Abstract
Although microRNAs (miRNA) have extensively been investigated in cancer research, less attention has been paid to their regulation by carcinogens and/or protective factors in early stages of the carcinogenesis process. The present study was designed to evaluate the modulation of mRNA expression as related to exposure of neonatal mice to environmental cigarette smoke (ECS) and to treatment with chemopreventive agents. Exposure to ECS started immediately after birth and for 2 weeks after weaning. Thereafter, groups of mice received daily either budesonide (BUD) or phenethyl isothiocyanate (PEITC) with the diet. The expression of 576 miRNAs was evaluated by miRNA microarray in liver and lung. In sham-exposed mice, the expression of miRNAs tended to be higher in liver than in lung. ECS downregulated the expression of a number of miRNAs in lung, whereas mixed alterations were observed in liver. PEITC and BUD did not substantially affect the physiological situation in lung, whereas both agents caused intense variations in liver, reflecting the occurrence of damage mechanisms, such as inflammation, DNA and protein damage, cellular stress, proliferation and apoptosis. PEITC and BUD protected the lung from ECS-induced alterations of miRNA expression but exhibited some adverse effects in liver.
Introduction
The regulation of gene expression is crucial for maintaining tissue homeostasis as well as for the development of pathological conditions, such as cancer, cardiovascular diseases, diabetes, neurological disorders and viral infections (1). MicroRNAs (miRNAs) constitute a family of small non-coding RNA molecules that negatively regulate gene expression by affecting protein translation either through inhibition of messenger RNA translation, initiation and elongation or through messenger RNA cleavage (2). Alike mRNAs, miRNAs are expressed in a tissue-specific manner, and human adult tissues have unique miRNA profiles (3). miRNAs interact with classic oncogene and tumor suppressor networks thereby contributing to the initiation and progression of many, if not all, human malignancies (4).
miRNAs have extensively been investigated in cancer research, but little is known regarding their response either to noxious agents or to protective agents in apparently healthy tissues, during early stages of the carcinogenesis process. Recently, we analyzed the expression of 484 miRNAs in the lung of Sprague–Dawley rats exposed to environmental cigarette smoke (ECS). Microarray and quantitative polymerase chain reaction (qPCR) analyses provided evidence that ECS causes an extensive downregulation in miRNA expression (5), which correlates with formation of bulky DNA adducts and with overexpression of a number of genes and proteins in the same tissues (6,7). Further studies in CD-1 mice confirmed that exposure to ECS produces dramatic changes in the lung, mostly in the sense of downregulation, which reflects both adaptive mechanisms and activation of pathways involved in the pathogenesis of pulmonary diseases (8). Interestingly, cigarette smoking causes downregulation of many miRNAs also in the human airway epithelium (9). A study that profiled 217 miRNAs across multiple types of lung cancer tissues found that most miRNAs were downregulated in tumors compared with normal tissues, and a signature of downregulated miRNAs was identified in lung cancer tissues relative to adjacent normal lung (10).
It is also of interest to evaluate whether pharmacological agents and dietary components are able to modulate the alterations of miRNA expression induced by carcinogens. Indole-3-carbinol was found to reduce miRNA expression in lung tumors induced by vinyl carbamate in mice (11). We evaluated the effects of several chemopreventive agents, including indole-3-carbinol, N-acetylcysteine, 5,6-benzoflavone and phenethyl isothiocyanate (PEITC), either individually or in combination, in the apparently healthy lungs from either ECS-free or ECS-exposed adult rats, taken after 4 weeks of treatment. None of the above chemoprevention regimens appreciably affected the baseline miRNA expression, indicating potential safety, while they attenuated ECS-induced alterations to a variable extent and with different patterns, indicating potential preventive efficacy (12).
The present study was designed to evaluate the early effects of ECS not only in the lung but also in the liver of neonatal mice and to investigate the effects of budesonide (BUD) and PEITC in both organs of either ECS-free or ECS-exposed mice. While it is well established that tobacco smoke is the dominant cause of lung cancer, its involvement in liver carcinogenesis is still controversial. Exposure of rats to mainstream cigarette smoke selectively induced the formation of DNA adducts in lung but not in liver (6,13). However, the most recent epidemiological studies have shown a moderate association between the tobacco smoking and the risk of liver cancer (14). BUD is a synthetic glucocorticoid that is commonly used for the treatment of chronic inflammatory diseases such as asthma and Crohn's disease. BUD inhibited the formation of lung tumors in mice treated with smoke compounds, such as benzo[a]pyrene (15–19) and vinyl carbamate (20). Moreover, BUD modulated the expression of a number of genes in the lung tumors developed in benzo[a]pyrene-treated mice (21). PEITC is a naturally occurring isothiocyanate contained in watercress (Nasturtium officinale), which has been shown to block the metabolic activation of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl-1-butanone) (22) and to inhibit nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl-1-butanone)-induced lung tumors in both mice and rats (22–25). Moreover, PEITC inhibited DNA alterations and other intermediate biomarkers (26) and modulated the expression of a number of genes (6) and miRNAs (12) in ECS-exposed rats. In the same mice used in the present study, both PEITC and BUD inhibited the damage to pulmonary DNA and, after 9 months, showed a moderate ability to inhibit the formation of ECS-induced lung tumors (27).
The results herein reported show that the baseline expression of miRNAs is generally higher in liver than in lung. Exposure of neonatal mice to ECS dysregulated miRNA expression in lung and, even more intensely, in liver. While BUD and especially PEITC protected the lung from ECS-induced miRNA alterations, dysregulation of miRNA expression in liver by both chemopreventive agents prevailed on modulation of ECS-related alterations.
Materials and methods
Mice
Twenty-five pregnant Swiss CD-1 albino mice were purchased from Harlan Italy (Correzzana, Milan, Italy), housed in Makrolon cages on sawdust bedding in a cabinet where filtered air was circulated and maintained on standard rodent chow (Teklad 2018, Harlan Italy) and tap water ad libitum. The housing conditions were as follows: temperature of 23 ± 2°C, relative humidity of 55%, 12 h day–night cycle. Housing and all treatments of mice were in accordance with National Institutes of Health guidelines and with our institutional guidelines.
Treatments
Immediately after birth, the mice were divided into six experimental groups, including sham-exposed mice, kept in filtered air (Group A), sham-exposed mice treated with PEITC after weaning (Group B), sham-exposed mice treated with BUD after weaning (Group C), mice exposed to ECS since birth (Group D), mice exposed to ECS since birth and treated with PEITC after weaning (Group E) and mice exposed to ECS since birth and treated with BUD after weaning (Group F). The mice used in the present study (eight per group) were part of larger sets of mice used for a subchronic toxicity study with PEITC and BUD and for a chemoprevention study in ECS-exposed mice (27).
Within 12 h after birth, the neonatal mice composing Groups D–F started to be exposed whole body to ECS, generated in a smoking machine (model TE-10c, Teague Enterprises, Davis, CA) adjusted to produce a combination of sidestream (89%) and mainstream smoke (11%). Burning five Kentucky 2R4F reference cigarettes (Tobacco Research Institute, Lexington, KY), having a declared content of 9.2 mg of tar and 0.8 mg of nicotine each, yielded on average a total suspended particulate matter of 63.3 mg/m3 in the exposure chambers. Exposure was daily, 6 h/day divided into two rounds with a 3 h interval and continued until the end of the experiment.
PEITC and BUD were purchased from Sigma Chemical Co. (St Louis, MO). Immediately after weaning (∼30 days after birth), the mice composing Groups B and E started receiving daily PEITC (1000 mg/kg diet) and those composing Groups C and F started receiving daily BUD (2.4 mg/kg diet) until the end of the experiment. The doses of PEITC and BUD were selected based on the results of a preliminary 6 week toxicity study (28).
Fifteen days after weaning, i.e. 45 days after birth, eight male mice per group were deeply anesthetized with diethyl ether and killed by cervical dislocation. Lung and livers were collected. A portion of the right lung was used for DNA extraction and analysis of bulky DNA adducts (27). Another portion of the right lung and the whole liver, to be used for RNA extraction and analysis of miRNAs (this study), were immersed in an RNA-stabilizing buffer and stored at −80°C.
RNA extraction and analysis
Two pools of liver and two pools of lung were prepared within each one of the six experimental groups, for a total of 24 samples to be subjected to miRNA analysis. RNA was extracted by means of TRIzol Plus RNA Purification Kit (Invitrogen, Carlsbad, CA). Briefly, the lung samples were homogenized in TRIzol (1 ml/50–100 mg tissue) and 200 μl chloroform were added to 1 ml TRIzol. The mixture was shaken and the samples were centrifuged at 12 000g for 15 min at 4°C. The colorless upper aqueous phase containing RNA was transferred to a fresh tube and an equal volume of 70% ethanol was added and mixed thoroughly. The sample was then dispensed into an RNA spin cartridge and centrifuged at 12 000g for 15 min. The flow-through was discarded and the cartridge retaining RNA was washed twice with a washing buffer and dried by centrifugation. The RNA was then eluted by adding 50 μl RNase-free water to the cartridge and by centrifuging at 12 000g for 2 min.
RNA amounts and quality were evaluated by fiber optic spectrophotometry using a fiber optic spectrophotometer (NanoDrop Technologies, Wilmington, DE). The results obtained indicated a good quality of all purified samples (260/230 absorbance ratio ≥ 1.95). RNA structural integrity was assessed by capillary electrophoresis by using a bioanalyzer (2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA) calculating the RNA integrity number, which takes into account the entire electrophoretic trace as well as its peaks (18S, 28S ribosomal RNA). Lung and liver RNA samples were dried by centrifuge evaporator and stored at −80°C.
miRNA microarray
Microarray analyses evaluated the expression of 576 mouse miRNAs. miRNA microarrays were prepared on UltraGAPS™ Coated Slides with amino-silanized surface (Corning Incorporated, Corning, NY). The miRNA library used was ‘miRCURY LNA™ microRNA Array ready-to-spot probe set, v.10.0—human, mouse & rat’ (Exiqon, Vedback, Denmark). Each capture probe (300 pmol) was dried in individual wells of a 4× 384-well microplates. Probes were dissolved in Epoxide Solution (Corning Incorporated) to a final concentration of 10 μM and spotted on microarrays (GeneMachine OmniGrid Microarrayer, Accent Digilab, Halliston, MA). Each capture probe was printed in triplicate. After spotting, the slides were dried in a vacuum evaporator, crosslinked by ultraviolet light at 600 mJ/cm2 and kept under vacuum until use.
Sample labeling and miRNA array hybridization
miRNA samples were labeled by using the Mirus miRNA Labeling Kit (Label IT® miRNA Labeling Kit, Cy™3/Cy™5; Mirus Bio LLC, Madison, WI). A common miRNA reference standard composed of synthetic RNA oligonucleotides (Ambion, Foster City, CA) was used to facilitate sample comparison. Each miRNA sample was labeled with Cy3, whereas the reference standard was labeled with Cy5. miRNA samples (1 μg) were added to 10× Labeling Buffer M, MB-grade Water, Label IT Cy™3 or Cy™5 Reagent. The labeling reaction was performed at 37°C for 1 h and stopped by adding 0.1 volumes of 10× STOP Reagent. Labeled miRNAs were purified by ethanol precipitation, dried and resuspended in a hybridization solution.
miRNA microarray hybridization was performed by using a hybridization station (HS4000-PRO; Tecan, Männedorf, Switzerland) at 56°C for 16 h. The hybridized slides were washed with buffer and dried under a nitrogen flow. The microarrays were scanned at 535 and 635 nm (AXON GENEPIX 4000B Scanner; Molecular Devices, Sunnyvale, CA).
Data analysis
For each microarray, the local spot background was subtracted from the raw spot intensity. The data were then log transformed, normalized and analyzed by GeneSpring® software version 7.2 (Agilent Technologies). miRNA expression data were normalized both per gene and per array by using the GeneSpring median-centered normalization option. Triplicate data generated for each miRNA were compared among the various experimental groups by volcano plot analysis. Two-fold variations, accompanied by statistical significance (P < 0.05), as evaluated by analysis of variance after Bonferroni multiple testing correction, were assumed as thresholds for positivity. Global miRNA expression profiles, as related to the various experimental conditions tested, were compared by hierarchical cluster analysis and bidimensional principal component analysis of variance (PCA).
Results
Comparison of miRNA expression in the lung and liver of sham-exposed mice
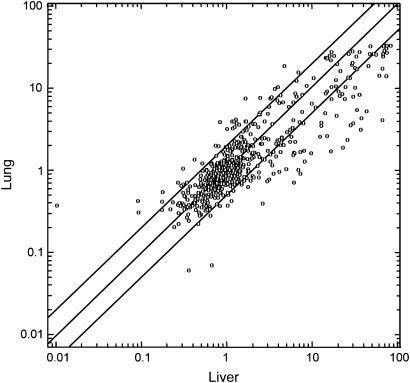
The comparative analysis of the lung and liver mixed-cell populations from sham-exposed mice revealed an evident organospecificity of miRNA profiles. Figure 1 shows a scatter plot relating the expression intensity of 576 miRNAs in mouse liver (x-axis) and lung (y-axis). Differences are evident between the two organs, with a general trend toward a higher expression of miRNAs in liver. In fact, the expression of 89 miRNAs (15.4%) was at least 2-fold higher in liver as compared with lung, whereas the expression of 39 miRNAs (6.8%) was at least 2-fold higher in lung as compared with liver. The greatest differences occurred with miR-122, which had a 7-fold higher baseline expression in liver than in lung. The complete list of differentially expressed miRNAs in mouse liver and lung is available in the Geo database (Geo number requested).
Fig. 1.
Scatter plot relating the expression of 576 miRNAs in the liver (x-axis) and in the lung (y-axis) of sham-exposed male mice. The scales indicate the normalized miRNA expression intensity. The central diagonal line indicates equivalence in the intensity of miRNA expression in liver and lung. The outer diagonal lines indicate 2-fold differences in miRNA expression between the two organs. The circles falling in the upper left area refer to miRNAs whose expression was >2-fold higher in lung as compared with liver, whereas the circles falling in the bottom right area refer to miRNAs whose expression was >2-fold higher in liver as compared with lung.
ECS-induced miRNA alterations in mouse lung and liver
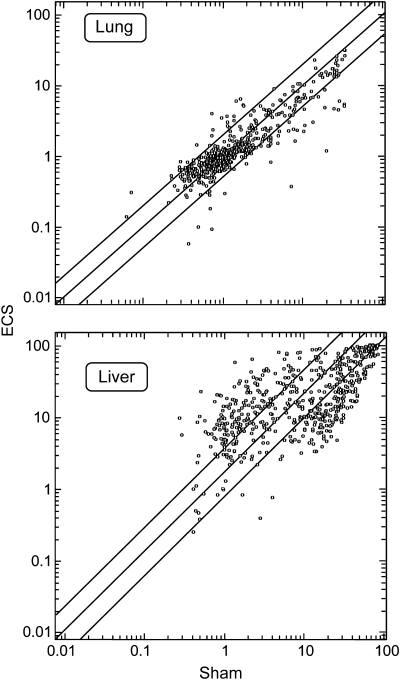
Figure 2 shows scatter plots relating the expression intensity of 576 miRNAs in the lung and liver of mice, either sham-exposed (x-axis) or ECS-exposed (y-axis). In lung, 43 miRNAs (7.5%) were downregulated and 29 (5.0%) were upregulated, whereas in liver, 90 miRNAs (15.6%) were downregulated and 76 (13.2%) were upregulated. The complete list of differentially expressed miRNAs in mouse liver and lung, as related to exposure to ECS, is available in the Geo database (Geo number requested).
Fig. 2.
Scatter plots relating the expression of 576 miRNAs in the lung and liver of male mice, either sham-exposed (x-axis) or ECS-exposed (y-axis). The scales indicate the normalized miRNA expression intensity. The central diagonal line indicates equivalence in the intensity of miRNA expression in sham-exposed and ECS-exposed mice. The outer diagonal lines indicate 2-fold differences in miRNA expression between the two exposure conditions. The circles falling in the upper left area refer to miRNAs whose expression was >2-fold higher in ECS-exposed mice as compared with sham-exposed mice, whereas the circles falling in the bottom right area refer to miRNAs whose expression was >2-fold higher in sham-exposed mice as compared with ECS-exposed mice.
Modulation by PEITC and BUD of miRNA expression in the lung and liver of sham-exposed mice
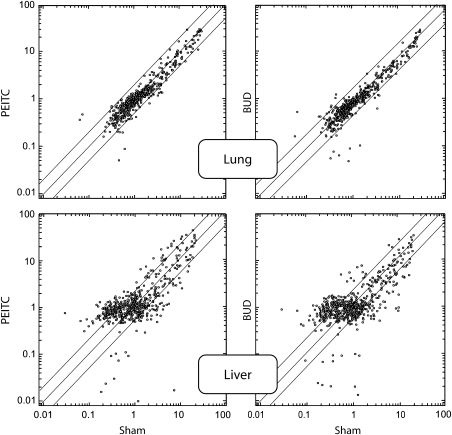
Figure 3 shows scatter plots relating the expression intensity of 576 miRNAs in the lung and liver of mice, either sham exposed (x-axis) or treated with either PEITC or BUD (y-axis). The effect of both PEITC and BUD on the baseline miRNA expression was negligible in lung, whereas intense variations were observed in liver. In particular, as shown in Table I and inferred from Venn diagrams and volcano-plot analyses (data not shown), several miRNAs were altered in their expression.
Fig. 3.
Scatter plots relating the expression of 576 miRNAs in the lung and liver of male mice, either untreated (sham, x-axis) or treated with either PEITC or BUD (y-axis). The central diagonal line indicates equivalence in the intensity of miRNA expression in untreated mice (sham) and in mice receiving either BUD or PEITC. The outer diagonal lines indicate 2-fold differences in miRNA expression between the two treatments. The circles falling in the upper left area refer to miRNAs whose expression was >2-fold higher in mice treated with either chemopreventive agent as compared with sham, whereas the circles falling in the bottom right area refer to miRNAs whose expression was >2-fold higher in sham as compared with mice treated with either chemopreventive agent.
Table I.
Significant variations of miRNA expression in the lung and liver of mice, as related to ECS and treatment with either BUD or PEITC
| miRNA identity | Lung |
Liver |
||||||||
| BUD/sham | PEITC/sham | ECS/sham | ECS + BUD/ECS | ECS + PEITC/ECS | BUD/Sham | PEITC/Sham | ECS/Sham | ECS + BUD/ECS | ECS + PEITC/ECS | |
| let-7a | — | — | ↓4.9 | ↑2.3 | ↑5.1 | ↓2.5 | — | — | — | — |
| let-7c | — | — | ↓4.3 | — | ↑4.2 | — | — | — | — | — |
| miR-15a | — | — | — | — | — | — | — | ↓2.6 | ↑2.4 | — |
| miR-21 | — | — | — | — | — | — | — | ↓2.0 | ↑2.2 | — |
| miR-26a | — | — | ↓2.0 | ↑2.2 | ↑2.0 | — | ↓2.0 | — | — | — |
| miR-27a | — | — | — | — | — | ↓2.0 | — | — | — | — |
| miR-29b | — | — | ↑2.0 | — | ↓2.9 | — | — | — | — | — |
| miR-31 | — | — | ↑2.1 | ↓2.2 | ↓3.0 | — | — | — | — | — |
| miR-34c | — | — | ↓2.2 | — | — | — | ↑2.8 | — | — | — |
| miR-100 | — | — | — | — | — | ↓2.0 | — | — | — | — |
| miR-106b | — | — | — | — | — | ↓2.8 | — | — | — | — |
| miR-125a | — | — | ↓2.2 | — | — | ↓2.1 | ↓2.6 | — | — | — |
| miR-125b | — | — | ↓2.6 | — | ↑2.2 | — | — | ↓2.8 | ↑2.6 | ↑2.2 |
| miR-133 | ↓2.0 | — | — | — | — | — | — | — | — | — |
| miR-135b | — | — | ↑2.6 | — | ↓3.1 | — | — | — | — | — |
| miR-142 | — | — | — | — | — | ↓2.0 | ↓2.3 | — | — | — |
| miR-153 | — | — | — | — | — | — | — | ↑2.1 | — | ↓2.2 |
| miR-181 | — | ↓2.0 | — | — | — | — | — | — | — | — |
| miR-200b | — | — | ↑2.0 | — | ↓2.5 | ↓2.5 | ↓2.6 | — | — | — |
| miR-292 | — | — | — | — | — | — | — | ↑2.0 | ↓2.0 | ↓2.1 |
| miR-297a | — | — | — | — | — | — | — | ↓3.8 | — | ↑3.1 |
| miR-297b | — | — | — | — | — | — | — | ↓4.2 | ↑3.5 | ↑3.1 |
| miR-299 | — | — | — | — | — | — | ↑3.3 | — | — | — |
| miR-300 | — | — | — | — | — | ↓2.2 | — | — | — | — |
| miR-320 | — | — | — | — | — | ↓2.7 | — | — | — | — |
| miR-322 | — | — | — | — | — | — | — | ↑2.0 | ↓2.1 | ↓2.3 |
| miR-323 | — | — | — | — | — | — | ↓2.9 | — | — | ↓2.5 |
| miR-331 | — | — | — | — | — | ↓2.5 | ↓2.7 | — | — | — |
| miR-338 | — | — | — | — | — | ↓2.1 | ↓2.0 | — | — | — |
| miR-376b | — | — | — | — | — | — | — | ↑2.1 | — | ↓2.0 |
| miR-382 | — | — | ↑3.7 | ↓2.8 | ↓3.0 | — | — | — | — | — |
| miR-452 | — | — | — | — | — | — | ↑2.4 | — | — | — |
| miR-463 | — | — | ↓2.0 | ↑2.1 | — | — | — | ↑3.2 | — | ↓2.1 |
| miR-466a | — | ↓2.0 | ↓2.2 | — | — | ↓3.0 | ↓2.1 | — | — | — |
| miR-466b | — | — | ↓2.7 | — | — | — | — | ↓2.0 | — | ↑2.5 |
| miR-466f | — | — | ↓2.5 | — | — | — | — | ↓2.9 | ↑3.6 | ↑3.7 |
| miR-467a | — | — | — | — | — | — | — | ↓3.4 | — | ↑3.9 |
| miR-467d | — | — | — | — | — | — | — | ↓3.2 | — | ↑3.7 |
| miR-467e | — | — | — | — | — | — | — | ↓4.3 | ↑4.2 | ↑3.8 |
| miR-470 | — | — | — | — | — | — | — | ↑2.1 | — | ↓2.0 |
| miR-483 | — | — | — | — | — | ↑2.1 | — | ↑4.2 | ↑3.7 | ↑3.6 |
| miR-539 | — | — | — | — | — | ↑3.4 | — | — | — | — |
| miR-551 | — | — | — | — | — | ↓3.8 | ↓4.0 | — | — | — |
| miR-666 | — | ↓2.0 | — | — | — | — | — | — | — | — |
| miR-687 | — | — | — | — | — | — | — | ↑2.3 | ↓2.4 | ↓2.3 |
| miR-690 | — | — | — | — | — | — | — | ↓4.8 | ↑3.6 | — |
| miR-697 | — | — | — | — | — | — | — | ↑2.2 | ↓2.5 | ↓2.4 |
| miR-706 | — | ↓2.1 | — | — | — | — | — | — | — | — |
| miR-708 | — | ↓2.9 | — | — | — | — | — | — | — | — |
| miR-709 | — | — | — | — | — | — | — | ↓4.7 | ↑3.5 | ↑4.0 |
| miR-710 | — | — | — | — | — | — | — | ↓4.8 | ↑3.1 | ↑3.8 |
| miR-719 | — | — | — | — | — | — | — | ↑2.6 | ↓3.0 | ↓2.9 |
| miR-742 | — | — | — | — | — | ↑2.2 | — | — | — | — |
| miR-763 | — | — | — | — | — | ↓3.9 | — | — | — | — |
| miR-874 | — | — | — | — | — | — | — | ↑3.2 | — | ↓2.5 |
| miR-883a | — | — | — | — | — | — | — | ↓4.3 | ↑3.6 | ↑3.3 |
Upward and downward arrows indicate significant upregulation and downregulation of miRNA expression, respectively. The numbers next to arrows indicate the fold variation. The dash means no significant variation. Only those miRNAs that were significantly dysregulated by the chemopreventive agents in either organ are reported. See the text for other miRNAs that were dysregulated by ECS only.
The identity of those miRNAs whose expression was altered by either PEITC and/or BUD in the lung and liver of sham-exposed mice is reported in Table I. Upward and downward arrows indicate the trend of the alteration, with the indication of the fold variation. The prevailing trend exerted by the two chemopreventive agents in both lung and liver was toward a downregulation of miRNA expression. In particular, in the lung, five miRNAs (0.9%) were downregulated by PEITC and one only (0.2%) was downregulated by BUD. In the liver, 12 miRNAs (2.1%) were altered by PEITC (3 upregulated and 9 downregulated) and 17 (3.0%) were altered by BUD (3 upregulated and 14 downregulated). Seven liver miRNAs were targeted by both chemopreventive agents.
The functions regulated by the altered miRNAs were inferred from the Targetscan database, by selecting miRNA target genes having a context score >0.31. As shown in Table II, the alteration of miRNA profiles by chemopreventive agents reflects the occurrence of a variety of mechanisms.
Table II.
Identity and functions of those miRNA that were affected by either BUD or PEITC in the lung and liver of Sham- and/or ECS-exposed mice
| miRNA | Regulated functions |
| let-7a/c | Cell proliferation, Ras activation, angiogenesis |
| miR-15a | Cell shape and motility, cell proliferation, stress response, protein secretion, inflammation |
| miR-21 | Protein repair, stress response, cell proliferation |
| miR-26a | Transforming growth factor expression |
| miR-27a | Cell proliferation, stress response, protein repair |
| miR-29b | Collagen production, inflammation |
| miR-31 | Protein synthesis and secretion, stress response |
| miR-34c | P53 effector |
| miR-100 | Apoptosis |
| miR-106b | Cell adhesion, TNF activation, stress response |
| miR-125a | Erbb2 activation |
| miR-125b | Stress response |
| miR-133 | Angiogenesis, linkage between muscle fiber and basement membrane |
| miR-135b | Regulation of Ras-related activities involved in cell membrane homeostasis, cell focal adhesion and gene transcription |
| miR-142 | Protein repair, DNA repair, prostaglandin-mediated platelet aggregation |
| miR-153 | Protein repair, protein synthesis, signal transduction |
| miR-181 | Stress response |
| miR-200b | Intracellular trafficking, protein repair |
| miR-292 | Hepatocyte growth factor-induced cell proliferation, angiogenesis |
| miR-297a/b | Protein repair, cell cycle progression |
| miR-299 | NF-κB activation, stress response, peroxisome activation |
| miR-300 | Protein repair, intracellular trafficking, cell proliferation |
| miR-320 | Cell adhesion, protein repair, intracellular trafficking, cell proliferation |
| miR-322 | Protein repair, cell proliferation |
| miR-323 | Peroxisome activation, protein repair |
| miR-331 | Stress response |
| miR-338 | Protein repair, stress response |
| miR-376b | Cell cycle progression, signal transduction, apoptosis, intracellular vesicle trafficking |
| miR-382 | Endocytosis, angiogenesis, protein repair |
| miR-452 | Stress response, cell cycle arrest in response to DNA damage |
| miR-463 | Cell proliferation, protein repair, stress response |
| miR-466a/b/f | Apoptosis, protein repair, cell proliferation |
| miR-467a | Cell proliferation, protein synthesis |
| miR-467d | NA |
| miR-467e | Cell proliferation, stress response |
| miR-470 | Ras activation, intracellular vesicle trafficking, xenobiotic metabolism |
| miR-483 | NA |
| miR-539 | Protein repair, intracellular trafficking |
| miR-551 | DNA repair, inflammation, cell proliferation |
| miR-666 | Protein repair, stress response |
| miR-687 | Tumor suppression by phosphatidylinositol catabolism, cell proliferation |
| miR-690 | Cell proliferation, cell adhesion |
| miR-697 | Protein repair, intracellular trafficking, cell adhesion |
| miR-706 | Intracellular trafficking, cell motility, TNF activation |
| miR-708 | Stress response, TNF and NF-κB activation |
| miR-709 | Stress response, inflammation, lysosome activation |
| miR-710 | Cell proliferation, collagen production, Ras activation |
| miR-719 | Inflammation |
| miR-742 | Protein repair, stress response |
| miR-763 | Cell membrane integrity, peroxisome biogenesis, stress response |
| miR-874 | Protein repair, intracellular vesicle trafficking, cell proliferation, P53-dependent apoptosis, inflammation, stress response |
| miR-879 | Peroxisome activation |
| miR-883a | Tumor suppressing activity through phosphatidylinositol catabolism, protein synthesis, intracellular vesicle trafficking, protein repair, cell proliferation |
NA, no target genes having a context score >0.30 is available in the Targetscan database; NF-κB, nuclear factor-kappaB; TNF, tumor necrosis factor.
Modulation by PEITC and BUD of miRNA expression in the lung of ECS-exposed mice
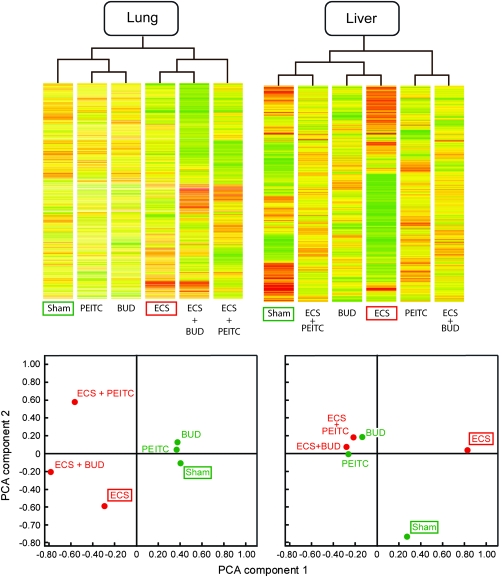
Figure 4A shows the results of supervised hierarchical cluster analysis for lung miRNAs. miRNA profiles in mice treated with either chemopreventive agent cluster with Sham. In contrast, ECS is in a separate cluster from Sham. Both ECS + BUD and ECS + PEITC are linked to ECS. However, the two chemopreventive agents behave differently, in that ECS + BUD directly clusters with ECS, whereas ECS + PEITC indirectly clusters with ECS. In addition, differences in color scales between ECS + BUD and ECS + PEITC are evident, which suggests that the two chemopreventive agents target different miRNAs in ECS-exposed mice.
Fig. 4.
Supervised hierarchical cluster analysis (upper panels) and PCA (lower panels) relative to miRNA profiles in mouse lung and liver, as related to exposure to ECS and treatment with the chemopreventive agents PEITC and BUD. Each column in the upper panels refers to a specific experimental condition, where the intensity of expression of each one of the 576 analyzed miRNAs is represented according to a color scale ranging from blue (lowest expression) to red (highest expression). Experimental conditions having similar miRNA expression profiles are linked together in the upper dendrogram. Each circle in the lower panel, corresponding to a specific experimental condition, is allocated in four quadrants according to the two main components of variance resulting from miRNA expression analysis.
These results were confirmed by bidimensional PCA showing the allocation in different quadrants of miRNA profiles in mouse lung, as related to exposure to ECS and treatment with the chemopreventive agents (Figure 4C). It is evident that ECS is allocated far away from Sham. Both chemopreventive agents, administered to ECS-free mice, are close to Sham. When administered to ECS-exposed mice, BUD and, more effectively, PEITC tend to depart from ECS alone, but both agents fail to approach the Sham situation. Again, the different allocation of ECS + BUD and ECS + PEITC reflects the fact that these agents target different miRNAs for attenuating ECS-induced miRNA alterations. As shown in Table I, in the case of ECS-downregulated miRNAs, this modulating effect reached the statistical significance threshold for four miRNAs in ECS + PEITC-treated versus ECS-exposed mice (9.3%) and for three miRNAs in ECS + BUD-treated versus ECS-exposed mice (7.0%). In the case of ECS-upregulated miRNAs, modulation by chemopreventive agents reached the statistical significance threshold for five miRNAs in ECS + PEITC-treated versus ECS-exposed mice (17.2%) and for two miRNAs in ECS + BUD-treated versus ECS-exposed mice (6.9%).
Table II shows the functions of those miRNAs that were affected by either PEITC or BUD in the lung of ECS-exposed mice.
Modulation by PEITC and BUD of miRNA expression in the liver of ECS-exposed mice
As shown in Figure 4B, supervised hierarchical cluster analysis provided evidence that, in liver, not only ECS but also PEITC and BUD, when administered to ECS-free mice, did not cluster with Sham. At variance with BUD, PEITC exhibited some protective effect, as demonstrated by the fact that ECS + PEITC clustered with Sham.
The results of PCA in liver (Figure 4D) showed that ECS is allocated far away from Sham. However, at variance with lung, PEITC and BUD are far away from both Sham and ECS, irrespective of exposure of mice to ECS. These patterns suggest that both PEITC and BUD alter the baseline miRNA profiles in liver and that the alterations induced in this organ by these two chemopreventive agents prevail on those induced by ECS. As shown in Table I, in the case of ECS-downregulated miRNAs modulation by chemopreventive agents reached the statistical significance threshold for 12 miRNAs in ECS + PEITC-treated versus ECS-exposed mice (13.3%) and for 11 miRNAs in ECS + BUD-treated versus ECS-exposed mice (12.2%). In the case of ECS-upregulated miRNAs, modulation by chemopreventive agents reached the statistical significance threshold for 11 miRNAs in ECS + PEITC-treated versus ECS-exposed mice (14.5%) and for five miRNAs in ECS + BUD-treated versus ECS-exposed mice (6.6%).
Table II shows the functions of those miRNAs that were affected by either PEITC or BUD in the liver of ECS-exposed mice.
Discussion
The present study evaluated miRNA expression profiles in the lung and liver of young mice and provided information on (i) physiological interorgan differences, (ii) dysregulation following exposure to ECS since birth, (iii) effects of BUD and PEITC, administered after weaning, on the baseline expression and (iv) modulation by the same agents of ECS-related alterations in both organs.
In the comparison between lung and liver, it should be taken into account that both organs are composed of multiple cell types. Hence, the results obtained reflect the average situation of mixed-cell populations. The general trend was toward a higher expression of miRNAs in liver than in lung, which presumably underlies the multiplicity and complexity of liver functions. The most typical tissue specificity was recorded with miR-122, the most abundant miRNA in the liver, which is known to be involved in cholesterol biosynthesis and in maintenance of the liver phenotype (3).
Dysregulation of miRNA expression in the lung of ECS-exposed mice was mainly oriented toward downregulation of a variety of miRNAs involved in important cellular functions, such as stress response, cell proliferation, activation of oncogenes, apoptosis and angiogenesis. These results agree with the conclusions of our previous studies in ECS-exposed rats (5,12) and mice (8). Similar indications were obtained by evaluating miRNA expression in the human airway epithelium of smokers (9). Even more intense was the dysregulation of miRNA expression produced by ECS in mouse liver, both in the sense of upregulation and downregulation. Several miRNAs associated with adaptive functions, such as stress response, protein synthesis, repair and secretion, xenobiotic metabolism and intracellular vesicle trafficking, changed their expression in the liver as a response to exposure of mice to ECS. As evaluated in our previous study (6), of 4858 genes whose expression was analyzed in ECS-exposed rats, only the 0.9% was upregulated and the 0.3% was downregulated. These data suggest that miRNA analysis is more sensitive than gene expression analysis in revealing ECS-related effects. Exposure of neonatal mice to ECS (28) and, even more efficiently, to mainstream cigarette smoke (29) resulted after 7–9 months in the formation of tumors in the lung, whereas in the liver, only signs of steatosis and parenchymal dystrophy were observed, in the absence of significant increases in tumor yield. This conclusion correlates with the present finding that, in liver, the ECS-dysregulated miRNAs were mainly associated with activation of adaptive functions. In fact, ECS induced high levels of bulky DNA adducts in rat lung but not in liver (6,13).
Administration of BUD to smoke-free mice had negligible effects on miRNA expression profiles in lung, whereas PEITC downregulated five miRNAs only, mainly targeting stress response and adaptive functions. The results obtained with PEITC in mouse lung are in agreement with those obtained with the same agent in rat lung, in which only a negligible number of miRNAs were found to be dysregulated (12).
In contrast, both agents induced profound alterations of miRNA expression in liver, mostly in the sense of downregulation. This organ specificity is probably to be related, at least in part, to first-pass effects after oral administration. In particular, PEITC downregulated five miRNAs targeting protein repair, whereas BUD downregulated five miRNAs targeting stress response, seven miRNAs targeting protein repair and six miRNAs targeting cell proliferation were downregulated by BUD. Interestingly, seven miRNAs were targeted by both chemopreventive agents. These patterns may reflect the induction of early adverse effects of BUD and PEITC in the liver, as it will be discussed below.
Likewise, the analysis of the expression of individual miRNAs and their global evaluation by means of hierarchical cluster analysis and PCA confirmed that both BUD and PEITC have differential effects in the lung and liver of ECS-exposed mice. In fact, in the lung, BUD and, more efficiently, PEITC tended to attenuate ECS-related miRNA alterations but both agents failed to fully restore the physiological situation recorded in sham-exposed mice. In particular, both BUD and PEITC counteracted the ECS-induced dysregulation in mouse lung of let-7a, miR-26a, miR-31 and miR-382. In addition, BUD tended to restore the expression of miR-463 and PEITC tended to restore the expression of let-7c, miR-29b, miR-125b, miR-135b and miR-200b. A similar situation had been observed with PEITC in the lung of ECS-exposed rats. In particular, PEITC effectively protected pulmonary let-7a, let-7c, miR-26a and miR-125(b) from ECS-related downregulation in both mice (this study) and rats (12).
As shown in Table II, the miRNAs modulated by PEITC and BUD are involved in a variety of functions playing a role in pulmonary carcinogenesis. The observed protective effects exerted by PEITC and BUD on ECS-induced miRNA alterations correlate both with inhibition of DNA damage in the lung of the same mice and with a moderate ability of both chemopreventive agents to inhibit the formation of ECS-induced lung tumors after 9 months (27).
In the liver, PEITC and especially BUD exhibited a poor ability in counteracting the effects of ECS on miRNA expression to such an extent that the miRNA alterations observed in ECS-exposed mice treated with these chemopreventive agents were comparable with those produced by the same agents in ECS-free mice. The great heterogeneity in the early response of miRNA expression to PEITC and BUD reflects the occurrence of both adaptive mechanisms and pathways triggering damage to the liver, consistently with the above discussed results in ECS-free mice. In neonatal mice exposed to ECS for 4 months, followed by 5 months of recovery in filtered air, administration of BUD after weaning was shown to protect the liver from ECS-related steatosis. However, in the same mice, both BUD and PEITC significantly increased the incidence of sinusoidal hyaline degeneration and BUD also increased the vascular hyaline degeneration of the liver (27).
The intense miRNA variations observed in the liver of mice treated with BUD could be ascribed to the fact that, after oral administration, this glucocorticoid undergoes extensive hepatic metabolism via CYP3A (30). Glucocorticoids are known to cause side effects in humans (16) and, at toxic doses, BUD even induced liver tumors in rats (31). The simultaneous occurrence of miRNA upregulation and downregulation in the liver of ECS-exposed mice treated with PEITC may reflect the fact that this isothiocyanate inhibits Phase I activities but at the same time induces Phase II activities (32). In vitro, PEITC has been shown to induce genotoxic effects mediated by the formation of reactive oxygen species (33).
In conclusion, the results of the present study show for the first time that ECS not only downregulates miRNA expression in lung but also dysregulates miRNA expression in liver. In addition, evidence is provided that BUD and especially PEITC protect the lung from ECS-induced miRNA alterations, whereas dysregulation of miRNA expression in liver by these chemopreventive agents prevails on modulation of ECS-related alterations. Thus, miRNA analysis in different organs appears to be suitable to evaluate, in early steps of the carcinogenesis process, both protective effects of chemopreventive agents and their possible adverse effects.
Funding
National Cancer Institute (contract N01-CN 53301); Associazione Italiana per la Ricerca sul Cancro (contract 8909).
Acknowledgments
We thank Dr Ilaria Righi for assistance in preparation of the manuscript and illustrative material.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BUD
budesonide
- ECS
environmental cigarette smoke
- miRNA
microRNAs
- PCA
principal component analysis of variance
- PEITC
phenethyl isothiocyanate
- qPCR
quantitative polymerase chain reaction
References
- 1.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 2.Meltzer PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435:745–746. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- 3.Girard M, et al. MiR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, et al. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 5.Izzotti A, et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izzotti A, et al. Modulation of multigene expression and proteome profiles by chemopreventive agents. Mutat. Res. 2005;591:212–223. doi: 10.1016/j.mrfmmm.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Izzotti A, et al. Chemoprevention of genome, transcriptome, and proteome alterations induced by cigarette smoke in rat lung. Eur. J. Cancer. 2005;41:1864–1874. doi: 10.1016/j.ejca.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Izzotti A, et al. MicroRNA expression in mouse lung, as related to age and exposure to cigarette smoke and light. FASEB J. 2009;23:3243–3250. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schembri F, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl Acad. Sci. USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, et al. Uncovering growth-suppressive microRNAs in lung cancer. Clin. Cancer Res. 2009;15:1177–1183. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melkamu T, et al. Alteration of microRNA expression in vinyl-carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2010;31:252–258. doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- 12.Izzotti A, et al. Chemoprevention of cigarette smoke-induced alterations of microRNA expression in rat lung. Cancer Prev. Res. 2010;3:62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izzotti A, et al. DNA alterations in rat organs following chronic exposure to cigarette smoke and/or ethanol ingestion. FASEB J. 1998;12:753–758. doi: 10.1096/fasebj.12.9.753. [DOI] [PubMed] [Google Scholar]
- 14.IARC. IARC Monographs on the Evaluation of Carcinogenic Risk to Humans. Tobacco Smoke and Involuntary Smoking. Vol. 83. Lyon: IARC; 2004. [PMC free article] [PubMed] [Google Scholar]
- 15.Wattenberg LW, et al. Chemoprevention of pulmonary carcinogenesis by aerosolized budesonide in female A/J mice. Cancer Res. 1997;57:5489–5492. [PubMed] [Google Scholar]
- 16.Wattenberg LW, et al. Chemoprevention of pulmonary carcinogenesis by brief exposures to aerosolized budesonide or beclomethasone dipropionate and by the combination of aerosolized budesonide and dietary myo-inositol. Carcinogenesis. 2000;21:179–182. doi: 10.1093/carcin/21.2.179. [DOI] [PubMed] [Google Scholar]
- 17.Estensen RD, et al. Effect of chemopreventive agents on separate stages of progression of benzo[alpha]pyrene induced lung tumors in A/J mice. Carcinogenesis. 2004;25:197–201. doi: 10.1093/carcin/bgg196. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Mice with alterations in both p53 and Ink4a/Arf display a striking increase in lung tumor multiplicity and progression: differential chemopreventive effect of budesonide in wild-type and mutant A/J mice. Cancer Res. 2003;63:4389–4395. [PubMed] [Google Scholar]
- 19.Balansky R, et al. Influence of FHIT on benzo[a]pyrene-induced tumors and alopecia in mice: chemoprevention by budesonide and N-acetylcysteine. Proc. Natl Acad. Sci. USA. 2006;103:7823–7828. doi: 10.1073/pnas.0601412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira MA, et al. Prevention of mouse lung tumors by budesonide and its modulation of biomarkers. Carcinogenesis. 2002;23:1185–1189. doi: 10.1093/carcin/23.7.1185. [DOI] [PubMed] [Google Scholar]
- 21.Yao R, et al. Budesonide exerts its chemopreventive efficacy during mouse lung tumorigenesis by modulating gene expressions. Oncogene. 2004;23:7746–7752. doi: 10.1038/sj.onc.1207985. [DOI] [PubMed] [Google Scholar]
- 22.Hecht SS. Chemoprevention of cancer by isothiocyanates, modifiers of carcinogen metabolism. J. Nutr. 1999;129:768S–774S. doi: 10.1093/jn/129.3.768S. [DOI] [PubMed] [Google Scholar]
- 23.Stoner GD, et al. Isothiocyanates and plant polyphenols as inhibitors of lung and esophageal cancer. Cancer Lett. 1997;114:113–119. doi: 10.1016/s0304-3835(97)04639-9. [DOI] [PubMed] [Google Scholar]
- 24.Conaway CC, et al. Phenethyl isothiocyanate and sulforaphane and their N-acetylcysteine conjugates inhibit malignant progression of lung adenomas induced by tobacco carcinogens in A/J mice. Cancer Res. 2005;65:8548–8557. doi: 10.1158/0008-5472.CAN-05-0237. [DOI] [PubMed] [Google Scholar]
- 25.Ye B, et al. Induction of lung lesions in Wistar rats by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and its inhibition by aspirin and phenethyl isothiocyanate. BMC Cancer. 2007;7:90. doi: 10.1186/1471-2407-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izzotti A, et al. Modulation of biomarkers by chemopreventive agents in smoke-exposed rats. Cancer Res. 2001;61:2472–2479. [PubMed] [Google Scholar]
- 27.D'Agostini F, et al. Modulation by phenethyl isothiocyanate and budesonide of molecular and histopathological alterations induced by environmental cigarette smoke in mice. Cancer Prev. Res. 2009;2:546–556. doi: 10.1158/1940-6207.CAPR-08-0235. [DOI] [PubMed] [Google Scholar]
- 28.D'Agostini F, et al. Preneoplastic and neoplastic lesions in the lung, liver, and urinary tract of mice exposed to environmental cigarette smoke and UV light since birth. Int. J. Cancer. 2008;123:2497–2502. doi: 10.1002/ijc.23836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balansky R, et al. Potent carcinogenicity of cigarette smoke in mice exposed early in life. Carcinogenesis. 2007;28:2236–2243. doi: 10.1093/carcin/bgm122. [DOI] [PubMed] [Google Scholar]
- 30.Edsbäcker S, et al. Pharmacokinetics of budesonide (Entocort EC) capsules for Crohn's disease. Clin. Pharmacokinet. 2004;43:803–821. doi: 10.2165/00003088-200443120-00003. [DOI] [PubMed] [Google Scholar]
- 31.Ryrfeldt A, et al. Liver tumors in male rats following treatment with glucocorticosteroids. Toxicol. Pathol. 1992;20:115–117. doi: 10.1177/019262339202000114. [DOI] [PubMed] [Google Scholar]
- 32.Conaway CC, et al. Isothiocyanates as cancer chemopreventive agents: their biological activities and metabolism in rodents and humans. Curr. Drug Metab. 2002;3:233–255. doi: 10.2174/1389200023337496. [DOI] [PubMed] [Google Scholar]
- 33.Kassie F, et al. Genotoxic effects of allyl isothiocyanate (AITC) and phenethyl isothiocyanate (PEITC) Chem. Biol. Interact. 2000;127:163–180. doi: 10.1016/s0009-2797(00)00178-2. [DOI] [PubMed] [Google Scholar]