Abstract
Context
Brain maturation starts well before birth and occurs as a unified process with developmental interaction among different brain regions. Gene and environment play large roles in such process. Studies of genetic disorders such as fragile X syndrome (FXS) which is a disorder caused by a single gene mutation resulting in abnormal dendritic and synaptic pruning, together with healthy individuals may provide valuable information.
Objective
To examine morphometric spatial patterns that differentiate between FXS from controls in early childhood.
Design
A cross-sectional in-vivo neuroimaging study
Setting
Academic medical centers
Participants
A total of 101 children of ages 1 to 3; 51 boys with FXS, 32 typically developing (TD) boys and 18 boys with idiopathic developmental delay (DD)
Main Outcome Measures
Regional gray matter volume as measured by voxel-based morphometry and manual tracing, supplemented by permutation analyses. Regression analyses between gray and white matter volumes and IQ and fragile X mental retardation protein (FMRP). Linear support vector machine analyses to classify group membership.
Results
In addition to aberrant brain structures reported previously in older individuals with FXS, we found reduced gray matter volumes in regions such as the hypothalamus, insula and medial and lateral prefrontal cortices. These findings are consistent with the cognitive and behavioral phenotypes of FXS. Further, multivariate pattern classification analyses discriminated FXS from TD and DD controls with over 90 % prediction accuracy. The spatial patterns that classified FXS from controls included those that may have been difficult to identify previously using other methods. These included medial to lateral gradient of increased and decreased regional brain volumes in the posterior vermis, amygdala and hippocampus.
Conclusions
These findings are critical in understanding interplay among gene, environment, brain and behavior, and signify the importance of examining detailed spatial patterns of healthy and perturbed brain development.
INTRODUCTION
Human brain maturation is a complex process that can now be examined in detail using in vivo neuroimaging techniques 1. Early neuroimaging investigations focused on region, lobe or total brain volumes 1. However, more recent studies have begun to examine brain structures using finer voxel-wise resolution and temporal dynamics. These new approaches have proven useful in advancing our understanding of associations between cognition and healthy brain development 2.
Pediatric neuroimaging studies to date have primarily focused on healthy children ages 4 years and older using univariate approaches. However, brain maturation starts well before birth and occurs as a unified process with inter-regional interactions and dependencies. It is therefore critical to investigate spatial neurodevelopmental patterns using multivariate approaches in children soon after birth. It is also important to examine abnormal brain states to more fully understand normal brain maturational processes. Studies of fragile X syndrome (FXS) in early childhood may provide such unique opportunities.
Unlike many developmental disorders that are symptomatically defined and therefore likely to be heterogeneous, FXS has a well-characterized genetic etiology. FXS, an X-linked disorder, is caused by a mutation of the fragile X mental retardation-1 (FMR1) gene leading to diminished production of the associated protein (FMRP). In turn, diminished FMRP negatively impacts synaptic maturation and plasticity and thus, cerebral development 3. The disorder results in impaired cognition and aberrant behavior, including deficits in executive and social function, learning and memory, visuo-spatial skills, and emotion regulation 4.
As a component of an ongoing longitudinal study of brain development in FXS, we examined magnetic resonance images (MRI) of 51 boys with FXS, 32 typically developing boys (TD) and 18 boys with idiopathic developmentally delay (DD) of ages one to three. The main goal of the study was to examine structural brain abnormalities in FXS compared to both TD and DD and to examine neurodevelopmental differences in these very young FXS subjects relative to findings reported in older FXS 4–7. Primary analytical methods included univariate between-group comparisons and multivariate pattern classification methods. We hypothesized that regions best distinguishing the young FXS group from controls such as the caudate would show associations with FMRP as well as with cognitive measures (IQ). Negative associations between FMRP and caudate volumes have been shown in youth and adults, but not in children of this age-range 5, 8. We further hypothesized that morphometric patterns would accurately classify participants into their respective groups. The study protocols were approved by the human subjects committees at Stanford University School of Medicine and University of North Carolina – Chapel Hill (UNC) and consent was obtained from parents.
METHODS
Participants
Subjects included 51 boys with FXS (mean age 35.0 months ± standard deviation, SD 7.6), 32 TD boys (29.7 ± 7.1 months) and 18 boys with idiopathic DD (34.8 ± 5.1 months). Five FXS and three TD participants have been included in a previous report 9.
Subjects in all groups were recruited by Stanford University and UNC. Recruitment of children with FXS was accomplished utilizing both the Stanford and UNC registry databases, frequent postings on the National Fragile X Foundation website and quarterly newsletter, and mailings to regional fragile X organizations. Children with DD and TD were recruited locally through early intervention programs, preschools, child care centers, community media, and state run agencies (Regional Center system in California and Child Development Service Agencies in North Carolina). Inclusion in the FXS group required DNA testing confirming the fragile X full mutation as diagnosed with standard Southern Blot technique. Participants in the DD group included children with developmental delays of unknown etiology who did not exhibit symptoms indicative of an autism spectrum disorder. All children in this group demonstrated a composite standard score below 85 (<1 SD) on the Mullen Scales of Early Learning 10. Exclusion criteria for all groups included pre-term birth (< 34 weeks), low birth weight (< 2000 grams), evidence of a genetic condition or syndrome, sensory impairments, and any serious medical or neurological condition affecting growth and development (e.g., seizure disorder, diabetes, congenital heart disease).
DNA testing for the typical FMR1 expansion mutation was performed to confirm the presence of the full mutation in all subjects with FXS for whom diagnostic status was uncertain and to rule out FXS in DD controls. Standard Southern blot was performed followed by FMR1-specific probe hybridization 11. FMRP expression was ascertained by calculating the percentage of peripheral lymphocytes containing FMRP using immunostaining techniques 12.
Subjects were given a standard battery of measures including the Mullen Scales of Early Learning 10, the Vineland Adaptive Behavior Scales 13, the Child Behavior Checklist (CBCL) 14, and the Repetitive Behavior Scales (RBS) 15, 16. There was a significant difference in age between FXS and TD (P = 0.001) and DD and TD (P = 0.015) but not between FXS and DD (P = 0.91). Vineland adaptive behavior composite standard scores (SS) were also significantly different between FXS and TD (P < 0.001) and DD and TD (P < 0.001) but not between FXS and DD (P = 0.33). Mullen composite SS were significantly different between FXS and TD (P < 0.001) and DD and TD (P < 0.001) but not between FXS and DD (P = 0.38). Please see Table 1 for details of the three groups and between group statistics.
Table 1.
Demographic Information
| FX (N=51) | TD (N=32) | DD (N=18) | ANOVA | |||||
|---|---|---|---|---|---|---|---|---|
| mean | SD | mean | SD | mean | SD | F | P | |
| Age (months) | 35.0 | 7.6 | 29.7 | 7.1 | 34.8 | 5.1 | 6.16 | 0.003 |
| Mullen Composite (SS) | 54.65 | 8.98 | 108.50 | 18.42 | 57.72 | 9.71 | 187.04 | <0.001 |
|
Vineland Adaptive Behavior Composite (SS) |
62.02 | 9.67 | 97.23 | 12.44 | 65.00 | 11.92 | 105.30 | <0.001 |
| FMRP (%) | 5.74 | 3.41 | ||||||
SS: Standard Score; SD: standard deviation
Vineland: TD N=31, FMRP: N=46
Overall, 50 subjects were recruited at Stanford University and 51 subjects at UNC. There were no significant differences between sites in the proportion of diagnoses (chi = 1.51, P = 0.47). For each diagnostic group, age (P = 0.31 – 0.73), Vineland SS (P = 0.20 – 0.94), Mullen SS (P = 0.43 – 0.61) and FMRP (P = 0.62, FXS only) did not differ between sites. Four FXS and two DD individuals were on medication (FXS: one on bethanechol, one on piracetam, and two on clonidine; DD: one on memantine and the other on lamotrigine).
Magnetic Resonance Imaging (MRI) Preparation and Acquisition
All subjects with FXS or DD were sedated during their MRI. A pediatric anesthesiologist administered and monitored the sedation throughout the scan. TD children were scanned while sleeping. For the sleep scans, parents were instructed to wake their child up a little earlier on the morning of the scan and to shorten their nap time. Scans were scheduled for after the participants’ normal bedtime to ensure the children would be able to sleep during the scan. Parents received a packet of preparation materials for the scan which included a CD of scanner sounds to desensitize the children to the noise of the scanner while sleeping. Further, these subjects participated in a simulated MRI situation to practice holding still and to alleviate fear should the child awaken during the MRI.
General Electric 1.5 Tesla Signa LX scanners (GE Imagine Systems, Milwaukee, WI) and standard transmit/receive 4 channel head coils were used for MR image acquisition. Scans were obtained from 2003 through 2007, and performed at Stanford - Lucile Packard Children’s Hospital and UNC – Brain Imaging and Analysis Center. Identical pulse sequence protocols were used at both scan-sites, which were designed to maximize contrast between gray, white and cerebro-spinal fluid (CSF) for the participants’ age-range. This included a coronal T1 –weighted sequence with the following parameters: inversion recovery preparation pulse, 300 ms; repetition time, 12 ms; echo time, 5 ms; flip angle, 20 °; thickness, 1.5 mm; number of excitations, 1; field of view, 20 cm; and matrix, 256 × 192. An MR Quality Control (QC) phantom (MRI/QC/P, Data Systems, Inc., Hillsboro, N.C.) was scanned after each subject at both sites to standardize assessments over sites, individuals and time.
Image Processing
VBM Analyses
VBM analyses of T1 MR images were performed using SPM5 (http://www.fil.ion.ucl.ac.uk/spm) and VBM5 (http://dbm.neuro.uni-jena.de/vbm). After bias correction, T1 images were segmented to gray matter (GM), white matter (WM) and CSF. Hidden Markov Random Field (prior probability weight 0.3) was used to encode spatial information through spatial constraints of neighboring voxels. Normalization was performed using the following parameters: 12 parameter affine transformation and three dimensional discrete cosine transformation (DCT) basis function (12 × 12 × * where * is calculated to yield a spatial cut-off that is 25 in all directions). Linear and non-linear Jacobian modulation was then applied (reflecting GM volume [GMV] and WM volume [WMV]), followed by smoothing with an isotropic Gaussian kernel with full-width at half-maximum (FWHM) of 8 mm. Smoothing was also performed at 4 mm and the same regions were detected. For example, the FXS > TD, DD contrast showed caudate, thalamus, hypothalamus, fusiform gyrus and occipital regions, and the TD, DD > FXS contrast showed superior temporal gyrus, hippocampus, insula, orbitofrontal and medial prefrontal cortices at P = 0.05 family-wise error (FWE) corrected. Customized GM and WM templates created from all subjects were used for VBM processing. Segmentation and normalization for each subject was confirmed by manual inspection of the images. Two DD participants were excluded as a result of poor segmentation and was excluded from this study (final N = 18).
Volumetric Measures
Left and right total caudate nuclei volumes were obtained using the Automated Segmentation of Caudate Nucleus algorithm 17 using BrainImageJava (BIJ) (http://cibsr.stanford.edu), a public domain program for morphological image processing originally developed as a component of the National Institutes of Health Human Brain Project. The validity of the algorithm was tested with 55 high-resolution T1 – weighted MRI datasets, and the segmentation results were overlaid onto the original image data for visual inspection. The algorithm was run on the registered and inhomogenity corrected images. The right and left caudate volumes for each subject were checked manually and were corrected if needed.
The amygdala was manually traced on high resolution T1 images aligned along the long axis of the HIP using the IRIS/SNAP tool at UNC. The protocol developed by the Center for Neuroscience and the M.I.N.D. at University of California (UC) Davis was used 18 (see http://www.psychiatry.unc.edu/autismresearch/mri/ROIs/Amygdala.pdf for details). We first established reliability with the UC Davis group (average inter-rater reliability = 0.92). Subsequently, reliability was established on scans from a sample of 18–35 month olds. Reliability was obtained by two raters who made independent measurements on a set of 15 images, which included 5 images repeated 3 times (in random order). Intra-rater and inter-rater reliabilities (intraclass correlation) were 0.90 and 0.78, respectively. Two raters (who had the reliability of 0.90) performed all the amygdala traces. There were 7 cases (4 TDs, 2 DDs and 1 FXS) that were of insufficient quality to obtain an amygdala trace.
The cerebellar vermis was manually delineated into anterior vermis (I-V lobules), mid vermis (VI - VII lobules) and the posterior vermis (VIII-X lobules) using BrainImage 3.7 (http://cibsr.stanford.edu). The protocol required selecting the best midsagittal slice, as defined by prioritizing clarity of the following structures: cerebellar vermis, cerebral aqueduct, corpus callosum and the brain stem. All the analyses were performed independently by two raters who were blinded on subject's diagnosis and achieved inter-rater reliability of 0.95. The raters made two tracings of the three vermal regions for each subject and the values were averaged.
Statistical Analysis
Cross-Site Analysis
We undertook a number of procedures to ensure compatibility of MR images across sites. First, we used the same pulse sequence and the same scanner type at each site. Second, to characterize scanner quality, we calculated signal-to-noise ratio (SNR) for 24 random phantom scans (12 per scanner, selected across the entire period of the study by a blind research assistant) using the method described in 19. Two SNR measurements were performed and were not significantly different across sites (P = 0.39 and P = 0.22, respectively).
Third, we examined whether brain volumes differed as a function of scan-site. GMV, WMV, CSF and total tissue volume (TTV, GMV + WMV) were not significantly different across sites (all P’s > 0.05). Volumetric measures of caudate nuclei, amygdala and vermis did not differ significantly across scan-sites (P’s > 0.05). All regions that we report below which showed significant differences in regional brain volumes between groups also did not show significant differences between scan-sites except for the following regions: right cerebellar GMV, (region ‘G’ in Table 2), and pre-, postcentral GMV (regions ‘C’, ‘d’, ‘m’ in Table 2) and WMV (region ‘a’ in Table 3). Fourth, we included scan-site as a nuisance variable in all analyses that we report because of the small (non-significant) differences found between sites. Finally, we repeated the VBM analyses for each scan-site separately. While this reduced the sample-size to half, the only regions that were no longer significant was a right anterior cerebellar GM region (region ‘G’ in Table 2), and regions that showed only trends for significant differences were two of the three right medial frontal WM regions (region ‘c’ and ‘i’ in Table 3).
Table 2.
GM Regions that Show Significant Differences between FXS and Control Groups.
| ROI Name |
Region | Brodmann Area |
Talairach Coordinates | T value | P value (FWE) |
Cluster Size |
|||
|---|---|---|---|---|---|---|---|---|---|
| X | Y | Z | |||||||
| Gray Matter: FXS > TD, DD | |||||||||
| A | Right Caudate Body, Head | 14 | 2 | 19 | 10.28 | <0.001 | 7120 | ||
| 14 | 19 | −2 | 7.07 | <0.001 | |||||
| B | Left Caudate Body, Head | −17 | 4 | 20 | 9.11 | <0.001 | 6699 | ||
| −12 | 21 | −1 | 7.29 | <0.001 | |||||
| C | Left Precentral, Inferior Frontal Gyri |
6 | −44 | −1 | 28 | 7.11 | <0.001 | 591 | |
| D | Right Fusiform Gyrus | 18, 19 | 37 | −76 | −12 | 6.77 | <0.001 | 1857 | |
| 41 | −86 | −12 | 6.22 | 0.001 | |||||
| 31 | −91 | −15 | 6.2 | 0.001 | |||||
| E | Bilateral Thalamus (Dorsal Medial, Ventral Anterior Nuclei), Hypothalamus |
6 | −10 | 13 | 6.71 | <0.001 | 1506 | ||
| −7 | −11 | 13 | 6.61 | <0.001 | |||||
| −8 | −14 | 5 | 6.39 | <0.001 | |||||
| F | Left Middle, Superior Occipital Gyri |
19 | −43 | −86 | 19 | 6.59 | <0.001 | 714 | |
| G | Right Anterior Cerebellum | 26 | −45 | −24 | 6.37 | <0.001 | 322 | ||
| H | Left Cingulate Gyrus | 23 | −5 | −26 | 32 | 5.57 | 0.01 | 235 | |
| Gray Matter: TD, DD > FXS | |||||||||
| a | Right Insula | 13 | 30 | 12 | 11 | 9.21 | <0.001 | 4951 | |
| 40 | −4 | 19 | 8.22 | <0.001 | |||||
| b | Left Insula | 13 | −39 | −3 | 19 | 9.17 | <0.001 | 3801 | |
| −30 | 14 | 13 | 8.05 | <0.001 | |||||
| j | Right Hippocampal Gyrus | 33 | −39 | 8 | 6.87 | <0.001 | 2514 | ||
| 30 | −57 | 17 | 5.55 | 0.004 | |||||
| c | Right Postcentral Gyrus, Inferior Parietal Lobule (IPL) |
40,1,3 | 57 | −27 | 20 | 6.74 | <0.001 | 5899 | |
| 58 | −16 | 43 | 6.62 | <0.001 | |||||
| 62 | −7 | 16 | 6.41 | <0.001 | |||||
| d | Right Postcentral Gyrus, IPL | 40 | 49 | −30 | 48 | 6.68 | <0.001 | 556 | |
| 62 | −28 | 41 | 5.54 | 0.011 | |||||
| e | Right Medial Frontal Gyrus | 10 | 13 | 52 | −6 | 6.48 | <0.001 | 484 | |
| f | Right Inferior Frontal Gyrus | 44,45 | 53 | 17 | 18 | 6.44 | <0.001 | 1162 | |
| 59 | 9 | 19 | 5.95 | 0.002 | |||||
| g | Left Middle Frontal Gyrus | 11 | −22 | 41 | −13 | 6.34 | <0.001 | 431 | |
| h | Left Medial Frontal Gyrus | −10 | 53 | 5 | 6.19 | 0.001 | 1372 | ||
| −12 | 44 | 13 | 5.95 | 0.002 | |||||
| −9 | 43 | 0 | 5.68 | 0.006 | |||||
| k | Left Hippocampal Gyrus | −33 | −37 | 6 | 6.14 | <0.001 | 468 | ||
| i | Left IPL, Postcentral Gyrus | 40 | −62 | −34 | 38 | 6.09 | 0.001 | 395 | |
| j | Right Medial Frontal Gyrus | 11 | 6 | 44 | −14 | 5.96 | 0.002 | 408 | |
| 15 | 39 | −15 | 5.8 | 0.004 | |||||
| k | Left Precentral Gyrus | 6,4 | −62 | −4 | 27 | 5.87 | 0.003 | 1083 | |
| −61 | −10 | 38 | 5.76 | 0.005 | |||||
| −65 | −19 | 42 | 5.15 | 0.045 | |||||
| l | Left Superior Temporal, Precentral Gyri |
22,4 | −62 | −8 | 3 | 5.59 | 0.009 | 232 | |
| −54 | −8 | 5 | 5.39 | 0.019 | |||||
Table 3.
WM Regions that Show Significant Differences between FXS and Control Groups.
| ROI Name |
Region | Talairach Coordinates | T value | P value (FWE) |
Cluster Size |
|||
|---|---|---|---|---|---|---|---|---|
| X | Y | Z | ||||||
| White Matter: FXS > TD, DD | ||||||||
| A | Right Insular Ventrolateral Prefrontal Region | 31 | 8 | 14 | 8.38 | <0.001 | 1283 | |
| 33 | −14 | 17 | 6.53 | <0.001 | ||||
| B | Left Insular Ventrolateral Prefrontal Region | −30 | 12 | 13 | 8.2 | <0.001 | 1175 | |
| −38 | −3 | 20 | 7.3 | <0.001 | ||||
| −33 | −14 | 19 | 6.88 | <0.001 | ||||
| C | Left Superior Temporal Region | −40 | −51 | 23 | 7.07 | <0.001 | 1374 | |
| D | Right Medial Temporal Region | 39 | −41 | −5 | 6.6 | <0.001 | 1022 | |
| 34 | −32 | 1 | 6.37 | <0.001 | ||||
| E | Left Medial Temporal Region | −37 | −45 | −6 | 6.1 | <0.001 | 723 | |
| F | Right Middle Occipital Region | 34 | −67 | 3 | 5.74 | 0.002 | 252 | |
| White Matter: TD, DD > FXS | ||||||||
| a | Left PrecentralVentrolateral Prefrontal Region |
−45 | −1 | 27 | 8.52 | <0.001 | 1762 | |
| b | Left Caudate, Medial Frontal Region | −14 | 20 | −1 | 8.35 | <0.001 | 2083 | |
| −20 | 16 | 13 | 7.53 | <0.001 | ||||
| −20 | 9 | 19 | 7.23 | <0.001 | ||||
| c | Right Caudate, Medial Frontal Region | 14 | 20 | 0 | 7.56 | <0.001 | 444 | |
| d | Right Precentral Ventrolateral Prefrontal Region |
46 | −3 | 27 | 6.95 | <0.001 | 2075 | |
| 36 | −6 | 39 | 5.88 | 0.001 | ||||
| e | Left Pre, Postcentral Region | −30 | −29 | 50 | 6.29 | <0.001 | 249 | |
| f | Right Medial Frontal Region | 24 | 39 | 11 | 6.06 | 0.001 | 413 | |
| g | Left Dorsolateral Prefrontal Region | −32 | 33 | 21 | 5.66 | 0.003 | 248 | |
| h | Right Postcentral, Inferior Parietal Region | 47 | −18 | 29 | 5.64 | 0.003 | 411 | |
| i | Right Superior, Medial Frontal Region | −21 | 40 | 19 | 5.49 | 0.005 | 224 | |
| −24 | 39 | 9 | 5.22 | 0.014 | ||||
Analyses of GMV, WMV and CSF
We examined total GMV, WMV and TTV obtained from VBM analyses using univariate general linear model (GLM) followed by post-hoc analyses. Diagnostic group and scan-site were included as fixed factors, and age as a covariate. Site by diagnosis interaction was not modeled as this factor was not significant (P’s > 0.10) for any comparisons performed. Further analyses were performed by including IQ (Mullen Composite SS) as a covariate.
VBM Analysis
We examined regional GM and WM differences between FXS and controls using whole-brain analysis of covariance (ANCOVA) covarying out age, scan-site, globals and total GMV / WMV (for regional GM and WM analyses, respectively). For this analysis, we initially combined the TD and DD groups (N = 50) and compared with the FXS group (N = 51) to match sample sizes. We confirmed the results by performing a one-way ANCOVA (same covariates as above) but by treating the FXS, TD and DD participants as three separate groups (this showed similar results). To ensure that the significant effects were not driven by one of the control groups, the following analyses were performed. First, mean values from significant brain regions in the whole brain VBM analyses above were extracted for each subject. These values were then adjusted for age, scan-site and total GMV and WMV for GM and WM analyses, respectively. Values were adjusted by performing linear multiple regression analysis with age, scan-site and total GMV/WMV as independent variables, and obtaining the residuals. These adjusted brain volumes were compared between FXS and TD and FXS and DD separately.
Statistical threshold of P = 0.05 FWE corrected with an extent threshold of 200 was employed, correcting for non-isotropic smoothness. Statistical images were overlaid onto normalized GM and WM images from a representative TD, DD or FXS subject. Brain regions with significant effects are reported based on their anatomical locations overlaid on the custom template and several sample FXS, TD and DD T1 images from this study. While it is not necessarily valid to report Talairach coordinates and Brodmann Areas because of the very young study sample and the use of custom templates, we report these as reference for comparisons with other studies. The reported Talairach coordinates were converted from MNI space using the mni2tal function (http://www.mrc-cbu.cam.ac.uk/Imaging/Common/mnispace.shtml) and Brodmann Areas were identified using Talairach Daemon (Research Imaging Center, University of Texas Health Science Center in San Antonio (RIC UTHSCSA, TX USA) which were confirmed with the Talairach atlas 20.
To examine the consistency of the results, probabilistic GM maps of the VBM results were constructed by performing leave-one-out permutation analysis. Aforementioned between-group VBM GM analysis was repeated 100 times leaving one subject out at a time. The statistical images were thresholded similarly, binarized and summed to create probabilistic maps. The results were overlaid on a custom GM template.
Volumetric Analysis of Caudate Nuclei, Amygdala and Vermis
We examined volumes of left and right caudate nuclei, left and right amygdala and anterior, middle and posterior vermis adjusting for age, scan-site and TTV or total GMV (for total and GM caudate volumes, respectively) using the same method as described above for GMV, WMV and TTV comparisons. The only difference was that we performed additional analyses including TTV as a covariate.
Covariation between brain volumes and FMRP or IQ
Based on previous studies in older FXS individuals, we hypothesized that caudate volumes would show significant negative correlation with FMRP 4 (note that lower FMRP is thought to be correlated with greater severity of FXS). We therefore correlated caudate GMV which showed significant differences between FXS and controls in VBM and caudate volumes from manual measurements with FMRP in the FXS group. Regression analysis with IQ (Mullen composite standard score, SS) as the dependent measure was also performed for TD, FXS and DD groups separately. Statistical threshold with a joint-expected probability 21 of P = 0.01 for height and extent was used.
Multivariate Pattern Classification Analyses
The final analysis was designed to identify regions where spatially distributed GMV patterns carried information specific to FXS using a machine learning approach, support vector machine (SVM) analysis 22–25. First, we resampled unsmoothed GMV maps to 4 × 4 mm voxels within the custom GM template, and computed a n-dimentional GMV vector vf,1…n for each FXS subject f and vc,1…n for each control subject except for one subject (leave-one-out cross-validation). These represent spatial GMV patterns for n voxels. This was transformed to normalized GMV vectors Vf,1…n and Vc,1…n. Next, we performed principle component analysis (PCA) to reduce the number of features and used the number of eigenvectors that accounted for variance above 80 %, which gave Ff,1…N and Fc,1…N for N (3) features. We assigned the vectors for n-1 subjects as a training data-set that was used to train a linear support vector pattern classifier (with fixed regularization parameter C = 1) to correctly identify GMV patterns of the nth subject which was repeated n times. Prediction accuracy, sensitivity, specificity and positive predictive values were calculated. Weight vectors were transformed back to voxel-space as described in 25. We repeated the analyses to classify FXS from TD and FXS from DD separately as well as to classify DD from TD. Because there were only 18 DD subjects (vs. 51 FXS subjects), we ensured that the low classification accuracy of DD from TD was not due to small sample-size by randomly selecting 18 FXS subjects and classifying these FXS subject from TD (which yielded high classification accuracy). Classification accuracies of FXS and TD, and DD and TD were statistically compared using permutation analyses.
We also performed identical SVM analyses but used different methods to define features. The aforementioned method including all GM 4×4 mm voxels are referred to as the Voxel-wise feature selection method. To compare with this method, we coregistered all 116 brain regions in the Automated Talairach Atlas Label (AAL) (http://www.cyceron.fr/freeware/) to subjects’ unsmoothed normalized images and extracted GMV from all regions and repeated the analysis (Region-wise). Classification accuracies of the Voxel-wise and Region-wise Methods were statistically compared using permutation analyses.
Since caudate volumes on their own were relatively good classifiers, we also repeated the ‘Voxel-wise’ analysis including all GM voxels but excluding voxels in the caudate (Voxel-wise excl. Caudate). Caudate was defined using the AAL. These methods were also performed in a leave-one-out fashion.
Discriminating volumes (weight vectors transformed back into brain-space) were presented in two ways. First, we show weights of all voxels without any thresholding. We also show voxels that exceeded a specified statistical threshold using permutation analysis (random assignment of classes 2000 times; P = 0.05, weight = ± 0.0054).
RESULTS
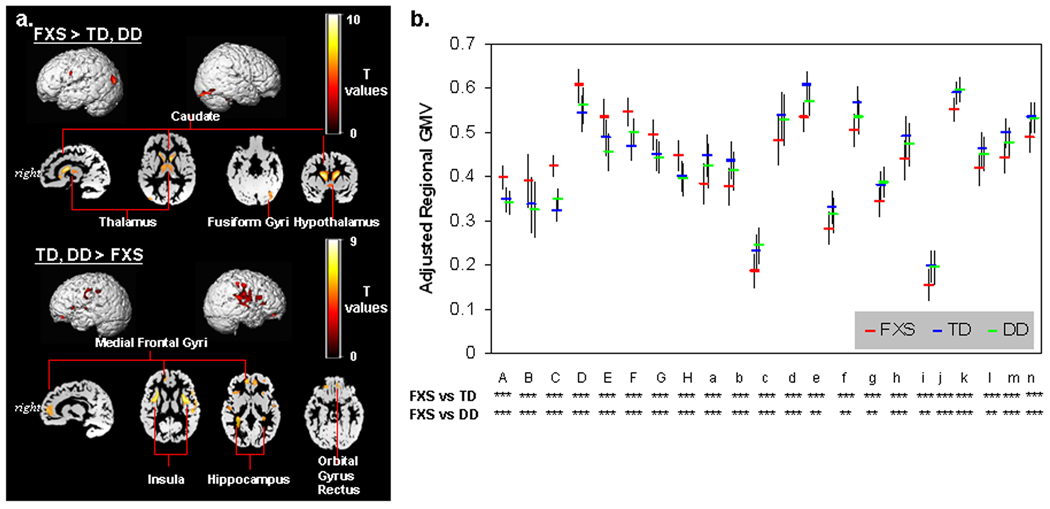
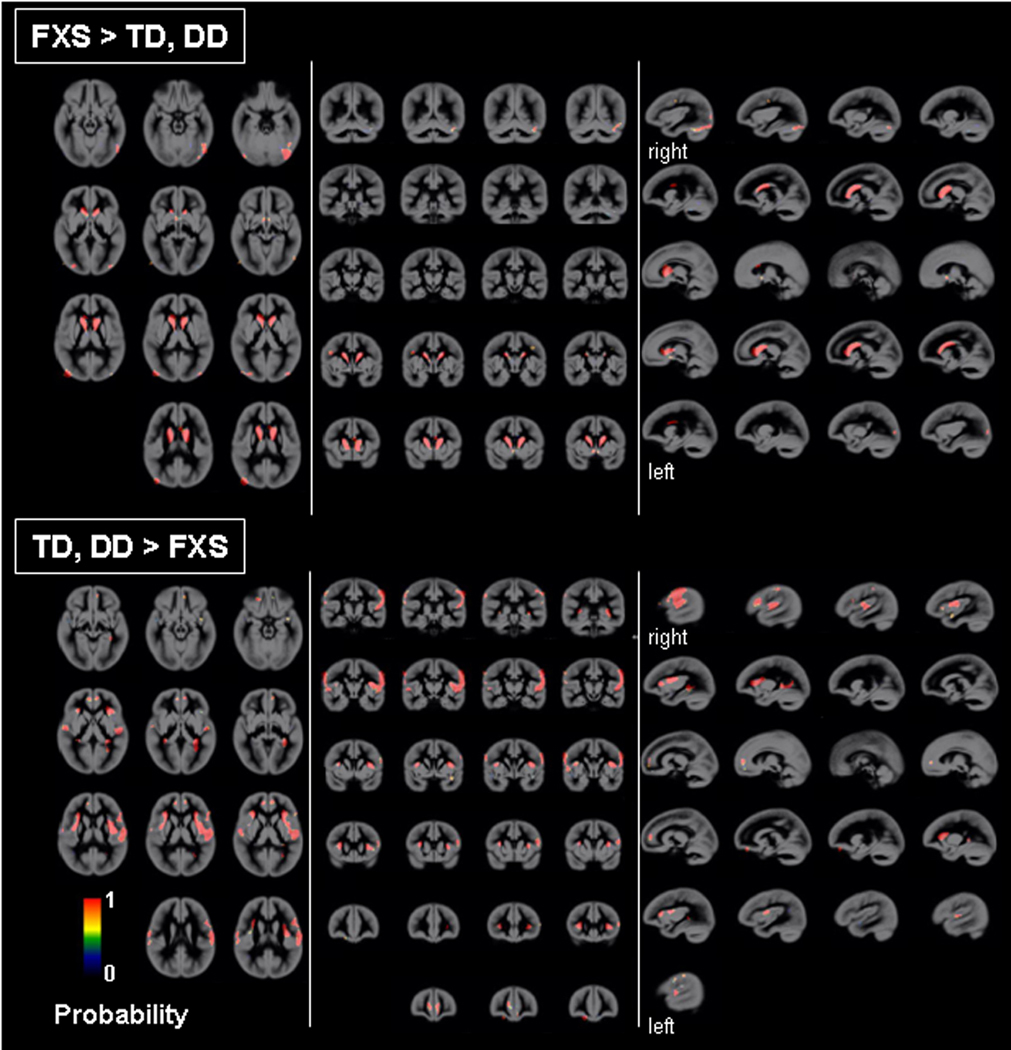
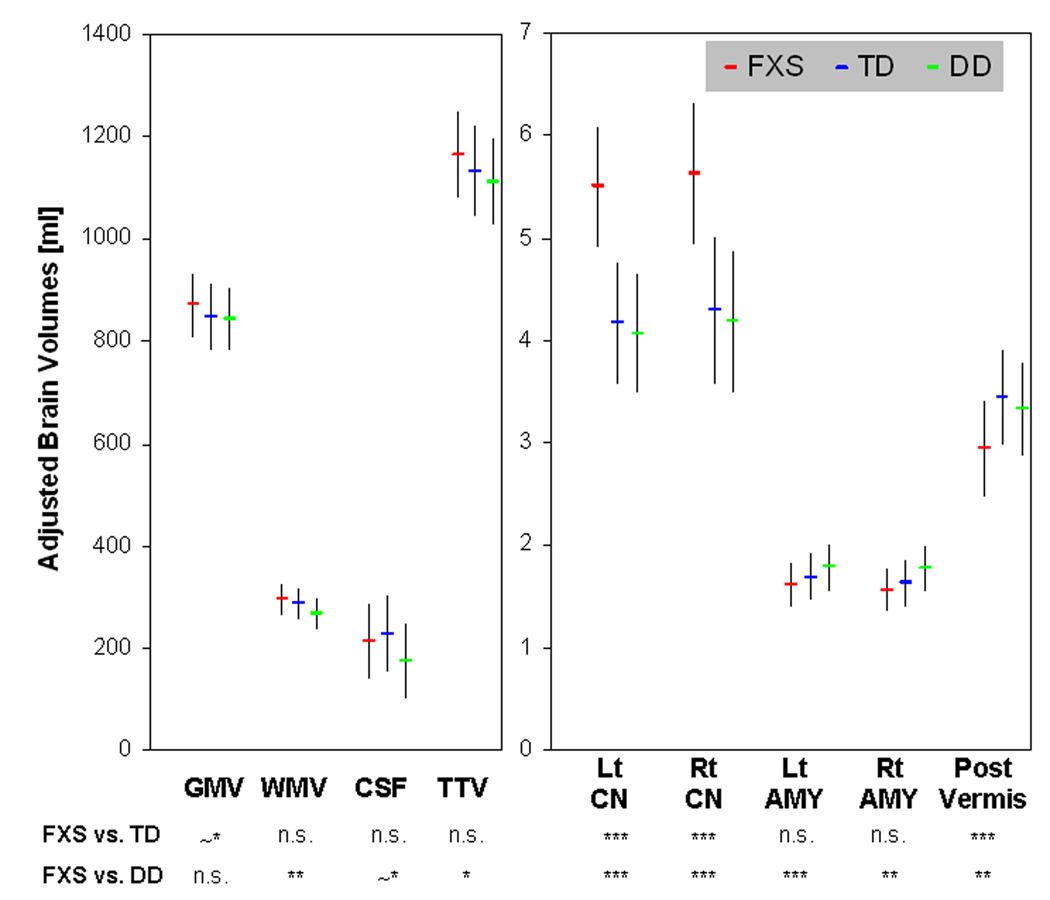
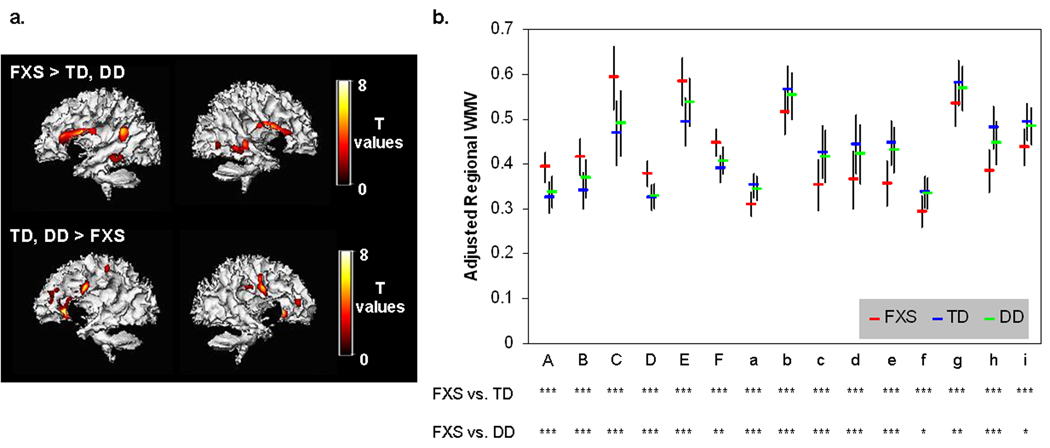
Univariate whole-brain voxel-wise analysis showed significantly greater GMV in FXS compared to TD and to DD controls in specific regions that included bilateral caudate, occipital cortex including fusiform gyrus (FG), hypothalamus and thalamus. In contrast, FXS showed significantly reduced GMV in superior temporal gyrus (STG), hippocampus, insula, orbitofrontal cortex (OFC) and medial prefrontal cortices (mPFC) compared to TD and DD (Fig. 1 a, b, Table 2). Probabilistic maps from permutation analysis show the highly reliable nature of the results (Fig. 2). Volumetric measures were consistent with the whole-brain results (Fig. 3, Table 4). Results from white matter (WM) regions showing significantly different white matter volumes between FXS and controls in the prefrontal and temporal regions are also reported in Fig. 4 and Table 3. Within FXS, FMRP levels were significantly (negatively) correlated with bilateral caudate volumes identified from both VBM and volumetric analyses (P’s < 0.05).
Fig. 1. Differences in GMV between groups.
a. Regions that show significant differences in GMV between FXS and controls. P=0.05 family-wise error corrected (FWE), extent threshold=200.
b. Adjusted GMVs are plotted to ensure that only one control group (TD or DD) is not driving the overall effect. Alphabets on x-axis correspond to ROI labels in Table 2. n.s.: P≥0.1, ~: P<0.1, *: P<0.05, **: P<0.01, ***: P<0.001. Error bars represent standard deviation.
Fig. 2. Probabilistic maps of regional GMV differences between groups.
GM regions that show significantly greater and less volumes in FXS compared to TD and DD groups was examined using leave-one out cross-validation analysis permuted 102 times. Scaling bars represent T-values. A value of 1 in a given voxel indicates that all permutations showed significant effects (at a threshold of P = 0.05 family-wise error corrected (FWE), extent threshold = 200). Age, scan-site and total GMV are entered as nuisance variables. Statistical maps are overlaid on a custom GM template. Most regions showing a value of 1 indicate the consistency of results. Left hemisphere is shown on left-side.
Fig. 3. Adjusted brain volumes for each group.
Total gray matter volume (GMV), total white matter volume (WMV), total cererbro-spinal fluid (CSF) and total tissue volume (TTV: GMV+WMV) are adjusted for age and scan-site. Left and right caudate nuclei volumes (Lt CN, Rt CN), left and right amygdala volumes (Lt AMY, Rt AMY) and posterior vermis volume (Post Vermis) are adjusted for age, scan-site and TTV. Error bars represent standard deviation. FXS: fragile X syndrome group, TD: typically developing group, DD: developmentally delayed group, n.s. P ≥ 0.10, ~*: P < 0.10, *: P < 0.05, **: P < 0.01, ***: P < 0.001.
Table 4.
Brain, Caudate, Amygdala and Cerebellar Vermis Volumes
| Brain Volume | Adjusted for | FXS (N=51) | TD (N=32) | DD (N=18) | P Values | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mean | SD | mean | SD | mean | SD | ANOVA | FXS vs. TD |
FXS vs. DD |
TD vs. DD |
||
| GMV (ml) | age, site | 872.0 | 62.0 | 847.3 | 63.8 | 844.9 | 61.3 | 0.13 | |||
| WMV (ml) | age, site | 294.7 | 30.3 | 287.2 | 31.2 | 267.3 | 30.0 | 0.005 | n.s. | ** | * |
| CSF (ml) | age, site | 212.8 | 71.1 | 228.6 | 73.2 | 175.3 | 70.3 | 0.05 | n.s. | ~* | * |
| TTV (ml) | age, site | 1166.7 | 85.0 | 1134.5 | 87.5 | 1112.2 | 84.0 | 0.04 | n.s. | * | n.s. |
| Left Caudate (ml) | age, site, TTV | 5.50 | 0.58 | 4.18 | 0.59 | 4.08 | 0.58 | <0.001 | *** | *** | n.s. |
|
Right Caudate Volume (ml) |
age, site, TTV | 5.63 | 0.69 | 4.30 | 0.71 | 4.19 | 0.69 | <0.001 | *** | *** | n.s. |
|
Left Amygdala Volume (ml) |
age, site, TTV | 1.62 | 0.21 | 1.70 | 0.23 | 1.79 | 0.22 | 0.002 (~*) | n.s. | ***(*) | * (~*) |
|
Right Amygdala Volume (ml) |
age, site, TTV | 1.57 | 0.21 | 1.63 | 0.22 | 1.78 | 0.21 | 0.02 (n.s.) |
n.s. | **(n.s.) | n.s. |
|
Anterior Vermis (ml) |
age, site, TTV | 4.84 | 0.55 | 4.81 | 0.56 | 4.76 | 0.54 | 0.86 | |||
| Middle Vermis (ml) | age, site, TTV | 2.84 | 0.51 | 2.98 | 0.52 | 2.99 | 0.51 | 0.42 | |||
|
Posterior Vermis (ml) |
age, site, TTV | 2.95 | 0.46 | 3.45 | 0.46 | 3.34 | 0.45 | <0.001 | *** | **(*) | n.s. |
GMV: gray matter volume, WMV: white matter volume, CSF: cerebro-spinal fluid, TTV: total brain volume (GMV+WMV)
SD: standard deviation, Amygdala (N): FX=50, TD=28, DD=16
n.s.: P ≥ 0.10,
P < 0.10,
P < 0.05,
P < 0.01,
P < 0.001
( ): Results adjusting for age and site are noted in brackets when different from results adjusting for age, site and TTV.
Fig. 4. Regional WMV differences between groups.
a. WM regions that show significantly greater and less volumes in FXS compared to TD and DD groups. P = 0.05 family-wise error corrected (FWE), extent threshold = 200. Age, scan-site and total WMV are entered as nuisance variables. Statistical maps are overlaid on a representative single FXS (top) and TD (bottom) subject’s segmented white matter.
b. Brain volumes adjusted for age, scan-site and total WMV from regions in (a) are extracted and plotted to ensure that one of the control groups (TD or DD) is not driving the effect. Alphabets on x-axis correspond to ROI labels in Table 3. n.s.: P ≥ 0.10, ~*: P < 0.10, *: P < 0.05, **: P < 0.01, ***: P < 0.001. †: Comparison between FXS and DD adjusted additionally for IQ.
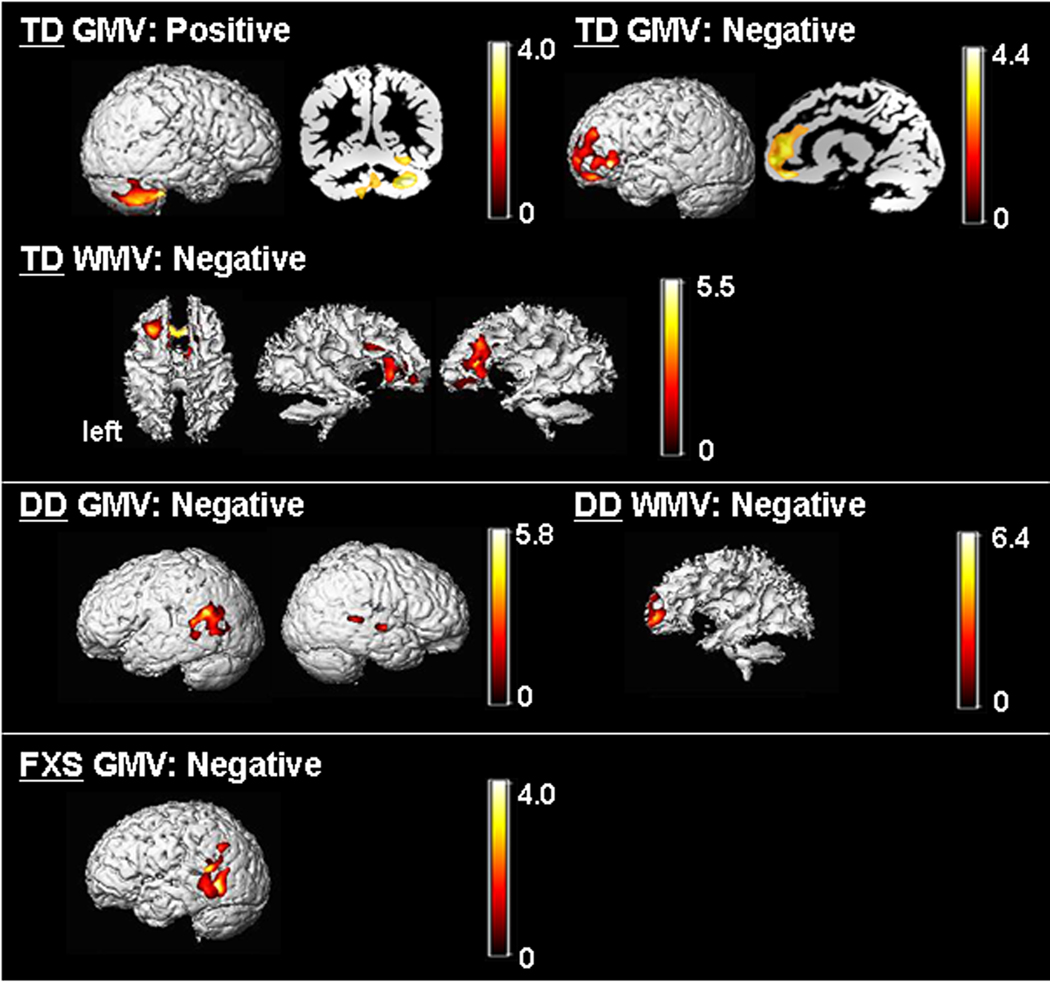
When covariation between cognitive functioning (IQ) and morphometric patterns was examined, in TD, IQ showed significant positive correlation with cerebellar hemispheres (left > right), vermis and FG GMV, and negative correlations with right dorsolateral PFC (DLPFC), bilateral mPFC/OFC GMV and mPF WMV (Fig. 5). DD showed significant positive correlation between IQ and GMV in bilateral parieto-temporal regions and negative correlation between IQ and WMV in a mPF region adjacent to that found in TD. In contrast, significant (negative) brain-IQ associations were observed in FXS in the left parieto-temporal region only, similar to DD, but not in other regions.
Fig. 5. Covariation of regional brain volumes and IQ.
GM and WM regions that show significant correlation with IQ (Mullen Composite Standard Score) in TD and DD groups (FXS group showed no significant correlation). P = 0.01 corrected. Controlled for age, scan-site and total GMV (for GM regions) / WMV (for WM regions). Statistical maps are overlaid on a representative single TD (for correlations in the TD group) or DD (for correlations in the DD group) subject’s segmented GM (for GM regions) or WM (for WM regions). Scaling bars represent T-values. Left side shows left hemispheres except when noted.
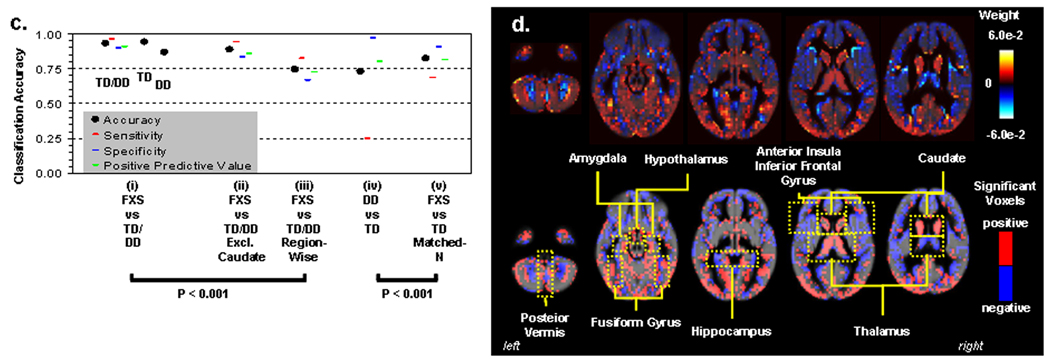
We adopted machine learning algorithms to examine whether whole-brain voxel-wise spatial patterns of GMV could discriminate between groups. While the spatial resolution of human neuroimaging is limited, hence leaving most studies to examine extended regions of the brain, it has recently been shown that MRI can be used to study fine-grained neural representations, even when they are encoded at a finer scale than the resolution of the measurement grid23. Using a leave-one-out linear SVM approach, FXS was discriminated from controls with 92.9 % classification accuracy (Fig. 6a(i)). Classification accuracy between FXS and TD was significantly better than between DD and TD even when matched for small sample-size of DDs (P < 0.001) (Fig. 6a(iv) and (v)). Comparisons across SVM methods showed the importance of voxel-wise spatial information in classification accuracy rather than using more coarse region-based measures (116 anatomical labels, http://www.cyceron.fr/freeware/) of GMV (P < 0.001) (Fig. 6a(iii)). This was presumably due to the mixture of voxels that showed both increased and decreased GMV in a given region (e.g. medial vs. lateral amygdala, hippocampus and posterior vermis, Fig. 6b). Finally, because enlarged caudate volume is the most characteristic morphometric feature in FXS, we repeated the classification analysis excluding caudate voxels, and still obtained high classification accuracy (88.9 %) (Fig. 6a(ii)).
Fig. 6. Pattern classification results.
a. Classification accuracy. SVM results classifying FXS and controls using all GM voxels (i), all GM voxels except caudate voxels (ii), and 116 brain regions (iii). To the right is SVM results classifying DD from TD using all GM voxels (iv), and FXS from TD in a subset of randomly selected FXS (N=18) to match sample-size of DD (v).
b. Whole-brain representation of pattern classification results from FXS vs. TD/DD using all GM voxels. Axial brain images of weight vectors from leave-one-out SVM for all voxels (top), and spatial patterns of most significant voxels when thresholded at P=0.05 (according to 2000 permutations) (bottom).
DISCUSSION
In this study, voxel-wise structural brain profiles obtained from healthy children ages 1 to 3 years were characterized and compared to profiles obtained from children with a specific neurodevelopmental disorder, FXS, and an idiopathic DD group. Findings of increased caudate, FG and thalamus GMV, and reduced STG, hippocampus and OFC GMV from the univariate voxel-wise analysis partially replicate results from studies of older individuals with FXS 4–7. Associations between larger caudate volume and reduced levels of FMRP (generally reflecting lower functioning in FXS) also replicates studies in older individuals with FXS 5, 8. Further, white matter abnormalities within the medial prefrontal region are in line with a diffusion tensor imaging (DTI) study in female adolescents with FXS 26 and supports the finding of executive function deficit in FXS. The detection of abnormal morphology of these regions in children as young as 18 months suggests that early genetic influences significantly influence selected components of neurodevelopment during the prenatal period or early infancy in FXS. Similar patterns of neuroanatomical differences observed between FXS and TD and FXS and DD also point to the specificity of early neurodevelopmental abnormalities resulting from the FMR1 full mutation as opposed to non-specific effects of developmental delay. Unlike previous observations in older subjects 5, abnormal size in regions such as the amygdala was not observed in FXS with voxel-based or volumetric analyses. Pattern classification analyses however, indicated that mixture of voxels exist, with both increased and decreased volumes in a medial/lateral fashion within such regions (note similar patterns in hippocampus and posterior vermis, Fig. 6b). The classification results indicate the complexity of genetic influences on brain morphology rather than a model in which genetic factors uniformly affect a given brain region. Replication of these results in longitudinal studies is warranted. Further, future research can benefit from the inclusion of larger samples of TD and DD controls.
On the other hand, several regions that were not identified as morphologically aberrant in previous studies of older FXS individuals were observed here; enlarged hypothalamus, and smaller insula, mPFC and lateral PFC. The hypothalamus finding is of particular interest in light of hypothalamic-pituitary-adrenal (HPA) dysfunction and abnormal stress responses found in both children with FXS 27 and in the animal model of this disorder 28. Abnormal insula and mPFC volume in FXS is consistent with observations of aberrant insular and mPFC activation during gaze processing (unpublished data), hyperarousal 29, and the shared social and cognitive features with autism 30. These findings indicate that the seeds of social cognitive dysfunction observed in FXS are likely to arise directly from neurodevelopmental effects of the mutation, later interacting with common environmental influences. Further, it is possible that abnormality of the lateral PFC, in concert with aberrant caudate morphology, may be responsible for the known executive function (response inhibition) deficit, hyperactivity and fronto-striatal dysfunction in FXS 4, 31, 32. Lastly, it is possible that some of the abnormalities observed in very young, but not older individuals with FXS ‘normalize’ over the course of development, at least in terms of volumes. Indeed, dendritic spine abnormalities in mouse models of FXS have been shown to diminish in certain brain regions during development 33. The exact mechanisms, however, remain to be elucidated.
Another finding of note is the association between regional brain volumes and IQ. The TD group showed a negative association between IQ and prefrontal GMV. This finding is consistent with what has been shown previously in healthy children of ages 3.8 – 8.4 years 2. We add to this and demonstrate that similar results are observed in younger TD children. We also present novel findings suggesting associations between anatomy and IQ for FG and cerebellar GM (TD group) and mPF WM in both TD and DD (but not in FXS). The negative association with the parieto-temporal region appeared non-specific and was found in both DD and FXS.
Finally, we illustrate that fine spatial patterns of GMV accurately predict whether a child has FXS. Similar analytic strategies as applied to functional neuroimaging data have been shown to be a powerful tool in decoding mental states within individuals where traditional massive univariate general linear models fail 23. Here we show its usefulness in classifying disease using structural neuroimaging data. These findings may inform future studies predicting cognitive outcome in FXS as well as research focused on children with other neurodevelopmental syndromes. Further, these findings have implications for understanding the genetic control of healthy brain development.
ACKNOWLEDGMENT
This study was funded by MH64708-05 (A.L.R., J.P.), MH61696 (J.P.) and HD03110-36 (J.P.) and Child Health Research Program (CHRP) from Stanford University School of Medicine (F.H.). The corresponding author had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. We sincerely thank all of the families who made this study possible. We also thank Chad Chappell, Nancy Garrett, Michael Graves, Cindy Hagan, Cindy Johnston, Arianna Martin, Robin Morris, Judy Morrow, Matthew Mosconi, Rachel G. Smith, Cristiana Vattuone, Christa Watson and Anh Weber for their invaluable work on the project.
REFERENCE
- 1.Toga AW, Thompson PM, Sowell ER. Mapping brain maturation. Trends Neurosci. 2006 Mar;29(3):148–159. doi: 10.1016/j.tins.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw P, Greenstein D, Lerch J, et al. Intellectual ability and cortical development in children and adolescents. Nature. 2006 Mar 30;440(7084):676–679. doi: 10.1038/nature04513. [DOI] [PubMed] [Google Scholar]
- 3.Greenough WT, Klintsova AY, Irwin SA, Galvez R, Bates KE, Weiler IJ. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss AL, Dant CC. The behavioral neurogenetics of fragile X syndrome: analyzing gene-brain-behavior relationships in child developmental psychopathologies. Dev Psychopathol. 2003;15(4):927–968. doi: 10.1017/s0954579403000464. Fall. [DOI] [PubMed] [Google Scholar]
- 5.Gothelf D, Furfaro JA, Hoeft F, et al. Neuroanatomy of fragile X syndrome is associated with aberrant behavior and FMRP. Annals of Neurology. 2007 Oct 11; doi: 10.1002/ana.21243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kates WR, Folley BS, Lanham DC, Capone GT, Kaufmann WE. Cerebral growth in Fragile X syndrome: review and comparison with Down syndrome. Microsc Res Tech. 2002 May 1;57(3):159–167. doi: 10.1002/jemt.10068. [DOI] [PubMed] [Google Scholar]
- 7.Lee AD, Leow AD, Lu A, et al. 3D pattern of brain abnormalities in Fragile X syndrome visualized using tensor-based morphometry. Neuroimage. 2007 Feb 1;34(3):924–938. doi: 10.1016/j.neuroimage.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiss AL, Abrams MT, Greenlaw R, Freund L, Denckla MB. Neurodevelopmental effects of the FMR-1 full mutation in humans. Nat Med. 1995 Feb;1(2):159–167. doi: 10.1038/nm0295-159. [DOI] [PubMed] [Google Scholar]
- 9.Gothelf D, Furfaro JA, Hoeft F, et al. Neuroanatomy of fragile X syndrome is associated with aberrant behavior and FMRP. Annals of Neurology. doi: 10.1002/ana.21243. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen EM. Mullen Scales of Early Learning AGS Edition. Circle Pines, MN: American Guidance Service, Inc; 1995. [Google Scholar]
- 11.Oberle I, Rousseau F, Heitz D, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5010):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 12.Willemsen R, Mohkamsing S, de Vries B, et al. Rapid antibody test for fragile X syndrome. Lancet. 1995 May 6;345(8958):1147–1148. doi: 10.1016/s0140-6736(95)90979-6. [DOI] [PubMed] [Google Scholar]
- 13.Sparrow SS, Balla DA, Cicche HV. Vineland Adaptive Behavior Scales-Interview Edition Survey Form Manual. Circle Pines: American Guidance Service, Inc; 1984. [Google Scholar]
- 14.Achenbach TM, Ruffle TM. The Child Behavior Checklist and related forms for assessing behavioral/emotional problems and competencies. Pediatr Rev. 2000 Aug;21(8):265–271. doi: 10.1542/pir.21-8-265. [DOI] [PubMed] [Google Scholar]
- 15.Bodfish JW, Crawford TW, Powell SB, Parker DE, Golden RN, Lewis MH. Compulsions in adults with mental retardation: prevalence, phenomenology, and comorbidity with stereotypy and self-injury. Am J Ment Retard. 1995 Sep;100(2):183–192. [PubMed] [Google Scholar]
- 16.Bodfish JW, Symons F, Lewis MH. The Repetitive Behavior Scales. Western Carolina Center Research Reports; 1999. [Google Scholar]
- 17.Xia Y, Bettinger K, Shen L, Reiss AL. Automatic segmentation of the caudate nucleus from human brain MR images. IEEE Trans Med Imaging. 2007 Apr;26(4):509–517. doi: 10.1109/TMI.2006.891481. [DOI] [PubMed] [Google Scholar]
- 18.Schumann CM, Hamstra J, Goodlin-Jones BL, et al. The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J Neurosci. 2004 Jul 14;24(28):6392–6401. doi: 10.1523/JNEUROSCI.1297-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henkelman RM. Measurement of signal intensities in the presence of noise in MR images. Med Phys. 1985 Mar-Apr;12(2):232–233. doi: 10.1118/1.595711. [DOI] [PubMed] [Google Scholar]
- 20.Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Thieme; 1988. [Google Scholar]
- 21.Poline JB, Worsley KJ, Evans AC, Friston KJ. Combining spatial extent and peak intensity to test for activations in functional imaging. Neuroimage. 1997 Feb;5(2):83–96. doi: 10.1006/nimg.1996.0248. [DOI] [PubMed] [Google Scholar]
- 22.Burges C. A tutorial on support vector machines for pattern recognition. Data Min.Knowl. Discov. 1998;2(2):144–152. [Google Scholar]
- 23.Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nat Rev Neurosci. 2006 Jul;7(7):523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- 24.Haynes JD, Sakai K, Rees G, Gilbert S, Frith C, Passingham RE. Reading hidden intentions in the human brain. Curr Biol. 2007 Feb 20;17(4):323–328. doi: 10.1016/j.cub.2006.11.072. [DOI] [PubMed] [Google Scholar]
- 25.Mourao-Miranda J, Reynaud E, McGlone F, Calvert G, Brammer M. The impact of temporal compression and space selection on SVM analysis of single-subject and multi-subject fMRI data. Neuroimage. 2006 Dec;33(4):1055–1065. doi: 10.1016/j.neuroimage.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Barnea-Goraly N, Eliez S, Hedeus M, et al. White matter tract alterations in fragile X syndrome: preliminary evidence from diffusion tensor imaging. Am J Med Genet B Neuropsychiatr Genet. 2003 Apr 1;118(1):81–88. doi: 10.1002/ajmg.b.10035. [DOI] [PubMed] [Google Scholar]
- 27.Hessl D, Rivera SM, Reiss AL. The neuroanatomy and neuroendocrinology of fragile X syndrome. Ment Retard Dev Disabil Res Rev. 2004;10(1):17–24. doi: 10.1002/mrdd.20004. [DOI] [PubMed] [Google Scholar]
- 28.Lauterborn JC. Stress induced changes in cortical and hypothalamic c-fos expression are altered in fragile X mutant mice. Brain Res Mol Brain Res. 2004 Nov 24;131(1–2):101–109. doi: 10.1016/j.molbrainres.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Miller LJ, McIntosh DN, McGrath J, et al. Electrodermal responses to sensory stimuli in individuals with fragile X syndrome: a preliminary report. Am J Med Genet. 1999 Apr 2;83(4):268–279. [PubMed] [Google Scholar]
- 30.Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci. 2006 Oct;9(10):1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- 31.Hoeft F, Hernandez A, Parthasarathy S, Watson CL, Hall SS, Reiss AL. Fronto-striatal dysfunction and potential compensatory mechanisms in male adolescents with fragile X syndrome. Hum Brain Mapp. 2007 Jun;28(6):543–554. doi: 10.1002/hbm.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menon V, Leroux J, White CD, Reiss AL. Frontostriatal deficits in fragile X syndrome: relation to FMR1 gene expression. Proc Natl Acad Sci U S A. 2004 Mar 9;101(10):3615–3620. doi: 10.1073/pnas.0304544101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005 May;6(5):376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]