important role in promoting the survival of myeloma cells and clonogenic precursors by reducing the rate of spontaneous and chemotherapy-induced apoptosis and might, therefore, represent attractive targets for novel myeloma-specific therapies.
Keywords: cancer-testis antigens, gene function, RNAi, apoptosis, tumor immunology, multiple myeloma, stem cell transplantation
Introduction
Multiple myeloma (MM), the second most common hematologic malignancy, is a B-cell neoplasia characterized by the clonal proliferation of malignant plasma cells in the bone marrow, with clinical consequences that include anemia, lytic bone lesions, renal dysfunction, hypercalcemia, hypogammaglobulinemia, and increased risk of infection.1 Clinical remissions of MM are commonly achieved by high-dose therapy followed by autologous stem cell transplantation as well as by the administration of recently introduced drugs such as bortezomib, thalidomide, and lenalidomide. However, the median survival has still not improved beyond 4–5 years,2 mainly because of the persistence of therapy-resistant minimal residual disease eventually leading to relapse and death in the vast majority of MM patients. Accordingly, alternative therapeutic modes, which more specifically and effectively target remaining myeloma cells, are needed.
The development of therapies specifically aiming at MM requires the identification of appropriate myeloma-specific structures and cancer-testis (CT) antigens have been suggested to represent a class of tumor proteins particularly suited for targeted therapies such as T-cell-mediated immunotherapy.3 CT antigens are typically expressed in a wide range of tumor types but not in any healthy tissues other than testis and placenta; the expression of these antigens is regulated by epigenetic mechanisms such as promoter methylation and histone acetylation.4 Among CT antigens, the melanoma antigen genes (MAGE) represent the largest gene family. The original MAGE genes included MAGE-A, -B, and –C, the so-called CT-X-MAGE proteins, which are localized in clusters on the X chromosome and encode proteins with 50 to 99% sequence identity to each other.5 In addition, a number of autosomal gene families named MAGE-D through MAGE-L appear to be more widely expressed in normal tissues.3 Generally, MAGE genes are defined by a shared MAGE homology domain, a well-conserved domain of about 200 amino acids.6 The MAGE homology domain does not contain any regions with significant homology with other known proteins, but it has been indicated that this domain represents an important site for protein-protein interactions.7
CT antigen expression is a rare event in most hematologic malignancies including leukemias and non-Hodgkin’s B-cell lymphomas.8,9 However, MM probably represents the malignancy with the richest CT antigen expression of all human cancers and the two CT antigens most frequently expressed in MM are MAGE-A3 and MAGE-C1/CT7.10–15 MAGE-A3 is also expressed in a wide variety of solid tumors. It is a cytoplasmic protein with a molecular weight of 35 kDa16 which was discovered analyzing CD8+ T-cell reactivity of a cancer patient against an autologous melanoma cell line.17 MAGE-C1/CT7 was identified simultaneously by representational difference analysis and serological analysis of recombinant cDNA expression libraries (SEREX).18,19 MAGE-C1/CT7 is about 800 amino acids longer than other MAGE proteins and contains a large number of unique short repetitive sequences in front of the MAGE homology domain.19 The MAGE-C1/CT7 gene is located on band Xq26 whereas the MAGE-A genes are located on Xq28.18–20 Like other MAGE genes, MAGE-C1/CT7 is specifically expressed in a variety of solid tumors, although less frequently than MAGE-A3.19
As in the case of most solid tumors, expression of CT antigens is increased in a coordinate manner in advanced stages of myeloma10,11,21,22 and we have recently shown that once CT antigen expression occurs in MM, it persists throughout the whole course of the disease.22 These observations point to a possible role of CT antigens in promoting the progression of MM, an idea which is further supported by recent microarray analyses showing an association between CT antigen expression, including the expression of MAGE genes, and a more aggressive course of the disease as well as reduced survival.14,21,23 Comparable results were described in a study focusing on the expression of CT antigens belonging to the SSX gene family in which the expression of these genes was related to reduced overall survival of MM patients.24 Notably, our findings and the observations of others indicate that, of all CT antigens, the expression of MAGE-A3 and, particularly, of MAGE-C1/CT7, has the strongest impact on the prognosis of myeloma.22,25
Although these collected data suggest that the expression of CT antigens might contribute to tumor progression, particularly in MM, their biological role in germ line cells and malignancies remains poorly understood. In this study, we performed the first investigation of the function of the two CT antigens most commonly expressed in myeloma, MAGE-C1/CT7 and MAGE-A3, describing an important role of these proteins in promoting the survival of myeloma cells. We believe that targeting such proteins might hit an Achilles’ heel of the malignancy leading to an improved outcome or even cures following standard therapy.
Design and Methods
Cell lines
Myeloma cell lines MOLP-8, RPMI-8226, KMS-12-BM, EJM; IM-9, NCI-H929, OPM-2, and LP-1 were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). Cell lines Brown and SK-007 were provided by the New York branch of the Ludwig Institute for Cancer Research. Lines were maintained in RPMI 1640 with 10% fetal calf serum (FCS) and penicillin/streptomycin.
Western blot
For some experiments myeloma cell lines were sorted based on their expression of CD138 using a FACSAria cell-sorting system (BD Biosciences, Heidelberg, Germany). Whole cell protein extracts were prepared from cell lines and western blotting was performed as previously described.26 For all target proteins analyzed, appropriate blocking studies were performed using recombinant proteins in order to confirm specificity of the staining.
Gene silencing using transfection with stealth RNA interference
BLOCK-iT fluorescein-labeled double-stranded DNA, scrambled control short interfering RNA (siRNA), and stealth siRNA targeting MAGE-C1/CT7 or MAGE-A3 were purchased from Invitrogen (Karlsruhe, Germany). Myeloma cell lines were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations and transfection efficiency was determined by fluorescence microscopy using fluorescein-labeled double-stranded DNA.
3-(4,5-dimethylthiaxol-2-yl)-2,5-diphenyltetrazolium bromide viability assay
Relative numbers of viable cells were assessed by a 3-(4,5-dimethylthiaxol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay27 72 h after transfection with CT antigen-specific siRNA or with scrambled control siRNA. Myeloma cells were cultured overnight and subsequently treated with bortezomib or melphalan 12 h before analyses. MTT was added to each well and cells were incubated for 4 h. Absorbance was read at 570 nm using a spectrophotometer.
Measurement of apoptosis
The terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was done according to the manufacturer’s recommendations (Millipore, Billerica, MA, USA). Briefly, cells were fixed in 1% paraformaldehyde, and frozen until staining at −20°C using 70% ethanol. For staining, cells were incubated sequentially in staining solution for 1 h, and propidium iodide/RNAse A solution for 30 min. For annexin V staining, cells were incubated with annexin V-fluorescein isothiocyanate in annexin binding buffer. Analysis by flow cytometry was performed within the next 3 h.
Colony formation assay
Myeloma cell lines Molp-8 and RPMI-8226 were plated at 1000 cells/mL of methylcellulose medium (StemCell Technologies, Cologne, Germany) in a 6-well culture dish. Plates were incubated at 37°C and colonies consisting of more than 40 cells were counted 7 to 10 days after the culture had been started.
Analysis of cell proliferation
Cell proliferation was measured by staining with carboxyfluorescein diacetate succinimidyl ester (CFSE) or using a plate-based system. Myeloma cells were pulsed with bromodeoxyuridine (BrdU) and peroxidase-labeled anti-BrdU was added to the cells. Resulting immune complexes were detected by a substrate reaction, and absorbance was read at 450 nm. For FACS analysis, myeloma cells were stained intracellularly with CFSE and CFSE-related green fluorescence intensity was measured by flow cytometry.
Cell adhesion assay
The myeloma cells were resuspended 72 h post-transfection in RPMI-1640 + 0.2% bovine serum albumin (adhesion medium) and were incubated for 30 min at 37°C. Cells were then resuspended in adhesion medium and were incubated for 4 h in 96-well plates coated with fibronectin. Unbound cells were removed by four washes and absorbance with Chromogen Substrate Solution (Alpco, Salem, NH, USA) was read at 450/620nm.
Statistical analysis
Statistical analyses were performed using SPSS software. The Mann-Whitney U test was used to calculate differences between different experimental conditions. Differences were regarded significant if the P value was less than 0.05.
The Design and Methods of the study are described in more detail in the Online Supplementary Appendix.
Results
Cancer-testis antigens MAGE-A3/A6 and MAGE-C1/CT7 are constitutively expressed in multiple myeloma cell lines and expression is specifically suppressed by transfection with interfering RNA
We have previously described very frequent expression of CT antigens in myeloma cell lines as well as in patients’ samples at the RNA level.10 As a prerequisite for the functional analysis of MAGE-A3 and MAGE-C1/CT7 expression in MM, in this study we investigated the protein expression of both genes in ten different MM cell lines. Considering high sequence homology of MAGE-A proteins, we first confirmed, in a western blot assay, specific binding of antibody M3H67 to recombinant full-length MAGE-A3 protein, but not to MAGE-A2, MAGE-A4, or MAGE-A12 recombinant proteins (data not shown). Recombinant MAGE-A6 protein was not available to us; however, as MAGE-A3 and MAGE-A6 show 95% protein sequence homology, binding of M3H67 to MAGE-A6 seems very likely and, therefore, the protein recognized in the respective immunoassays was designated MAGE-A3/A6. Western blot analyses revealed that MAGE-A3/A6 and MAGE-C1/CT7 were constitutively expressed in every single cell line (Figure 1A) further supporting a functional role of these genes in myeloma. A comparably high transfection efficiency (70–80%) was generally achieved using the MAGE-A3/A6- and MAGE-C1/CT7-expressing cell lines Molp-8 and RPMI-8226; these two cell lines were, therefore, preferably used for subsequent experiments.
Figure 1.
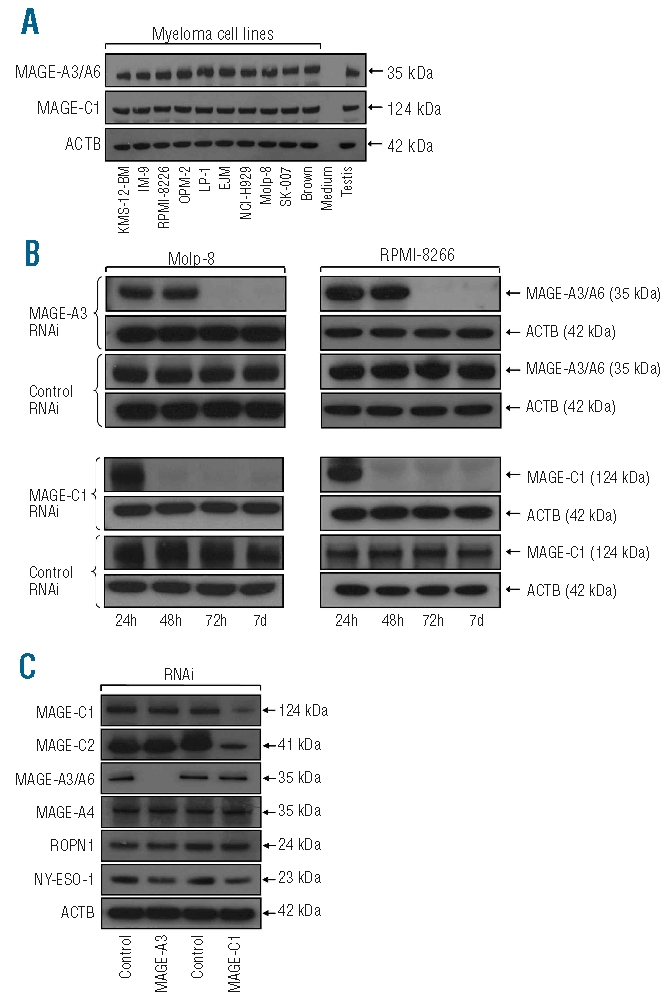
Cancer-testis genes MAGE-A3 and MAGE-C1/CT7 are constitutively expressed in myeloma cells and transfection with inhibitory RNAi results in gene-specific down-regulation of both proteins. (A) ten myeloma cell lines were investigated for the expression of MAGE-A3 and MAGE-C1/CT7 protein by western blot. β-actin (ACTB) served as an internal control for protein quality and protein lysate of normal human testis was used as a positive control. Notably, all ten myeloma cell lines showed constitutive expression of both CT antigens. (B) Myeloma cell lines Molp-8 (left column) and RPMI-8226 (right column) were transfected using two RNAi constructs specific for CT genes MAGE-A3 and MAGE-C1/CT7, respectively, or with scrambled control RNAi. Treatment resulted in knockdown of the expression of MAGE-A3 and MAGE-C1/CT7 protein in both cell lines starting 48–72 h after transfection as indicated by immunoblotting. Gene-silencing lasted at least until day 7 after transfection. (C) Importantly, treatment with gene-specific RNAi only affected the expression of the target genes and not the expression of other CT antigens tested (MAGE-A4, Ropporin-1, NY-ESO-1). The only exception was CT gene MAGE-C2/CT10, the expression of which was suppressed following transfection with RNAi targeting MAGE-C1/CT7. Comparable findings were observed following gene-silencing in cell line RPMI-8226 and using a second gene-specific RNAi construct (data not shown).
Since MAGE-A3/A6 and MAGE-C1/CT7 were present in all MM cell lines available to us, we could not investigate the function of these genes transfecting them into MAGE-negative MM lines. We, therefore, decided to perform knockdown experiments using interfering RNA and observed that two of the three siRNA constructs targeting different sequences of the respective gene’s mRNA showed a strong inhibitory effect leading to down-regulation of MAGE-A3/A6 and MAGE-C1/CT7 protein expression (Figure 1B). The two respective siRNA constructs showing an inhibitory effect were used in all subsequent experiments and the effects of transfection of myeloma cell lines Molp-8 and RPMI-8226 with one interfering RNA (RNAi) and scrambled control RNAi, respectively, are shown as examples in each figure. Knock-down efficiency for both genes reached its maximum at 72 h after transfection and lasted for at least 7 days. Importantly, analyzing whether silencing of MAGE-A3 or MAGE-C1/CT7 would have an effect on other MAGE family members or non-MAGE CT antigens, we found that RNAi transfection specifically affected the expression of the target genes and not the presence of other CT antigens such as MAGE-A4, ropporin-1, or NY-ESO-1 (Figure 1C). However, knockdown of MAGE-C1/CT7 seemed to have a slight down-regulating effect on the expression of MAGE-C2/CT10, a protein that shows a significant degree of homology with MAGE-C1/CT7.28
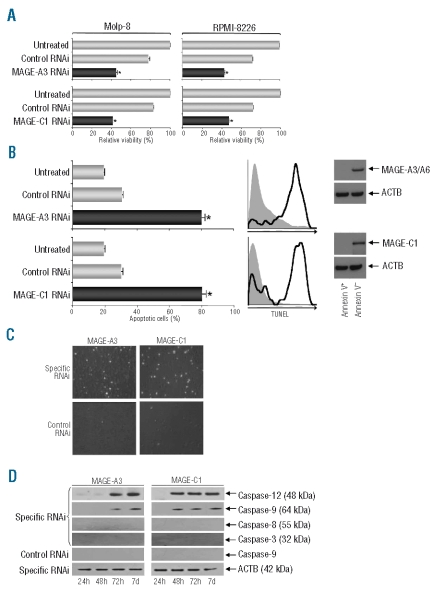
Cancer-testis antigens MAGE-A3 and MAGE-C1/CT7 protect myeloma cells from undergoing spontaneous apoptosis
Performing knockdown experiments we noticed a significant growth delay in cultures of both myeloma cell lines after transfection with both MAGE-specific RNAi constructs. Performing a classical MTT assay we confirmed that the numbers of viable cells were significantly lower in the cultures containing MM cells transfected with MAGE-A3- and MAGE-C1/CT7-specific RNAi but not with scrambled control RNAi (Figure 2A). In order to determine whether this was due to an anti-proliferative effect of gene knockdown or based on a pro-apoptotic effect of MAGE silencing, we next performed a TUNEL assay to demonstrate the proportion of apoptotic cells in cultures 72 h post-transfection. This assay revealed that, compared to untreated cells and myeloma cells transfected with control RNAi, silencing of MAGE-A3 and MAGE-C1/CT7 did indeed lead to the induction of spontaneous apoptosis in the malignant cells (Figure 2B). Remarkably, loss of the anti-apoptotic effect of MAGE-A3 and MAGE-C1/CT7 resulted in rates of apoptosis as high as 80% (Figure 2B). This finding was confirmed by microscopic analysis of MM cells 72 h post-transfection (Figure 2C). In order to determine whether MAGE genes had been efficiently downregulated in viable cells we transfected cell lines Molp-8 and RPMI-8226 with siRNA constructs specific for either MAGE gene. After 72 h we sorted viable and apoptotic cells according to annexin V-staining by FACS. Analyzing both populations by immunoblot we detected high levels of both MAGE genes in viable cells but no expression in apoptotic cells (Figure 2B). This suggests that the survival of a minority of myeloma cells is most likely based on the fact that not all cells could efficiently be transfected with the MAGE-specific siRNA. This idea is also supported by the average efficiency of siRNA transfection, which we had determined earlier.
Figure 2.
Silencing of CT genes MAGE-A3 or MAGE-C1/CT7 exerts a strong pro-apoptotic effect on myeloma cells. (A) Myeloma cell lines Molp-8 (left column) and RPMI-8226 (right column) were transfected with two RNAi constructs specific for CT genes MAGE-A3 or MAGE-C1/CT7. Numbers of viable myeloma cells transfected with RNAi specific for the target genes or of cells transfected with scrambled control RNAi were normalized to the number of viable untreated cells. Silencing of MAGE-A3 or MAGE-C1/CT7 resulted in significantly decreased cell viability at 72 h after transfection as indicated by an MTT assay. Results show mean values [+ standard error of means (SEM)] of three separate experiments and asterisks indicate statistically significant differences between untreated cells and cells transfected with scrambled control or one gene-specific RNAi (*P<0.05). (B) Percentages of apoptotic cells in Molp-8 cultures were analyzed in a flow cytometry-based TUNEL assay at 72 h after transfection with RNAi specific for CT genes MAGE-A3 or MAGE-C1/CT7. Bars indicate mean values (+SEM) of three separate experiments. Percentages of apoptotic myeloma cells transfected with specific or scrambled control RNAi were compared to those of untreated cells and asterisks indicate statistically significant differences (*P<0.05). Silencing of MAGE-A3-or MAGE-C1/CT7, resulted in significantly and specifically increased rates of apoptosis. Histograms indicate findings of a representative experiment analyzing TUNEL expression in cells transfected with control RNAi (gray area) or gene-specific RNAi (black line). Cell sorting according to annexin V-staining and subsequent immunoblot indicated that, in contrast to the apoptotic population, viable cells showed significant protein expression of either target gene following specific siRNA transfection. Comparable findings were observed following gene-silencing in cell line RPMI-8226 (data not shown). (C) Microscopic analysis of one representative experiment shows strong expression of green fluorescence in TUNEL-positive myeloma cells following silencing of MAGE-A3 or MAGE-C1/CT7. (D) Increased apoptosis in Molp-8 myeloma cells was accompanied by an increased expression of caspases 12 and 9, but not caspase-8 or caspase-3, in a western blot assay at 72 h after MAGE-A3 or MAGE-C1/CT7 knockdown.
When we analyzed which factors might be involved in mediating spontaneous apoptosis following knockdown of MAGE-A3 and MAGE-C1/CT7, we observed specific up-regulation of caspase-9 and caspase-12 expression starting 48 h after transfection with gene-specific RNAi but not with control RNAi (Figure 2D). Interestingly, we did not detect upregulated expression of caspases 3 or 8, pointing to a dominant role of the former factors in mediating apoptosis in myeloma cells following silencing of MAGE-A3 and MAGE-C1/CT7.
Cancer-testis antigens MAGE-A3/A6 and MAGE-C1/CT7 are expressed in clonogenic myeloma precursors and protect them from apoptosis
It has long been suggested that in the case of MM, a small number of cycling precursors generates and replenishes a large number of non-dividing myeloma cells. Dividing precursor cells are thought to be resistant to chemotherapy and to be responsible for relapses commonly seen in this malignancy. Clonogenic precursors are present in the patients’ bone marrow and also among myeloma cell lines and can be sorted based on the fact that they, in contrast to the bulk of malignant cells, do not express surface molecule CD138.29 We analyzed the expression of CT antigens in CD138-positive MM cells and CD138-negative myeloma precursors sorted from bulk cultures of cell lines Molp-8 and KMS-12-BM and we found that both fractions strongly expressed MAGE-A3/A6 and MAGE-C1/CT7 (Figure 3A). Importantly, following silencing of MAGE-A3/A6 and MAGE-C1/CT7 in two different myeloma cell lines using two inhibitory RNAi constructs, we observed a significantly reduced number of cell colonies in a standard assay measuring clonogenic growth of myeloma precursors (Figure 3B). These results demonstrate that both MAGE genes not only have a promoting effect on the survival of common myeloma cells but also support anti-apoptotic effects in myeloma precursor cells replenishing the bulk of myeloma cells.
Figure 3.
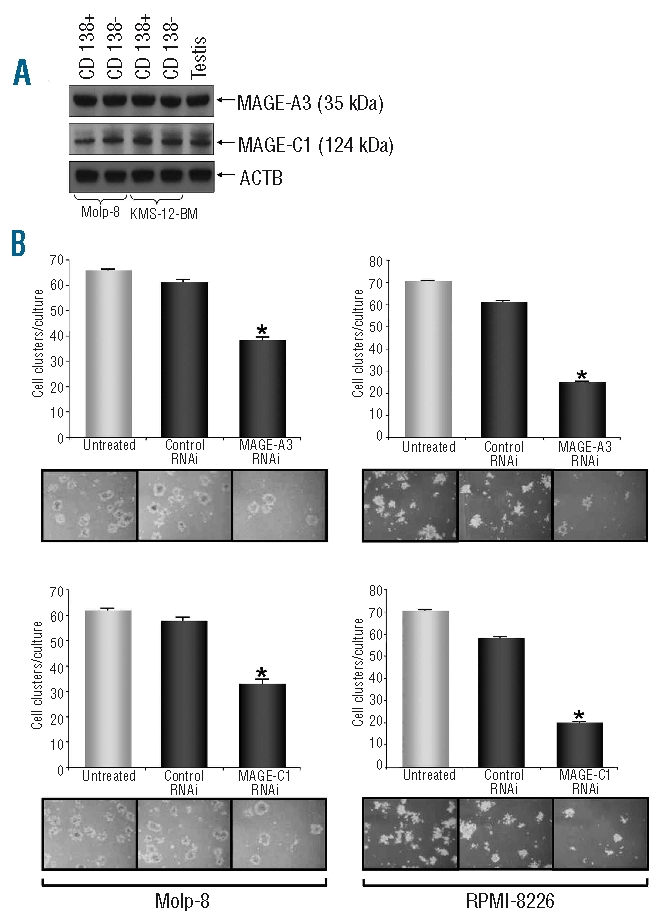
Silencing of CT genes MAGE-A3 or MAGE-C1/CT7 results in a decreased outgrowth of myeloma precursors in a colony formation assay. Previous studies have shown that myeloma precursors are also present in the bulk population of myeloma cell lines and that these cells can be identified by their lack of CD138 expression.29 FACS-sorting CD138+ and CD138- subpopulations we show here by western blot analysis that myeloma precursors derived from cell lines Molp-8 and KMS-12-BM as well as conventional myeloma plasma cells express MAGE-A3 and MAGE-C1/CT7 (A). In a clonogenic growth assay, colonies of myeloma precursors were counted 7–10 days after culture initiation using myeloma cell lines Molp-8 (left column) and RPMI-8226 (right column) separately transfected with two RNAi constructs specific for CT genes MAGE-A3 or MAGE-C1/CT7, respectively (B). Bars indicate mean values (+SEM) of three separate experiments. Numbers of colonies produced by myeloma cells transfected with RNAi specific for the target genes or of cells transfected with scrambled control RNAi were compared with colony numbers in cultures with untreated cells and asterisks indicate statistically significant differences (*P<0.05). Photos show results of representative analyses. Silencing of MAGE-A3- or MAGE-C1/CT7 resulted in significantly and specifically decreased outgrowth of myeloma precursors.
MAGE-A3 and MAGE-C1/CT7 promote multiple myeloma growth mainly by preventing apoptosis and not by promoting proliferation or cell adhesion
Next, we investigated whether effects on the proliferative function of myeloma cells or on their adhesion capacity might contribute to the growth-promoting effect of MAGE-A3 and MAGE-C1/CT7. CT antigens, such as CAGE, promote the motility of tumor cells30 and it has been shown that knockdown of CT antigens belonging to the SSX family of genes in a melanoma cell line results in reduced migratory capacity of the tumor cells.31 However, performing an adhesion assay following silencing of MAGE-A3 or MAGE-.C1/CT7 in the myeloma cell line Molp-8 we did not observe any difference in adhesion capacity between cells transfected with gene-specific RNAi and cells transfected with control RNAi (data not shown).
Analyzing the proliferative activity of myeloma cells following knockdown of MAGE-A3 or MAGE-C1/CT7 (Figure 4) we observed only a slight suppressive effect of gene-specific RNAi transfection on cellular growth. However, we cannot completely rule out the possibility that increased rates of apoptosis during the culture period might have contributed to this effect on cell proliferation. In agreement with this idea, we found that gating on non-apoptotic cells in flow cytometry-based assay, myeloma cells transfected with MAGE-specific RNAi or control RNAi had both undergone multiple cell divisions at 72 h after transfection (data not shown).
Figure 4.
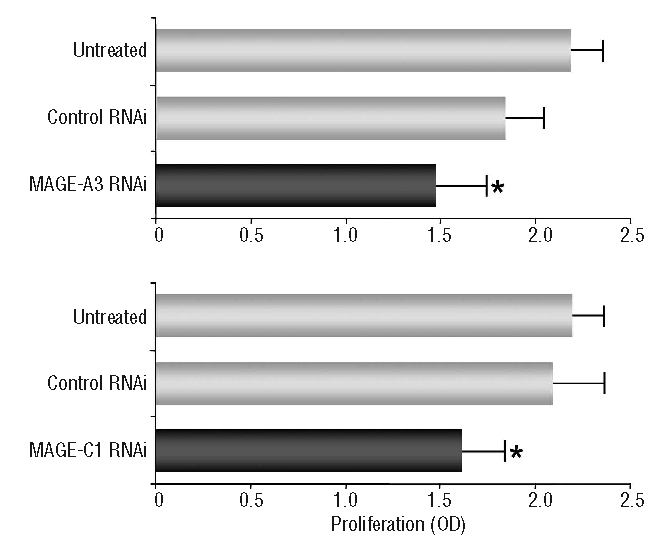
Silencing of CT genes MAGE-A3 or MAGE-C1/CT7 only exerts a minor influence on the proliferation of myeloma cells. Proliferation of myeloma cell line Molp-8 following silencing of MAGE-A3- or MAGE-C1/CT7 was assessed by a BrdU incorporation assay. Proliferation was measured 72 h after transfection with RNAi specific for the target gene or with scrambled control RNAi and results were compared to untreated cells. Bars show mean values (+SEM) of three separate experiments and asterisks indicate statistically significant differences between untreated cells and scrambled control or gene specific RNAi (*P<0.05).
Targeting of MAGE-A3 and MAGE-C1/CT7 further increases the therapeutic efficacy of conventional modes of therapy for multiple myeloma
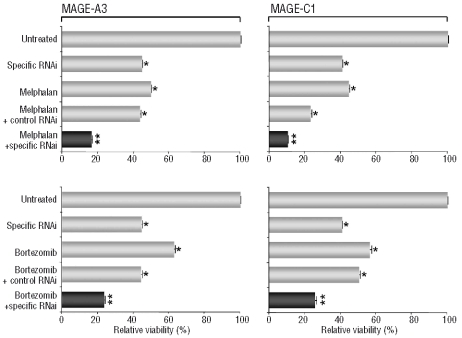
Resistance to conventional myeloma-specific therapies represents a severe problem in this hematologic malignancy. In order to explore whether MAGE-specific interventions could complement established modes of therapy for MM, we combined silencing of MAGE-A3 and MAGE-C1/CT7 with chemotherapeutic treatment or application of a ‘novel’ drug, namely the proteasome inhibitor bortezomib. A slight decrease of the relative number of viable cells following transfection with scrambled control RNAi was likely caused by an additional cytotoxic effect of the lipofection reagent used in this assay. Nevertheless, we observed that specific targeting of MAGE-A3 or MAGE-C1/CT7 more than doubled the therapeutic efficacy exerted by the cytotoxic agent melphalan (Figure 5A) or bortezomib (Figure 5B) alone in comparison to control RNAi. These findings indicate that a MAGE-specific therapy could represent an ideal adjunct to conventional modes of therapy for MM, preventing recurrences and leading to persistent responses in higher proportions of patients with this disease.
Figure 5.
Silencing of MAGE-A3- or MAGE-C1/CT7 has an additive effect on cell death when combined with conventional myeloma-specifif therapies. The myeloma cell line Molp-8 was transfected with two RNAi constructs specific for CT genes MAGE-A3 (left column) or MAGE-C1/CT7 (right column) or with scrambled control RNAi. Starting 72 h after transfection, myeloma cells were treated with conventional myeloma-specific therapies melphalan (A) or bortezeomib (B). Twelve hours later, myeloma cell viability was assessed by an MTT assay and was normalized using untreated cells. Results show mean values (+SEM) of three separate experiments and asterisks indicate statistically significant differences between untreated cells and scrambled control or gene-specific RNAi (*P<0.05, **P<0.01). Comparable findings were observed following gene-silencing in the RPMI-8226 cell line and using a second gene-specific RNAi construct (data not shown).
Discussion
In this study we performed the first analysis of the function of CT antigens in MM. Observing that knockdown of MAGE-A3/A6 and MAGE-C1/CT7 led to a reduced number of viable myeloma cells in the culture, we first asked whether this might have been due to diminished tumor proliferation after CT gene silencing. This seemed likely since in vitro studies had indicated an association between the expression of CT antigens, such as CAGE, and the proliferation of cell lines derived from solid tumors.32,33 In addition, recent findings had suggested that an increased expression of MAGE-C1/CT7 or MAGE-A3 in MM patients was associated with a higher proportion of proliferating plasma cells in the bone marrow.11,15 Finally, microarray analyses of MM samples had suggested an association between CT antigen expression and the expression of proliferation-associated genes.14 However, although it proved technically difficult to assess the proliferation of myeloma cells on the background of a strong pro-apoptotic effect of MAGE-C1/CT7 and MAGE-A3 silencing, our results indicate that, if anything, both genes only have a modest effect on cell proliferation. Similar findings were observed in another study demonstrating that suppression of MAGE genes had only slight effects on cell cycle progression in a mast cell leukemia cell line.34
We show here that CT antigens, particularly MAGE-A3 and MAGE-C1/CT7, play an important role in protecting myeloma cells from apoptosis. This finding is in agreement with a study indicating that MAGE genes might act as anti-apoptotic factors in melanoma.35 Furthermore, in a murine myeloblast cell line MAGE-A3 was shown to bind to pro-caspase-12 and inhibit its activation under conditions of toxic stress to the endoplasmic reticulum, thereby blocking activation of caspase-9 and downstream activation of caspase-3.36 The authors concluded that MAGE-A3 overexpression renders murine tumor cells resistant to endoplasmic reticulum stress by suppressing the activation of caspase-12. However, it remained unclear whether MAGE genes played any role in caspase regulation in human cells.
Although preliminary, our results indicate that the pro-apoptotic effect of MAGE silencing is related to capases -12 and -9 also in human tumor cells. This observation is in agreement with previous findings that MAGE-A3 does not bind to any caspases other than caspase-12.36 While we cannot definitely prove that downstream effector caspase 3 is not induced at some point following knockdown of MAGE-A3 and MAGE-C1/CT7, it might be that in human myeloma cells the pro-apoptotic effect of MAGE silencing is indeed independent of this otherwise central caspase.
CT genes might theoretically interact with each other and, accordingly, it was recently suggested that the CT gene NY-ESO-1 is a binding partner for MAGE-C1/CT7.37 When we knocked down MAGE-C1/CT7 we did not observe an effect on NY-ESO-1 expression in MM. However, silencing MAGE-C1/CT7 had an effect on the expression of MAGE-C2/CT10, a protein which shows 56% homology to MAGE-C1/CT7. Database analyses of the RNAi sequences used did not support a direct inhibitory effect of RNAi transfection on the expression of the second member of the MAGE-C subfamily and, therefore, a possible interaction between both genes should be investigated further.
A small fraction of MM cells usually escapes chemotherapy and/or treatment with novel agents such as bortezemib and thalidomide/lenalidomide and remains present in the patient’s bone marrow despite a ‘complete remission’. It has been hypothesized that CT antigens might contribute to therapy-resistance and persistence of residual disease in human cancers. Accordingly, studies have shown that expression of MAGE and GAGE genes in cancer cell lines induces a chemotherapy-resistant phenotype in vitro26 and tumor expression of MAGE-A1 seems to correlate with clinical responses to chemotherapy in gastric cancer.38 Our finding of an additive effect of MAGE knockdown on the cytotoxicity exerted by drugs used for myeloma therapy, melphalan and bortezomib, indicates that targeting CT genes might open a new therapeutic route for patients with disease resistant to conventional therapies.
Most malignancies probably arise from a rare population of “cancer stem cells” having the ability to self-renew and maintain the tumor. In the case of MM the malignant plasma cells seem to arise from less differentiated cells that resemble post-germinal center, CD138-negative B cells.29 Novel anti-myeloma agents bortezomib and lenalidomide inhibit malignant plasma cells but, at least in vitro, have little activity against these myeloma precursors.27 This differential activity may explain why these compounds yield significant clinical responses but not cures.39 Interestingly, it has been hypothesized that CT genes, which are often heterogeneously expressed by only few cells within the tumor mass, might represent a hallmark of cancer stem cells40 and we demonstrate here for the first time expression of CT genes MAGE-A3 and MAGE-C1/CT7 in CD138+ myeloma cells and their 138-negative precursors.
Colony growth assays have previously been used to assess the number and function of myeloma precursors present in the tumor bulk since these precursors represent an independent predictive factor for poor prognosis in MM.41 We found that knockdown of CT genes MAGE-A3 or MAGE-C1/CT7 led to a significantly delayed outgrowth of clonogenic myeloma precursors. These findings suggest that targeting CT antigens might not only have the potential to affect fully differentiated myeloma cells but might also contribute to eradicating their clonogenic precursors in MM, a malignancy which is still incurable due to inevitable relapse following standard therapy.
How could CT antigens be targeted in MM? Myeloma is a malignancy which is thought to be controlled, at least to a certain extent, by the adaptive immune system, a view which is supported by the fact that the therapeutic effect of allogeneic stem cell transplantation is partly mediated by donor-derived T cells and that infusions of donor T cells are capable of inducing remission in relapsed MM patients.42,43 We and others have recently shown that spontaneous antibody and T-cell immunity against MAGE-A3 and MAGE-C1/CT7 is present in patients with MM10,44–46 and that the presence of such immunity might be associated with an improved prognosis.44 Our finding that immune responses against CT antigens are induced by allogeneic stem cell transplantation10 suggests that these tumor antigens might indeed represent natural targets for donor-derived allo-immune or even spontaneous anti-myeloma immune responses. Collectively, these findings indicate that T-cell-based immunotherapies might represent one therapeutic route for effective targeting of CT antigens. Importantly, immunization with MAGE-A3 protein has previously been performed in melanoma and lung cancer patients and has proven to be safe, to be capable of evoking specific humoral and T-cell responses,47,48 and to have some clinical activity in patients in the adjuvant setting.49 However, in addition to applying immunotherapy directed against MAGE-C1/CT7 or MAGE-A3 these genes might also be targeted by more “immediate” therapeutic modes such as small molecules, antisense oligonucleotides, or classical gene therapy.
While the exact molecular mechanisms mediating effects of CT genes on myeloma cells still need to be elucidated, our findings strongly suggest that MAGE-C1/CT7 and/or MAGE-A3 are important for the survival and progression of myeloma cells and maybe also of other malignancies. Together with our previous observation that myeloma represents the malignancy with the richest expression of CT antigens, the persistence and prognostic relevance of CT gene expression in MM, and the ability of CT antigens to induce spontaneous and therapy-induced immune responses, our current study provides a strong rationale for employing MAGE-C1/CT7 and MAGE-A3 as therapeutic targets in MM. We believe that such a strategy might prove particularly fruitful when CT antigen-specific approaches are used in a setting of minimal residual disease following conventional myeloma therapy.
Footnotes
Funding: this work was supported by grants from the Erich und Gertrud Roggenbuck-Stiftung, Eppendorfer Krebs- und Leukämiehilfe e.V., José Carreras Leukämie-Stiftung, and from the Cancer Research Institute (to DA) and from Deutsche Krebshilfe and José Carreras Leukämie-Stiftung (to NK)
Authorship and Disclosures
DA designed research, analyzed data, and wrote the paper; YH contributed vital new analytical tools, performed research, analyzed data, and wrote the paper; AJ performed research; YC performed research; TL designed research and analyzed data; SM performed research; SK analyzed data; KB performed research; CP analyzed data; NL performed research; MJ performed research; TS performed research; ARZ analyzed data; CB analyzed data;. NK designed research and analyzed data.
The authors declare no potential conflicts of interest.
References
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. N Engl J Med. 2004;351(18):1860–73. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- 2.Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111(5):2516–20. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 4.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4(5):339–49. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 5.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61(14):5544–51. [PubMed] [Google Scholar]
- 6.Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67(6):705–12. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 7.Taniura H, Kobayashi M, Yoshikawa K. Functional domains of necdin for protein-protein interaction, nuclear matrix targeting, and cell growth suppression. J Cell Biochem. 2005;94(4):804–15. doi: 10.1002/jcb.20345. [DOI] [PubMed] [Google Scholar]
- 8.Chambost H, van Baren N, Brasseur F, Olive D. MAGE-A genes are not expressed in human leukemias. Leukemia. 2001;15(11):1769–71. doi: 10.1038/sj.leu.2402278. [DOI] [PubMed] [Google Scholar]
- 9.Lim SH, Austin S, Owen-Jones E, Robinson L. Expression of testicular genes in haematological malignancies. Br J Cancer. 1999;81(7):1162–4. doi: 10.1038/sj.bjc.6690824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atanackovic D, Arfsten J, Cao Y, Gnjatic S, Schnieders F, Bartels K, et al. Cancer-testis antigens are commonly expressed in multiple myeloma and induce systemic immunity following allogeneic stem cell transplantation. Blood. 2007;109(3):1103–12. doi: 10.1182/blood-2006-04-014480. [DOI] [PubMed] [Google Scholar]
- 11.Jungbluth AA, Ely S, DiLiberto M, Niesvizky R, Williamson B, Frosina D, et al. The cancer-testis antigens CT7 (MAGE-C1) and MAGE-A3/6 are commonly expressed in multiple myeloma and correlate with plasma-cell proliferation. Blood. 2005;106(1):167–74. doi: 10.1182/blood-2004-12-4931. [DOI] [PubMed] [Google Scholar]
- 12.Pellat-Deceunynck C, Mellerin MP, Labarriere N, Jego G, Moreau-Aubry A, Harousseau JL, et al. The cancer germline genes MAGE-1, MAGE-3 and PRAME are commonly expressed by human myeloma cells. Eur J Immunol. 2000;30(3):803–9. doi: 10.1002/1521-4141(200003)30:3<803::AID-IMMU803>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.De Vos J, Thykjaer T, Tarte K, Ensslen M, Raynaud P, Requirand G, et al. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene. 2002;21(44):6848–57. doi: 10.1038/sj.onc.1205868. [DOI] [PubMed] [Google Scholar]
- 14.Condomines M, Hose D, Raynaud P, Hundemer M, De Vos J, Baudard M, et al. Cancer/testis genes in multiple myeloma: expression patterns and prognosis value determined by microarray analysis. J Immunol. 2007;178(5):3307–15. doi: 10.4049/jimmunol.178.5.3307. [DOI] [PubMed] [Google Scholar]
- 15.Tinguely M, Jenni B, Knights A, Lopes B, Korol D, Rousson V, et al. MAGE-C1/CT-7 expression in plasma cell myeloma: sub-cellular localization impacts on clinical outcome. Cancer Sci. 2008;99(4):720–5. doi: 10.1111/j.1349-7006.2008.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaugler B, Van den Eynde B, van der Bruggen P, Romero P, Gaforio JJ, De Plaen E, et al. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179(3):921–30. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocher T, Schultz-Thater E, Gudat F, Schaefer C, Casorati G, Juretic A, et al. Identification and intracellular location of MAGE-3 gene product. Cancer Res. 1995;55(11):2236–9. [PubMed] [Google Scholar]
- 18.Chen YT, Gure AO, Tsang S, Stockert E, Jager E, Knuth A, et al. Identification of multiple cancer/testis antigens by allogeneic antibody screening of a melanoma cell line library. Proc Natl Acad Sci USA. 1998;95(12):6919–23. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucas S, De Smet C, Arden KC, Viars CS, Lethe B, Lurquin C, et al. Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res. 1998;58(4):743–52. [PubMed] [Google Scholar]
- 20.Kruger S, Ola V, Feller AC, Fischer D, Friedrich M. Expression of cancer-testis antigen CT7 (MAGE-C1) in breast cancer: an immunohistochemical study with emphasis on prognostic utility. Pathol Oncol Res. 2007;13(2):91–6. doi: 10.1007/BF02893483. [DOI] [PubMed] [Google Scholar]
- 21.Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67(7):2982–9. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 22.Atanackovic D, Luetkens T, Hildebrandt Y, Arfsten J, Bartels K, Horn C, et al. Longitudinal analysis and prognostic effect of cancer-testis antigen expression in multiple myeloma. Clin Cancer Res. 2009;15(4):1343–52. doi: 10.1158/1078-0432.CCR-08-0989. [DOI] [PubMed] [Google Scholar]
- 23.Mattioli M, Agnelli L, Fabris S, Baldini L, Morabito F, Bicciato S, et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene. 2005;24(15):2461–73. doi: 10.1038/sj.onc.1208447. [DOI] [PubMed] [Google Scholar]
- 24.Taylor BJ, Reiman T, Pittman JA, Keats JJ, de Bruijn DR, Mant MJ, et al. SSX cancer testis antigens are expressed in most multiple myeloma patients: co-expression of SSX1, 2, 4, and 5 correlates with adverse prognosis and high frequencies of SSX-positive PCs. J Immunother. 2005;28(6):564–75. doi: 10.1097/01.cji.0000175685.36239.e5. [DOI] [PubMed] [Google Scholar]
- 25.Andrade VC, Vettore AL, Felix RS, Almeida MS, Carvalho F, Oliveira JS, et al. Prognostic impact of cancer/testis antigen expression in advanced stage multiple myeloma patients. Cancer Immun. 2008;8:2. [PMC free article] [PubMed] [Google Scholar]
- 26.Duan Z, Duan Y, Lamendola DE, Yusuf RZ, Naeem R, Penson RT, et al. Overexpression of MAGE/GAGE genes in paclitaxel/doxorubicin-resistant human cancer cell lines. Clin Cancer Res. 2003;9(7):2778–85. [PubMed] [Google Scholar]
- 27.Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68(1):190–7. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gure AO, Stockert E, Arden KC, Boyer AD, Viars CS, Scanlan MJ, et al. CT10: a new cancer-testis (CT) antigen homologous to CT7 and the MAGE family, identified by representational-difference analysis. Int J Cancer. 2000;85(5):726–32. doi: 10.1002/(sici)1097-0215(20000301)85:5<726::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 29.Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, et al. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103(6):2332–6. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cilensek ZM, Yehiely F, Kular RK, Deiss LP. A member of the GAGE family of tumor antigens is an anti-apoptotic gene that confers resistance to Fas/CD95/APO-1, inter-feron-gamma, taxol and gamma-irradiation. Cancer Biol Ther. 2002;1(4):380–7. [PubMed] [Google Scholar]
- 31.Cronwright G, Le Blanc K, Gotherstrom C, Darcy P, Ehnman M, Brodin B. Cancer/testis antigen expression in human mesenchymal stem cells: down-regulation of SSX impairs cell migration and matrix metalloproteinase 2 expression. Cancer Res. 2005;65(6):2207–15. doi: 10.1158/0008-5472.CAN-04-1882. [DOI] [PubMed] [Google Scholar]
- 32.Cho B, Lim Y, Lee DY, Park SY, Lee H, Kim WH, et al. Identification and characterization of a novel cancer/testis antigen gene CAGE. Biochem Biophys Res Commun. 2002;292(3):715–26. doi: 10.1006/bbrc.2002.6701. [DOI] [PubMed] [Google Scholar]
- 33.Shim E, Shim H, Bae J, Lee H, Jeoung D. CAGE displays oncogenic potential and induces cytolytic T lymphocyte activity. Biotechnol Lett. 2006;28(7):515–22. doi: 10.1007/s10529-006-0008-5. [DOI] [PubMed] [Google Scholar]
- 34.Yang B, O’Herrin S, Wu J, Reagan-Shaw S, Ma Y, Nihal M, et al. Select cancer testes antigens of the MAGE-A, -B, and -C families are expressed in mast cell lines and pro-mote cell viability in vitro and in vivo. J Invest Dermatol. 2007;127(2):267–75. doi: 10.1038/sj.jid.5700548. [DOI] [PubMed] [Google Scholar]
- 35.Yang B, O’Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007;67(20):9954–62. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 36.Morishima N, Nakanishi K, Takenouchi H, Shibata T, Yasuhiko Y. An endoplasmic reticulum stress-specific caspase cascade in apoptosis. Cytochrome c-independent activation of caspase-9 by caspase-12. J Biol Chem. 2002;277(37):34287–94. doi: 10.1074/jbc.M204973200. [DOI] [PubMed] [Google Scholar]
- 37.Cho HJ, Caballero OL, Gnjatic S, Andrade VC, Colleoni GW, Vettore AL, et al. Physical interaction of two cancer-testis antigens, MAGE-C1 (CT7) and NY-ESO-1 (CT6) Cancer Immun. 2006;6:12. [PubMed] [Google Scholar]
- 38.Suzuki T, Yoshida K, Wada Y, Hamai Y, Sentani K, Oue N, et al. Melanoma-associated antigen-A1 expression predicts resistance to docetaxel and paclitaxel in advanced and recurrent gastric cancer. Oncol Rep. 2007;18(2):329–36. [PubMed] [Google Scholar]
- 39.Huff CA, Matsui W, Smith BD, Jones RJ. The paradox of response and survival in cancer therapeutics. Blood. 2006;107(2):431–4. doi: 10.1182/blood-2005-06-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costa FF, Le Blanc K, Brodin B. Concise review: cancer/testis antigens, stem cells, and cancer. Stem Cells. 2007;25(3):707–11. doi: 10.1634/stemcells.2006-0469. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi T, Lim B, Jamal N, Tritchler D, Lockwood G, McKinney S, et al. Colony growth and self renewal of plasma cell precursors in multiple myeloma. J Clin Oncol. 1985;3(12):1613–23. doi: 10.1200/JCO.1985.3.12.1613. [DOI] [PubMed] [Google Scholar]
- 42.Tricot G, Vesole DH, Jagannath S, Hilton J, Munshi N, Barlogie B. Graft-versus-myeloma effect: proof of principle. Blood. 1996;87(3):1196–8. [PubMed] [Google Scholar]
- 43.Orsini E, Bellucci R, Alyea EP, Schlossman R, Canning C, McLaughlin S, et al. Expansion of tumor-specific CD8+ T cell clones in patients with relapsed myeloma after donor lymphocyte infusion. Cancer Res. 2003;63(10):2561–8. [PubMed] [Google Scholar]
- 44.Goodyear OC, Pratt G, McLarnon A, Cook M, Piper K, Moss P. Differential pattern of CD4+ and CD8+ T-cell immunity to MAGE-A1/A2/A3 in patients with mono-clonal gammopathy of undetermined significance (MGUS) and multiple myeloma. Blood. 2008;112(8):3362–72. doi: 10.1182/blood-2008-04-149393. [DOI] [PubMed] [Google Scholar]
- 45.Goodyear O, Piper K, Khan N, Starczynski J, Mahendra P, Pratt G, et al. CD8+ T cells specific for cancer-germline gene antigens are found in many patients with multiple myeloma and their frequency correlates with disease burden. Blood. 2005;106(13):4217–24. doi: 10.1182/blood-2005-02-0563. [DOI] [PubMed] [Google Scholar]
- 46.Curioni-Fontecedro A, Knights AJ, Tinguely M, Nuber N, Schneider C, Thomson CW, et al. MAGE-C1/CT7 is the dominant cancer-testis antigen targeted by humoral immune responses in patients with multiple myeloma. Leukemia. 2008;22(8):1646–8. doi: 10.1038/leu.2008.43. [DOI] [PubMed] [Google Scholar]
- 47.Atanackovic D, Altorki NK, Stockert E, Williamson B, Jungbluth AA, Ritter E, et al. Vaccine-induced CD4+ T cell responses to MAGE-3 protein in lung cancer patients. J Immunol. 2004;172(5):3289–96. doi: 10.4049/jimmunol.172.5.3289. [DOI] [PubMed] [Google Scholar]
- 48.Atanackovic D, Altorki NK, Cao Y, Ritter E, Ferrara CA, Ritter G, et al. Booster vaccination of cancer patients with MAGE-A3 protein reveals long-term immunological memory or tolerance depending on priming. Proc Natl Acad Sci USA. 2008;105(5):1650–5. doi: 10.1073/pnas.0707140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brichard VG, Lejeune D. GSK’s antigen-specific cancer immunotherapy programme: pilot results leading to phase III clinical development. Vaccine. 2007;25 (Suppl 2):B61–71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]