Abstract
Traumatic spinal cord injury (SCI) causes marked neuropathological changes in the spinal cord, resulting in limited functional recovery. Currently, there are no effective treatments, and the mechanisms underlying these neuropathological changes are not completely understood. In this study, two-dimensional gel electrophoresis coupled with mass spectrometry was used to investigate injury-related changes in the abundance (SYPRO Ruby stain) and phosphorylation (Pro-Q Diamond stain) of proteins from the soluble fraction of the lesion epicenter at 24 h following SCI. Over 1500 SYPRO Ruby-stained spots and 100 Pro-Q Diamond-stained spots were examined. We identified 26 unique proteins within 38 gel spots that differentially changed in abundance, phosphorylation, or both in response to SCI. Protein redundancies among the gel spots were likely due to differences in proteolysis, post-translational modifications, and the existence of isoforms. The proteins affected were blood-related proteins, heat-shock proteins, glycolytic enzymes, antioxidants, and proteins that function in cell structure, cell signaling, DNA damage, and protein degradation. These protein changes post injury may suggest additional avenues of investigation into the underlying molecular mechanisms responsible for the pathophysiological consequences of SCI.
Key words: contusion; phosphoproteome; proteome; spinal cord injury, 2-DE
Introduction
Traumatic spinal cord injury (SCI) is a major public health problem, often leaving patients with lifelong disabilities. Currently, there are more than 250,000 individuals in the United States living with a SCI, and approximately 11,000 additional persons are injured each year (www.spinalcord.uab.edu). At the present time, there are no effective treatments for SCI (Wuermser et al., 2007), and functional recovery is limited. Maximization of functional recovery after SCI depends on rapid diagnosis of the severity of the injury and initiation of appropriate clinical management. Use of high-cost imaging methods (e.g., computed tomography scans and magnetic resonance imaging) may help define injury location and severity (Kaji and Hockberger, 2007). However, these imaging technologies cannot be used to elucidate the biological mechanisms underlying the progressive cell death after SCI, and the current lack of reliable biomarkers for monitoring molecular changes over time following SCI makes an accurate evaluation of the progression of SCI difficult.
SCI causes marked neuropathology – following the initial insult, there is a delayed and prolonged period of secondary damage due to the activation of several destructive pathophysiological and pathobiochemical cascades. Secondary injury is characterized in part by increased permeability of the blood–spinal cord barrier (Noble and Wrathall, 1987, 1988), inflammatory responses (Ahn et al., 2006; Bareyre and Schwab, 2003), oxidative stress (Hall and Springer, 2004; Springer et al., 1997), and cell death (Grossman et al., 2001; Ling and Liu, 2007; Springer, 2002), but the exact molecular mechanisms and biochemical pathways mediating secondary damage remain elusive. This incomplete understanding of disease pathogenesis has greatly impeded the development of therapeutic strategies.
Microarrays are beginning to reveal global changes in gene transcripts following SCI (Aimone et al., 2004; Bareyre and Schwab, 2003), but this work has not yet extended into a global analysis of protein changes. Proteomic approaches are important tools for the study of SCI because the initial injury initiates a complicated sequence of cellular events that affects many aspects of cell signaling, translational regulations, post-translational modifications, and steady-state protein levels (Cox et al., 2005; Denslow et al., 2003; Fagan et al., 2007; Kobeissy et al., 2006; Ottens et al., 2007; Siman et al., 2004, 2005; K.K. Wang et al., 2004). Although the proteomic profile of contusive brain injury has been well documented (Denslow et al., 2003; Kobeissy et al., 2006; Ottens et al., 2007; Siman et al., 2004, 2005; K.K. Wang et al., 2004), similar studies following SCI have been limited.
Many protein kinases and phosphatases are expressed in high levels in the mammalian central nervous system (CNS). To date, virtually no phosphoproteomic approaches have been used in studies of SCI. Reversible phosphorylation of serine, threonine, and tyrosine residues is one of the most significant post-translational modifications of proteins, which plays an important role in various types of metabolic regulation and signal transduction (Cohen, 1982, 1992; Hunter, 2000; Pawson and Scott, 2005) and, as such, is likely to be involved in the mechanisms contributing to secondary injury following the initial insult. Studying changes in the phosphoproteome should lead to a more complete understanding of the pathogenesis of the secondary injury of SCI. Two-dimensional gel electrophoresis (2-DE) coupled with mass spectrometry (MS) was used in this study to investigate injury-related changes in the proteome and phosphoproteome of the rat spinal cord at 24 h following a mild-to-moderate spinal cord contusion. This approach may provide a better understanding of the complex molecular mechanisms of SCI and may eventually lead to the identification of novel biomarker candidates.
Methods
Subjects
Subjects were 22 Long-Evans, young adult, female rats (Harlan Laboratories, Inc., Indianapolis, IN) that weighed approximately 200 g at the time of surgery. The rats were housed in the vivarium of the University of Kentucky College of Medicine on a 12:12 h light/dark cycle with food and water available ad libitum. The animal unit of the College of Medicine at the University of Kentucky Medical Center is fully accredited by AAALAC. The care of animals used in this experiment complied with standards set forth in the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All experimental procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee and conformed to guidelines of the Society for Neuroscience and the Society for Neurotrauma.
Surgical procedures
All surgical techniques were performed under aseptic conditions and were conducted as previously described (McEwen et al., 2007; Ravikumar et al., 2007). Rats were anesthetized with 40 mg/kg (i.p.) sodium pentobarbital (Nembutal; Abbott Laboratories, North Chicago, IL) and dorsal incisions were made in the skin and underlying muscles. The muscles were retracted and a partial laminectomy was performed at thoracic segment 10 (T10). Eleven rats received no further surgery and served as the sham controls (laminectomy only). The muscles at the incision site were then closed with 5-0 absorbable braided suture (Ethicon, Somerville, NJ) and the skin was closed with 9-mm wound clips (Becton Dickinson, Sparks, MD).
The vertebral column of the 11 remaining rats was stabilized by clamping the vertebrae at T9 and T11. The spinal cord was then injured at T10 with the Infinite Horizons impactor (Precision Systems and Instrumentation, Lexington, KY), which has been described in detail (Q. Cao et al., 2005; McEwen et al., 2007; Ravikumar et al., 2007; Scheff et al., 2003). Briefly, a stainless-steel-tipped probe (2.5 mm in diameter) was rapidly lowered onto the dorsal surface of the spinal cord until an impact force of 150 kdynes was achieved. This injury parameter produced a mild-to-moderate SCI. A data acquisition program subsequently displayed the actual force applied to the spinal cord, the maximum spinal cord displacement, and the velocity of the probe at the time of peak force and peak displacement. The incision site was then closed in layers as described for the sham controls. All rats were placed into an incubator for 2 to 3 h to recover from anesthesia and then returned to their home cages in the colony room.
Protein extraction and quantification
At 24 h after surgery, 12 rats (six injured and six sham controls) were deeply anesthetized with 100 mg/kg sodium pentobarbital (i.p.). An 18-gauge needle was attached to a 30-ml syringe that had been filled with ice-cold PBS. The needle tip was inserted into the caudal-most region of the vertebral column, and the buffer was rapidly injected in order to force the spinal cord out through the rostral foramen. A 5-mm segment of spinal cord tissue at T10, centered on the injury epicenter or the laminectomy site of the sham controls, was rapidly excised and immediately homogenized in a lysis buffer (8 M urea, 4% CHAPS (w/v), 0.5% IPG buffer [pH 3–10]) that was supplemented with a protease inhibitor cocktail (1:100; Sigma-Aldrich, St. Louis, MO). The spinal cord samples were then sonicated at 100 W for 30 s and centrifuged at 12,000g for 1 h. The supernatants were carefully removed, and the protein concentrations were analyzed in triplicate with a 2-D Quant Kit (GE Healthcare, Piscataway, NJ). The supernatant samples were then aliquot and stored at −70°C until analyzed further.
Two-dimensional gel electrophoresis
For each sample, 500 μg protein was combined with 2 μg ovalbumin (internal standard), and the samples were passively rehydrated onto 24-cm immobilized pH gradient (IPG) strips (pH 3–10; GE Healthcare). Isoelectric focusing was then performed using an Ettan IPGphore II System (GE Healthcare). Specifically, IPG strips were incubated for 15 min in an equilibration buffer (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, 2% SDS, and trace amounts of bromophenol blue) supplemented with 1% DTT, followed by a 15-min incubation in a fresh equilibration buffer supplemented with 2.5% iodoacetamide. Each IPG strip was then placed on top of a large format 12.5% SDS-polyacrylamide gel (25.5 cm × 21.2 cm; Jule, Inc., Milford, CT), along with a peppermint-stick phosphoprotein molecular weight standard (Invitrogen, Carlsbad, CA), and subjected to 2-DE at 5 W per gel for 30 min followed by 17 W per gel for 5 h, using an Ettan DALT-6 vertical system (GE Healthcare). Gels were stained with Pro-Q Diamond phosphoprotein gel stain according to the manufacturer's instructions (Invitrogen) in order to reveal phosphoprotein expression. The gels were then scanned with a Typhoon 9400 laser scanner (GE Healthcare) at excitation and emission wavelengths of 532 nm and 560 nm respectively. Gels were subsequently stained with SYPRO Ruby protein gel stain according to the manufacturer's instructions (Invitrogen) in order to reveal total protein expression, and rescanned at excitation and emission wavelengths of 457 nm and 610 nm respectively.
ImageMaster 2D Platinum software (V6.0; GE Healthcare) was used to normalize, quantify, and compare the gel spots. Gels were normalized based on matched spots between gel pairs. Only spots that were consistently present in the gels from at least five of the six rats in each experimental group were analyzed further, in order to guard against spot differences that were due either to gel-to-gel variation or biological variation inherent among the rats. The final spot matches among the gels were verified manually, and the quality of the gel spots of interest was further examined with a three-dimensional view of these gel regions. The x and y axes represented the isoelectric point (pI), which ranged from 3 to 10, and the molecular weight (MW), which ranged from 10 to 100 kDa respectively. The z-axis corresponded to the pixel intensity.
ImageMaster software was then used to quantify the amount of phosphorylation (Pro-Q Diamond staining) in each gel spot using the algorithm for volume (Vol). The amount of protein (SYPRO Ruby staining) in each spot was calculated as a function of the total protein in the gel using the percent volume (%Vol) algorithm. Subsequently, the relative volume ratio (Vol. Ratio), the ratio of the Pro-Q Diamond signal to the SYPRO Ruby signal (VolPro-Q/VolSYPRO Ruby), was used to calculate the relative phosphorylation level at each gel spot in order to relate the amount of phosphorylation to the amount of protein in each gel spot. Scatter plots for each gel set were produced in order to improve the validity of these algorithms. Fold increases in the amount of phosphorylation or total protein at each gel spot following SCI were calculated from the ratio between the injured and sham control samples (SCI/sham). Fold decreases in phosphorylation or total protein following SCI were calculated by dividing one by the ratio of the injured to sham samples. Unpaired Student's t-tests were used to compare the volumes of the gel spots of the injured and the control samples, and p values ≤0.05 were considered statistically significant.
In-gel digestion and mass spectrometric analyses
The gel spots that differed between the injured and sham samples were subsequently excised and digested with 150 ng porcine modified trypsin protease (Promega, Madison, WI). The tryptic peptides were extracted from the gel plugs, reconstituted in 10 μl of 0.1% trifluoroacetic acid (TFA), and bound to ZipTipSCX pipette tips according to the manufacturer's instructions (Millipore, Bedford, MA). Samples were then directly spotted onto stainless-steel matrix-assisted laser desorption/ionizing (MALDI) target plates by eluting them from the pipette tips with a solution composed of 30% methanol, 5% fresh ammonium hydroxide, and 0.1% TFA. Once dry, the spots were overlaid with 0.6 μl of α-cyano-4-hydroxycinnamic acid matrix (recrystallized from product obtained from Sigma).
MS spectra were generated with an ABI 4800 MALDI TOF/TOF Analyzer (Applied Biosystems, Foster City, CA) using 400 laser shots. The 10 largest peaks from each spot were individually selected for tandem mass spectrophotometric (MS/MS) analysis using collision-induced dissociation in order to fragment each peptide. The spectral data were searched against the SwissProt database 52.4, as of 5/1/2007 (species Rodentia), using GPS Explorer software (V3.5; Applied Biosystems) with Mascot 2.0 software (Matrix Science, Ltd.). Mass tolerance was set at 100 ppm for MS and 0.4 Da for MS/MS. Fixed modifications (carbamidomethylation) and variable modifications (deamidation, methionine oxidation, N-terminal acetylation, pyroglutamation [Glu or Gln]) were allowed.
The MS results from each spectrum were analyzed using the peptide mass fingerprint (PMF) module of the Mascot algorithm (www.matrixscience.com), which identified the protein(s) in each spot, based on the tryptic peptide mass, yielding both the protein score and the protein score confidence interval percentage (CI%). Similarly, the 10 tandem MS/MS spectra from selected spots (PMF protein score <400) were further analyzed using the MS/MS ion search module of the Mascot algorithm. This algorithm identified the protein(s) in a spot based on the accurate parent peptide mass (from the MS spectrum) and the fragment MS/MS spectrum of any single peptide mass, yielding both the total ion score and the total ion score CI%. These calculations represent the combined score and CI% for all the peptide MS/MS spectra from the spot that correspond to the identified protein. For both sets of data, CI% of at least 95% was considered statistically significant at the 0.05 level.
Western blotting
Antibodies against enolase, dihydropyrimidinase-related protein-2 (DRP-2), and glial fibrillary acidic protein (GFAP) were used with Western blotting to corroborate some of the results of the 2-DE analysis. The remaining 10 rats were deeply anesthetized at 24 h post surgery (five injured and five sham controls), and the spinal cords were rapidly extracted from the vertebral column via pressure injection as already described above. A 5-mm segment of spinal cord tissue at T10, centered on the injury epicenter or the laminectomy site of the sham controls, was rapidly excised and homogenized in fresh RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.5% SDS) supplemented with a protease inhibitor cocktail (1:100; Sigma). The samples were sonicated at 100 W for 30 s and then centrifuged at 12,000g for 1 h. The protein concentrations of the supernatants were then measured in triplicate with a 2-D Quant Kit (GE Healthcare). The samples were divided into aliquots and stored at −70°C until analyzed further.
Samples (10 μg protein) were subsequently loaded onto 4–20% Tris-HCl Criterion gradient gels (Bio-Rad Laboratories, Hercules, CA), along with 15 μl of the Precision-Plus dual-color molecular weight marker (Bio-Rad). The proteins were separated by molecular weight and transferred to polyvinylidene difluoride (PVDF) membranes using standard techniques. The membranes were blocked in 5% nonfat dry milk for 1 h and then incubated overnight at 4°C in blocking solution that contained the primary antibody of interest. Rabbit polyclonal antienolase (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-GFAP (Sigma) antibodies were both used at dilutions of 1:10,000; the mouse monoclonal anti-DRP-2 antibody (Immune-Biological Laboratories, Takasaki-Shi, Gunma, Japan) was diluted 1:15,000. Membranes were washed and then incubated for 2 h at room temperature in a blocking solution that contained the appropriate secondary antibody: horseradish peroxidase-conjugated affinity-pure goat anti-rabbit or goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted 1:50,000. Each membrane was then washed and incubated in ECL-Plus detection reagent (GE Healthcare), and the chemiluminescent signal was detected on Hyblot CL autoradiography film (Denville Scientific, Metuchen, NJ). Films were scanned with a tabletop scanner, and images were saved for subsequent analysis of band densities.
Each membrane was subsequently striped with Pierce Restore-Plus western blot stripping buffer according to the manufacturer's instructions (Thermo Fisher Scientific, Rockford, IL). The membranes were reblocked in 5% nonfat dry milk for 1 h and then incubated overnight at 4°C with a mouse monoclonal antibody to β-actin (1:2500) (Sigma), which served as an internal load control. Membranes were washed and incubated in horseradish peroxidase-conjugated affinity-pure goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) diluted 1:50,000 as already described. Membranes were then washed and incubated in a ECL-Plus detection reagent (GE Healthcare), and the films were developed and scanned as described.
Statistical analysis
ImageQuant TL image analysis software (GE Healthcare) was used to quantify the volume of the immunopositive bands identified with each of the primary antibodies. Band volumes for enolase, DRP-2, and GFAP for each subject were normalized to the volume of the corresponding actin band, and unpaired Student's t-tests were used to compare the relative band volumes of samples from the injured rats to the sham control rats, and p values ≤0.05 were considered statistically significant.
Results
The biomechanical parameters of the injury (peak force, peak displacement, and impact velocity) did not differ between rats utilized for the 2-DE or the Western blot analyses (data not shown). The volumes of the ovalbumin-positive gel spots (2-DE) or the actin-positive bands (Western blotting) did not significantly differ between the injured and control samples (data not shown), suggesting that similar amounts of protein for each sample were loaded onto the gels.
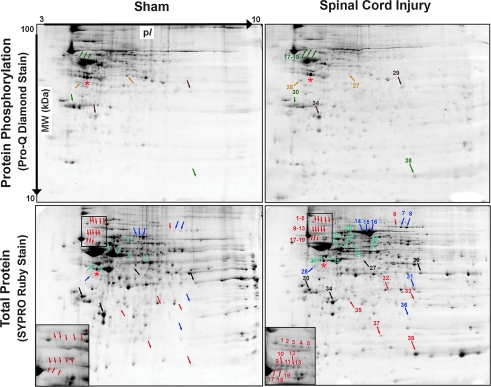
The 2-DE analysis revealed 26 unique proteins within 38 gel spots from the soluble fraction that were differentially regulated in abundance, phosphorylation, or both at 24 h following SCI. Figure 1 shows representative Pro-Q Diamond-stained (protein phosphorylation) and SYPRO Ruby-stained (total protein) 2-DE spot maps from one injured rat and one sham control rat. Nineteen SYPRO Ruby-stained gel spots were detected only in the gels from the SCI rats (Table 1). Fifteen SYPRO Ruby-stained gel spots that were present in the gels from both groups of rats either increased (eight spots) or decreased (seven spots) in volume following SCI (Table 2). Four gel spots contained proteins that were phosphorylated only following SCI (Table 3A). Four additional gel spots that contained proteins phosphorylated in the samples from both groups of rats either increased (two spots) or decreased (two spots) in phosphorylation level following SCI (Table 3B). The Western blot analysis corroborated the findings of the 2-DE analysis (Fig. 2).
FIG. 1.
Representative images of gels from a sham-operated rat (left column) and a spinal cord-injured rat (right column), which were mapped according to isoelectric point (pI) and molecular weight (MW). Thirty-eight gel spots were significantly different between the injured and sham spinal cords with respect to protein abundance, phosphorylation, or both (colored numbers and arrows). SYPRO Ruby Stain (bottom row): 19 gel spots were identified only in the injured samples (orange; see Table 1); 15 gel spots were identified in both groups but protein abundance either increased (dark blue) or decreased (light blue) after injury (see Table 2). The insets on the SYPRO Ruby-stained gels provide higher magnification of corresponding regions of the sham and injured samples, illustrating several gel spots that were identified only after the injury. The black numbers and arrows on the SYPRO Ruby-stained gels indicate spots that did not differ in abundance between the injured and sham cords; however, the phosphorylation state of these spots changed with injury and is included for comparison with the Pro-Q Diamond-stained gels. Pro-Q Diamond Stain (top row): five phosphorylated spots were identified in the injured cords only (green; see Table 3A); four phosphorylated spots were identified in both groups of rats but phosphorylation level either increased (brown) or decreased (tan) after injury (see Table 3B). Ovalbumin (red asterisks) is the internal standard.
Table 1.
SYPRO Ruby-Stained Gel Spots Present Only in the 24-h-Injured Spinal Cord
| Gel spot number and associated protein isoformsa | NCBI acces. no. | Seq. cov. (%)b | No. peptides (PMF)c/(MS/MS)d | Theoretical MW/pIe | Functions |
|---|---|---|---|---|---|
| 1. 14-3-3 protein delta/zeta | P63102¶ | 25 | –/5 | 28/4.7 | Adapter protein |
| 2. Heat-shock protein HSP 90-alpha | P82995‡ | 19 | 15/1 | 85/4.9 | Stress protein |
| 3. Heat-shock protein HSP 90-alpha | P82995‡ | 15 | 14/– | 85/4.9 | Stress protein |
| 4. Heat-shock protein HSP 90-alpha | P82995ঠ| 17 | 15/2 | 85/4.9 | Stress protein |
| 5. Heat-shock protein HSP 90-alpha | P82995ঠ| 12 | 10/1 | 85/4.9 | Stress protein |
| 6. Transferrin | P12346‡ | 54 | 71/– | 76/7.1 | Plasma protein |
| 9. Heat-shock cognate 71 kDa protein | P63018ঠ| 23 | 12/3 | 71/5.4 | Stress protein |
| Kininogen 2 precursor | P08932ঠ| 16 | 7/2 | 49/5.9 | Plasma protein |
| Kininogen 1 precursor | P01048ঠ| 16 | 8/1 | 49/6.1 | Plasma protein |
| Heat-shock 70 kDa protein 1A/1B | Q07439ঠ| 19 | 7/1 | 70/5.6 | Stress protein |
| Heat-shock 70 kDa protein 1L | P55063† | 19 | 7/– | 71/6.0 | Stress protein |
| 10. Kininogen-1 precursor | P08934¶ | 13 | 8/1 | 72/6.3 | Plasma protein |
| 11. Heat-shock cognate 71 kDa protein | P63018ঠ| 38 | 21/1 | 71/5.4 | Stress protein |
| Serum albumin precursor | P02770ঠ| 20 | 9/1 | 71/6.1 | Plasma protein |
| Kininogen-1 precursor | P01048ঠ| 41 | 15/3 | 49/6.1 | Plasma protein |
| Kininogen 2 precursor | P08932† | 30 | 14/– | 49/5.9 | Plasma protein |
| 12. Heat shock-related 70 kDa protein 2 | P14659‡ | 19 | 8/– | 70/5.4 | Stress protein |
| Serum albumin precursor | P02770† | 17 | 7/– | 71/6.1 | Plasma protein |
| 13. Heat-shock cognate 71 kDa protein | P63017ঠ| 8 | 3/1 | 71/5.4 | Stress protein |
| 17. Neurofilament light chain | P19527ঠ| 42 | 25/2 | 61/4.6 | Axonal cytoskeleton |
| 18. Fetuin A | P24090ঠ| 30 | 6/2 | 39/6.1 | Plasma protein |
| 19. Probable coiled-coil domain-containing protein 8 | P62521† | 8 | 7/– | 70/10 | DNA damage |
| 32. Triosephosphate isomerase | P48500ঠ| 46 | 7/1 | 27/6.5 | Glycolytic enzyme |
| 33. Glutathione S-transferase Yb-3 | P08009†¶ | 30 | 5/1 | 26/7.3 | Oxidative stress |
| 35. Apolipoprotein A-I precursor | P04639ঠ| 57 | 13/3 | 30/5.5 | Plasma protein |
| Peroxiredoxin-6 | O35244ঠ| 32 | 7/1 | 25/5.7 | Oxidative stress |
| 37. Galectin | P47967¶ | 60 | –/7 | 16/6.2 | Erythrocyte differentiation |
| 38. Beta-globin | Q6PDU6‡ | 76 | 16/– | 16/6.8 | Erythrocyte protein |
| Hemoglobin subunit alpha-1/2 | P01946‡ | 63 | 10/– | 15/7.9 | Erythrocyte protein |
refers to the spot numbers in Figure 1; brefers to the total percentage of the sequence covered by peptide mass fingerprinting (PMF); crefers to the number of peptides that match the sequence for PMF; drefers to the number of peptides used to identify a protein by tandem mass spectrometry (MS/MS) (CI% >95%); erefers to the theoretical molecular weight (MW) and the theoretical isoelectric point (pI); *Protein Score CI% >95%, ‡Protein Score CI% >99%; ¶Total Ion Score CI% >99%.
Table 2.
SYPRO Ruby-Stained Gel Spots Differentially Upregulated or Downregulated after Spinal Cord Injury
| Gel spot number and associated protein isoformsa | %Vol SCI | %Vol Sham | Fold change | NCBI acces. no. | Seq. cov. (%)b | No. of peptides (PMF)c/(MS/MS)d | Theoretical MW/pIe | Functions |
|---|---|---|---|---|---|---|---|---|
| 7. Transferrin | ↑ 0.31 ± 0.17** | 0.05 ± 0.03 | 6.20 | P12346‡ | 57 | 83/– | 76/7.1 | Plasma protein |
| 8. Transferrin | ↑ 0.39 ± 0.15** | 0.06 ± 0.06 | 6.50 | P12346‡ | 34 | 24/2 | 79/7.1 | Plasma protein |
| 14. Albumin | ↑ 1.10 ± 0.28** | 0.15 ± 0.05 | 7.33 | P02770‡ | 38 | 26/– | 69/6.1 | Plasma protein |
| 15. Albumin | ↑ 1.90 ± 0.39** | 0.36 ± 0.15 | 5.28 | P02770‡ | 73 | 97/– | 69/6.1 | Plasma protein |
| 16. Albumin | ↑ 7.45 ± 1.49** | 0.84 ± 0.29 | 8.87 | P02770‡ | 62 | 125 | 69/6.1 | Plasma protein |
| 28. Neurofilament light chain | ↑ 0.06 ± 0.03* | 0.02 ± 0.01 | 3.00 | P19527†¶ | 20 | 11/1 | 61/4.6 | Axonal cytoskeleton |
| 31. Triosephosphate isomerase | ↑ 0.22 ± 0.07* | 0.11 ± 0.09 | 2.00 | P48500‡ | 93 | 31/– | 27/7.1 | Glycolytic enzyme |
| 36. Alpha crystalline B-chain | ↑ 0.05 ± 0.02* | 0.03 ± 0.01 | 1.67 | P23928‡¶ | 52 | 10/2 | 20/6.8 | Stress protein |
| 20. Dihydropyrimidinase-related protein-2 | ↓ 0.05 ± 0.02* | 0.10 ± 0.04 | −2.00 | P47942‡¶ | 46 | 17/4 | 62/6.0 | Synaptic plasticity |
| 21. Glial fibrillary acidic protein | ↓ 0.09 ± 0.05* | 0.17 ± 0.05 | −1.89 | P47819†¶ | 5 | 2/1 | 50/5.4 | Cytoskeleton of astrocyte |
| 22. Glial fibrillary acidic protein | ↓ 1.15 ± 0.13* | 2.31 ± 0.85 | −2.01 | P47819‡¶ | 83 | 44/1 | 50/5.4 | Cytoskeleton of astrocyte |
| 23. Creatine kinase B-type | ↓ 0.21 ± 0.02** | 0.53 ± 0.07 | −2.52 | P07335‡¶ | 24 | 6/3 | 43/5.4 | Energy transduction |
| 24. Alpha-enolase | ↓ 0.42 ± 0.10* | 0.61 ± 0.14 | −1.45 | P04764‡ | 73 | 67/– | 47/6.2 | Glycolytic enzyme |
| 25. Gamma-enolase | ↓ 1.02 ± 0.12** | 1.27 ± 0.07 | −1.25 | P07323‡ | 47 | 16/– | 48/5.0 | Glycolytic enzyme |
| 26. Gamma-enolase | ↓ 0.21 ± 0.06* | 0.31 ± 0.06 | −1.48 | P07323‡¶ | 7 | 2/2 | 48/5.0 | Glycolytic enzyme |
%Vol (percent volume) was used to quantify the expression level of each SYPRO Ruby-stained gel spot. Positive fold changes (increases) were calculated from the ratio of SCI/Sham; negative fold changes (decreases) were calculated by dividing one by the ratio of the injured to sham samples.
refers to the spot numbers in Figure 1; brefers to the total percentage of the sequence covered by peptide mass fingerprinting (PMF); crefers to the number of peptides that match the sequence for PMF; drefers to the number of peptides used to identify a protein by tandem mass spectrometry (MS/MS) (CI% > 95%); erefers to the theoretical molecular weight (MW) and the theoretical isoelectric point (pI); *p < 0.05; **p < 0.01; †Protein Score CI% >95%, ‡Protein Score CI% > 99%; ¶Total Ion Score CI% >99%.
Table 3A.
Gel Spots Phosphorylated Only in the 24-H-Injured Spinal Cord
| Gel spot number and associated protein isoformsa | NCBI acces. no. | Seq. cov. (%)b | No. of peptides (PMF)c/(MS/MS)d | Theoretical MW/pIe | Functions |
|---|---|---|---|---|---|
| 17. Neurofilament light chain | P19527ঠ| 42 | 25/2 | 61/4.6 | Axonal cytoskeleton |
| 18. Fetuin-A | P24090ঠ| 30 | 6/2 | 39/6.1 | Plasma protein |
| 19. Probable coiled-coil domain-containing protein 8 | P62521† | 8 | 7/– | 70/10 | DNA damage |
| 30. 14-3-3 protein alpha/beta | P35213‡ | 35 | 20/– | 28/4.8 | Adapter protein |
| 14-3-3 protein delta/zeta | P63102‡ | 48 | 21/– | 28/4.7 | Adapter protein |
| 38. Beta-globin | Q6PDU6‡ | 76 | 16/– | 16/6.8 | Erythrocyte protein |
| Hemoglobin subunit alpha-1/2 | P01946‡ | 63 | 10/– | 15/7.9 | Erythrocyte protein |
Table 3B.
Gel Spots Differentially Phosphorylated after Spinal Cord Injury
| Gel spot number and associated protein isoformsa | Vol. Ratio SCI | Vol. Ratio sham | Fold change | NCBI acces. no. | Seq. cov. (%)b | No. of peptides | Theoretical MW/pIe | Functions |
|---|---|---|---|---|---|---|---|---|
| 29. Glyceraldehyde-3-phosphate dehydrogenase | ↑ 1.23 ± 0.34** | 0.56 ± 0.16 | 2.20 | P04797‡ | 54 | 33/– | 36/8.4 | Glycolytic enzyme |
| 34. Ubiquitin carboxyl-terminal hydrolase isozyme L1 | ↑ 1.50 ± 0.66** | 0.24 ± 0.14 | 6.25 | Q9R0P9‡ | 71 | 36/– | 25/5.1 | Deubiquitinating enzyme |
| 27. Cathepsin D precursor | ↓ 3.23 ± 1.24* | 7.41 ± 3.30 | −2.29 | P24268¶ | 6 | –/2 | 45/6.7 | Aspartyl protease |
| 28. Neurofilament light chain | ↓ 2.74 ± 0.81* | 6.77 ± 3.80 | −2.47 | P19527†¶ | 20 | 11/1 | 61/4.6 | Axonal cytoskeleton |
The volume ratio (Vol. Ratio) of the Pro-Q Diamond and SYPRO Ruby signals (calculated from VolPro-Q/VolSYPRO Ruby) was used to quantify the relative phosphorylation level of each gel spot; Mean Fold Change was calculated from the ratio of SCI/sham; fold decreases were calculated by dividing 1 by the ratio of the injured to sham samples; arefers to the spot numbers in Figure 1; brefers to the total percentage of the sequence covered by peptide mass fingerprinting (PMF); crefers to the number of peptides that match the sequence for PMF; drefers to the number of peptides used to identify a protein by tandem mass spectrometry (MS/MS) (CI% > 95%); erefers to the theoretical molecular weight (MW) and the theoretical isoelectric point (pI); *p < 0.05; **p < 0.01; †Protein Score CI% >95%, ‡Protein Score CI% > 99%; ¶Total Ion Score CI% >99%.
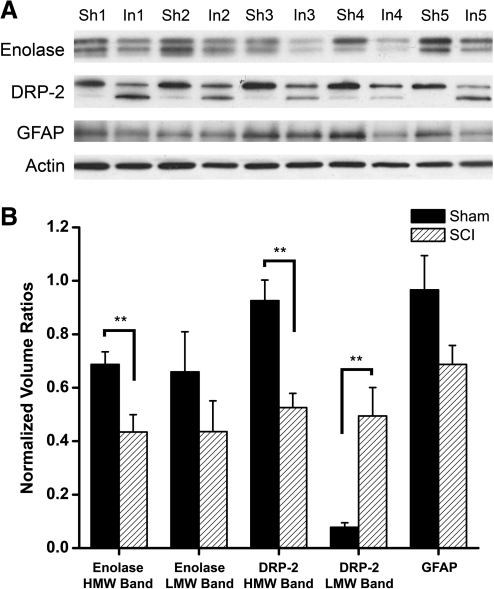
FIG. 2.
Western blot analysis of enolase, DRP-2, GFAP, and β-actin expression following SCI. (A) Immunoblot images of the enolase-, DRP-2-, GFAP-, and actin-positive bands for the five subjects of each group. (B) Graphic representation of the effects of SCI on mean band volume for each of the antibodies of interest, which were normalized to the volume of the corresponding actin band. Sh = Sham, In = 24 h SCI, **p < 0.01.
Vascular damage
Several blood-related proteins were either found only in the injured tissue (Table 1) or highly upregulated in the injured tissue compared to the sham controls (Table 2; p < 0.05). These proteins included albumin precursor, albumin, transferrin, kininogen precursor, fetuin A, apolipoprotein A, galectin, β-globin, and hemoglobin. The phosphorylation of fetuin A, β-globin, and hemoglobin also increased at 24 h following SCI (Table 3; p < 0.05).
Cellular damage
GFAP is the major intermediate filament protein of mature astrocytes. The results of the 2-DE analysis (Fig. 1, Table 2) demonstrate that GFAP protein levels at the injury epicenter significantly decreased by 24 h following the injury (p < 0.05). There was a similar trend with the Western blotting analysis (Fig. 2), but the effect fell short of statistical significance (p = 0.09). DRP-2, which is associated with synaptic plasticity, decreased in abundance by 24 h following SCI (p < 0.05). Western blotting revealed two DRP-2-immunopositive bands with slightly different MW (Fig. 2). The volume of the higher MW band (approximately 60 kDa) significantly decreased as a result of the injury (p < 0.01), whereas the volume of the lower MW band (approximately 55 kDa) significantly increased (p < 0.01). Neurofilament light chain, which is a major component of the neural cytoskeleton, was detected in two gel spots. One spot was present and phosphorylated only in the samples from the injured spinal cords (p < 0.05). The other spot was present in the gels from rats of both experimental groups, but the MW was approximately 20 kDa lower than the theoretical value. The protein in this latter gel spot decreased in abundance (p < 0.05) but increased in phosphorylation at 24 h after SCI (p < 0.05). Finally, probable coiled-coil domain-containing protein 8 (CCDC 8), which functions in DNA damage, was phosphorylated only in the injured spinal cord (p < 0.05).
Energy metabolism
The abundance or phosphorylation of three glycolytic enzymes was differentially regulated within the first 24 h after SCI. The 2-DE analysis revealed that the abundance of the α-and γ-enolase isoforms significantly decreased at 24 h following SCI (p < 0.05). These isoforms have theoretical molecular weights of 47 and 48 kDa, respectively. An enolase-positive doublet was identified via Western blotting (Fig. 2) at approximately 50 kDa, and may represent α- (lower band) and γ-enolase (higher band) because this antibody will recognize both isoforms (Santa Cruz antibody insert sheet). The volumes of both enolase-positive bands tended to decrease following SCI, but the effect was statistically significant for the higher MW band only (p < 0.01). In contrast, the volumes of the 2-DE gel spots that contained triosephosphate isomerase and phosphorylated glyceraldehyde-3-phosphate dehydrogenase, two other glycolytic enzymes, both increased at 24 h post injury compared to sham controls (p < 0.05). The tissue levels of creatine kinase B-type, which functions as an intracellular energy buffer, decreased at the lesion epicenter at 24 h post contusion (p < 0.05).
Cellular stress and antioxidant systems
Several gel spots from the injured spinal cords contained heat-shock-related proteins – HSP 70, HSP 90, or a heat-shock cognate protein of 71 kDa – that were not detected in the spinal cords of the sham controls (p < 0.05). Another cell stress-related protein, α-crystalline B-chain, was detected in the spinal cords from rats of both experimental groups but the abundance of this protein was significantly upregulated by 24 h following SCI (p < 0.05). The volumes of gel spots that contained peroxiredoxin-6 and glutathione-S-transferase (GST), which function in the endogenous antioxidant systems, increased at 24 h following SCI (p < 0.05).
Cell signaling and protein degradation
The abundance (p < 0.05) and phosphorylation (p < 0.05) of the adaptor protein 14-3-3 increased at 24 h after SCI. Phosphorylation of ubiquitin carboxyl terminal hydrolase L1, which is part of the ubiquitin-proteasome system that is involved in the degradation and removal of damaged, excess, or altered proteins, increased after SCI (p < 0.05). Cathepsin D, which is a prominent aspartic protease that is highly expressed in neuronal lysosomes and involved in the degradation of proteins, decreased in phosphorylation level at 24 h following SCI compared to sham controls (p < 0.05).
Discussion
The purpose of this study was not only to examine the types of proteins that changed in abundance in the spinal cord following acute SCI but also to examine injury-related changes in protein phosphorylation. To our knowledge, this is the first proteomic and phosphoproteomic study in which the protein expression profile was characterized following a mild-to-moderate contusion to the rat spinal cord. Previous studies have analyzed the proteomic profile following severe spinal cord contusion (Tsai et al., 2008) or spinal cord transection (Ding et al., 2006; Kang et al., 2006; Yan et al., 2009). We limited our analysis to the soluble protein fraction from the lesion epicenter at 24 h post SCI for this initial study, because we were interested in identifying proteins that would move easily between compartments (e.g., to the cerebrospinal fluid or serum) and could be assessed in future studies for their biomarker potential.
MALDI-TOF and tandem MS (TOF/TOF) analyses provided sensitive and accurate mass spectral data for database interrogation. Over 1500 SYPRO Ruby-stained spots (total protein) and 100 Pro-Q Diamond-stained spots (phosphoprotein) were examined, resulting in the identification of 26 unique proteins within 38 gel spots from the soluble fraction that were differentially regulated in abundance, phosphorylation, or both at 24 h following SCI. Protein redundancies among the gel spots were likely due to differences in proteolysis, post-translational modifications, and the existence of isoforms. Unfortunately, use of the Pro-Q Diamond phosphoprotein stain was incompatible with using the CyDye DIGE Fluor minimal dyes (GE Healthcare) for quantifying total protein. Because of this limitation, we had to use the less sensitive SYPRO Ruby protein stain for quantifying total protein, and we acknowledge that post-injury changes in other less-abundant proteins most likely occurred.
We chose to study the phosphoproteome because protein phosphorylation is an important post-translational step that defines protein function. In some instances, phosphorylation may activate the protein; in other situations, phosphorylation may inhibit protein activity. Phosphorylation may cause a protein to become bound to other proteins with specific recognition domains, effectively activating or inhibiting a particular signaling pathway. Phosphorylation may also serve as a signal that the phosphorylated protein should be degraded by the ubiquitin-proteosome system. Given the numerous events that occur after SCI, we were surprised to find that only nine protein spots were differentially phosphorylated at 24 h following SCI. This finding maybe due to the specific sample studied (soluble proteins at the lesion epicenter), the time point studied post injury (24 h), or technical limitations with the Pro-Q Diamond assay. Subsequent experiments will be necessary to determine any site-specific changes in protein phosphorylation following SCI.
Traumatic injury to the spinal cord causes significant damage to the vasculature at the site of the impact (Maikos and Shreiber, 2007; Noble and Wrathall, 1987, 1988, 1989), causing the extravasation of blood into the spinal cord tissue. Cells and their processes at the impact site are destroyed, which initiates cascades of events causing additional cell dysfunction and death that spreads distally from the injury site (Crowe et al., 1997; Li et al., 1999; X.Z. Liu et al., 1997; McEwen and Springer, 2005; Shuman et al., 1997; Springer et al., 1999; Yong et al., 1998). Not surprisingly, our 2-DE analyses revealed that the tissue levels of several blood-related proteins were significantly upregulated following SCI (albumin precursor, albumin, transferrin, kininogen precursor, fetuin A, apolipoprotein A-I precursor, galectin, β-globin, and hemoglobin). Kang and associates (2006) and Tsai and associates (2008) also reported the upregulation of several of these proteins in the soluble fraction from the spinal cord at 24 h following SCI. In addition, we observed that the phosphorylation level of three of those proteins – fetuin A, β-globin, and hemoglobin – also increased after the injury. The significance of the latter finding is not currently known.
GFAP, the major intermediate filament protein of mature astrocytes, decreased at 24 h after SCI in the present experiment, which supports previous reports of decreases in astrocyte number and GFAP mRNA at 24 h after spinal cord contusion (Grossman et al., 2001; Wu et al., 2005). However, upregulation of GFAP was reported following a more severe spinal cord contusion (Tsai et al., 2008) and following spinal cord transection (Kang et al., 2006). These differences may be due to the nature of the injury model or because a larger segment of spinal cord was examined in those studies than in the present study; the severity or type of injury may alter GFAP expression levels in the tissues adjacent to the injury epicenter. The abundance and phosphorylation of probable CCDC 8 also increased at 24 h following SCI in the present study. Although the function of this occurrence is not understood, phosphorylation of CCDC 8 is known to occur after DNA is damaged (Matsuoka et al., 2007).
Neurofilament proteins are major structural components of the axonal cytoskeleton and a useful marker of neurons; phosphorylation is important in the transport of neurofilament subunits and neurofilament assembly (Grant and Pant, 2000; Yates et al., 2009). In the present experiment, the abundance and phosphorylation of neurofilament light chain primarily increased at 24 h post injury (also see Kang et al., 2006). However, a second gel spot contained neurofilament light chain with a MW that was 20 kDa below the theoretical value. This protein increased in abundance and decreased in phosphorylation following injury and may be a degradation product. DRP-2 (also known as collapsin response mediator protein-2 [CRMP-2] or turned on after division, 64 kD protein [TOAD-64]) is a cytosolic phosphoprotein involved in axonal growth and guidance as well as synaptic plasticity and maturation (Goshima et al., 1995; L.H. Wang and Strittmatter, 1996). Our proteomic findings demonstrate that DRP-2 levels in the soluble fraction decreased at 24 h post SCI, suggesting that axonal growth and synaptic plasticity is inhibited in the acute period following SCI. Western blot analysis further suggested that intact DRP-2 protein (60 kDa) decreased and the putative degradation product (55 kDa) increased in abundance at 24 h post injury (c.f., Kobeissy et al., 2006). Tsai and associates (2008) and Kang and associates (2006) both reported an increase in DRP-2 levels in the soluble fraction at 24 h following SCI. However, it is not clear from their proteomic analysis whether or not the identified protein corresponded to the intact protein or the putative degradation product.
Mitochondrial function (McEwen et al., 2007; Sullivan et al., 2007) and energy metabolism (Anderson et al., 1980) are known to be impaired following SCI. In the present study, three glycolytic enzymes were differentially altered at 24 h following SCI: the abundance of α- and γ-enolase decreased, the abundance of triosephosphate isomerase increased, and the phosphorylation of glyceraldehyde-3-phosphate dehydrogenase increased. In addition, creatine kinase B-type, which functions in energy transduction, decreased after injury. Others have reported that neuron-derived enolase (F. Cao et al., 2008; Loy et al., 2005) and creatine kinase B-type (Nordby and Urdal, 1982) leak from neurons into the serum and cerebrospinal fluid, respectively, following CNS injury, which would decrease the tissue concentrations of these enzymes, as found here.
The adaptor protein 14-3-3 has been implicated in the regulation of numerous cell signaling pathways (Jin et al., 2004; Schoonheim et al., 2007), and phosphorylation may be important for regulating the interaction of 14-3-3 with its binding partners (Jin et al., 2004; Schoonheim et al., 2007; Springer et al., 2000). Results of the present study indicate that the abundance and phosphorylation of adaptor protein 14-3-3 increased at 24 h following SCI, which may serve as a compensatory mechanism to improve cell signaling post-injury. Interestingly, one gel spot (in at least five of the six gels) contained the protein 14-3-3 with a MW approximately 60 kDa higher than the theoretical value. However, the importance of this finding is not currently known.
HSPs increase when cells are exposed to stressors (Brown, 2007; Gutierrez and Guerriero, 1995; Q. Liu and Hendrickson, 2007; Nishimura and Sharp, 2005; Robinson et al., 2005). In the present study, gels of the injured spinal cord samples also contained several spots identified as HSP 70, HSP 90, or a heat-shock cognate protein of 71 kDa, which were not present in the gels of the sham controls. Many of these gel spots differed only in the pI value, which may indicate post-translational modifications to these proteins upon injury (Gutierrez and Guerriero, 1995). However, de novo generation of these proteins remains a possibility. Alpha-crystalline B-chain, another stress-related protein, also increased in abundance after SCI. Although the roles of these stress proteins in the context of SCI are not known, upregulation of these stress proteins may protect other proteins from stress-induced aggregation or facilitate protein refolding as described for other pathological conditions (Brown, 2007; Ganea, 2001; Gutierrez and Guerriero, 1995; Horwitz, 1992; Q. Liu and Hendrickson, 2007; McClellan et al., 2007; Nishimura and Sharp, 2005; Robinson et al., 2005).
Glutathione functions as part of the endogenous antioxidant system and is thought to contribute to neuronal survival after SCI (Lucas et al., 1998). Glutathione-S-transferase (GST) functions as an antioxidant enzyme that conjugates the reduced form of glutathione to a wide number of hydrophobic electrophiles. Peroxiredoxin-6 is an antioxidant enzyme that is expressed abundantly in glial cells in the CNS (Power et al., 2008). In the present study, the abundance of both GST and peroxiredoxin-6 increased at 24 h following SCI, suggesting upregulation of antioxidant systems following the injury. This finding supports previous research from this laboratory showing that glutathione levels increased at 24 h following a mild-to-moderate spinal cord contusion (Azbill et al., 1997). In support, Kang and associates (2006) also reported upregulation of peroxiredoxin-6 at 24 h following spinal cord transection. Tsai and associates (2008) reported a decrease in peroxiredoxin-6 at 24 h following spinal cord contusion, but these differences may reflect differences due to the severity of the injuries.
The ubiquitin-proteasome system is one of the main systems involved in the degradation and removal of damaged, excess, or altered proteins. In this study, the phosphorylation of ubiquitin carboxyl terminal hydrolase L1 significantly increased by 24 h post SCI. Cathepsin D is a prominent aspartic protease that is highly expressed in neuronal lysosomes, which are also involved in the degradation of proteins. In the present experiment, phosphorylation of cathepsin D precursor decreased after injury. Others have shown that dephosphorylation plays a role in the activation of cathepsin D (Fialho et al., 2005) and cathepsin D activity increases 24 h after SCI (Banik et al., 1986). The findings of the present experiment also suggest that cathepsin D is activated, and protein degradation is possibly enhanced, at 24 h following SCI.
In summary, 2-DE-based proteomic approaches have a limited dynamic range for identifying relatively high-abundant proteins that differ between the injured and control spinal cords within particular MW and pI ranges (Denslow et al., 2003; Gygi et al., 2000). Differential proteomic analysis of prefractionated samples may increase the likelihood of identifying injury-related changes in less abundant proteins (Kislinger et al., 2006; Taylor et al., 2003; Yates et al., 2005). In addition, bioinformatics software for pathway analysis may be useful for identifying signaling pathways affected following SCI, as well as identify potential low abundance protein targets. Regardless, the results of the current study provide a first step in characterizing protein and protein phosphorylation changes that occur in the spinal cord following injury.
Acknowledgments
The authors would like to thank Anne Stanley and Dr. Bruce A. Stanley in the Proteomics/Mass Spectrometry Core Facility at Pennsylvania State University College of Medicine for helping with the protein identification. This work was supported by PHS grant NS46380 (J.E.S.), an endowment from the Cardinal Hill Rehabilitation Hospital (J.E.S.), and a grant from the Craig H. Neilsen Foundation (M.L.M.).
Author Disclosure Statement
No conflicting financial interests exist.
References
- Ahn Y.H. Lee G. Kang S.K. Molecular insights of the injured lesions of rat spinal cords: Inflammation, apoptosis, and cell survival. Biochem. Biophys. Res. Commun. 2006;348:560–570. doi: 10.1016/j.bbrc.2006.07.105. [DOI] [PubMed] [Google Scholar]
- Aimone J.B. Leasure J.L. Perreau V.M. Thallmair M. Spatial and temporal gene expression profiling of the contused rat spinal cord. Exp. Neurol. 2004;189:204–221. doi: 10.1016/j.expneurol.2004.05.042. [DOI] [PubMed] [Google Scholar]
- Anderson D.K. Means E.D. Waters T.R. Spears C.J. Spinal cord energy metabolism following compression trauma to the feline spinal cord. J. Neurosurg. 1980;53:375–380. doi: 10.3171/jns.1980.53.3.0375. [DOI] [PubMed] [Google Scholar]
- Azbill R.D. Mu X. Bruce-Keller A.J. Mattson M.P. Springer J.E. Impaired mitochondrial function, oxidative stress and altered antioxidant enzyme activities following traumatic spinal cord injury. Brain Res. 1997;765:283–290. doi: 10.1016/s0006-8993(97)00573-8. [DOI] [PubMed] [Google Scholar]
- Banik N.L. Hogan E.L. Powers J.M. Smith K.P. Proteolytic enzymes in experimental spinal cord injury. J. Neurol. Sci. 1986;73:245–256. doi: 10.1016/0022-510x(86)90149-8. [DOI] [PubMed] [Google Scholar]
- Bareyre F.M. Schwab M.E. Inflammation, degeneration and regeneration in the injured spinal cord: insights from DNA microarrays. Trends Neurosci. 2003;26:555–563. doi: 10.1016/j.tins.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Brown I.R. Heat shock proteins and protection of the nervous system. Ann. N. Y. Acad. Sci. 2007;1113:147–158. doi: 10.1196/annals.1391.032. [DOI] [PubMed] [Google Scholar]
- Cao F. Yang X.F. Liu W.G. Hu W.W. Li G. Zheng X.J. Shen F. Zhao X.Q. Lv S.T. Elevation of neuron-specific enolase and S-100beta protein level in experimental acute spinal cord injury. J. Clin. Neurosci. 2008;15:541–544. doi: 10.1016/j.jocn.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Cao Q. Zhang Y.P. Iannotti C. DeVries W.H. Xu X.-M. Shields C.B. Whittemore S.R. Functional and electrophysiological changes after graded traumatic spinal cord injury in adult rat. Exp. Neurol. 2005;191:S3–S16. doi: 10.1016/j.expneurol.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Cohen P. The role of protein phosphorylation in neural and hormonal control of cellular activity. Nature. 1982;296:613–620. doi: 10.1038/296613a0. [DOI] [PubMed] [Google Scholar]
- Cohen P. Signal integration at the level of protein kinases, protein phosphatases and their substrates. Trends Biochem. Sci. 1992;17:408–413. doi: 10.1016/0968-0004(92)90010-7. [DOI] [PubMed] [Google Scholar]
- Cox B. Kislinger T. Emili A. Integrating gene and protein expression data: Pattern analysis and profile mining. Methods. 2005;35:303–314. doi: 10.1016/j.ymeth.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Crowe M.J. Bresnahan J.C. Shuman S.L. Masters J.N. Beattie M.S. Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys. Nat. Med. 1997;3:73–76. doi: 10.1038/nm0197-73. [DOI] [PubMed] [Google Scholar]
- Denslow N. Michel M.E. Temple M.D. Hsu C.Y. Saatman K. Hayes R.L. Application of proteomics technology to the field of neurotrauma. J. Neurotrauma. 2003;20:401–407. doi: 10.1089/089771503765355487. [DOI] [PubMed] [Google Scholar]
- Ding Q. Wu Z. Guo Y. Zhao C. Jia Y. Kong F. Chen B. Wang H. Xiong S. Que H. Jing S. Liu S. Proteome analysis of up-regulated proteins in the rat spinal cord induced by transection injury. Proteomics. 2006;6:505–518. doi: 10.1002/pmic.200500296. [DOI] [PubMed] [Google Scholar]
- Fagan A. Culhane A.C. Higgins D.G. A multivariate analysis approach to the integration of proteomic and gene expression data. Proteomics. 2007;7:2162–2171. doi: 10.1002/pmic.200600898. [DOI] [PubMed] [Google Scholar]
- Fialho E. Nakamura A. Juliano L. Masuda H. Silva-Neto M.A. Cathepsin D-mediated yolk protein degradation is blocked by acid phosphatase inhibitors. Arch. Biochem. Biophys. 2005;436:246–253. doi: 10.1016/j.abb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Ganea E. Chaperone-like activity of alpha-crystallin and other small heat shock proteins. Curr. Protein Pept. Sci. 2001;2:205–225. doi: 10.2174/1389203013381107. [DOI] [PubMed] [Google Scholar]
- Goshima Y. Nakamura F. Strittmatter P. Strittmatter S.M. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Grant P. Pant H.C. Neurofilament protein synthesis and phosphorylation. J. Neurocytol. 2000;29:843–872. doi: 10.1023/a:1010999509251. [DOI] [PubMed] [Google Scholar]
- Grossman S.D. Rosenberg L.J. Wrathall J.R. Temporal-spatial pattern of acute neuronal and glial loss after spinal cord contusion. Exp. Neurol. 2001;168:273–282. doi: 10.1006/exnr.2001.7628. [DOI] [PubMed] [Google Scholar]
- Gutierrez J.A. Guerriero V., Jr Chemical modifications of a recombinant bovine stress-inducible 70 kDa heat-shock protein (Hsp70) mimics Hsp70 isoforms from tissues. Biochem. J. 1995;305:197–203. doi: 10.1042/bj3050197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi S. P. Corthals G. L. Zhang Y. Rochon Y. Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall E.D. Springer J.E. Neuroprotection and acute spinal cord injury: A reappraisal. NeuroRx. 2004;1:80–100. doi: 10.1602/neurorx.1.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Signaling – 2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- Jin J. Smith F.D. Stark C. Wells C.D. Fawcett J.P. Kulkarni S. Metalnikov P. O'Donnell P. Taylor P. Taylor L. Zougman A. Woodgett J.R. Langeberg L.K. Scott J.D. Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Kaji A. Hockberger R. Imaging of spinal cord injuries. Emerg. Med. Clin. North Am. 2007;25:735–750. doi: 10.1016/j.emc.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Kang S.K. So H.H. Moon Y.S. Kim C.H. Proteomic analysis of injured spinal cord tissue proteins using 2-DE and MALDI-TOF MS. Proteomics. 2006;6:2797–2812. doi: 10.1002/pmic.200500621. [DOI] [PubMed] [Google Scholar]
- Kislinger T. Cox B. Kannan A. Chung C. Hu P. Ignatchenko A. Scott M.S. Gramolini A. O. Morris Q. Hallett M.T. Rossant J. Hughes T.R. Frey B. Emili A. Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Kobeissy F.H. Ottens A.K. Zhang Z. Liu M.C. Denslow N.D. Dave J.R. Tortella F.C. Hayes R.L. Wang K.K. Novel differential neuroproteomics analysis of traumatic brain injury in rats. Mol. Cell. Proteomics. 2006;5:1887–1898. doi: 10.1074/mcp.M600157-MCP200. [DOI] [PubMed] [Google Scholar]
- Li G.L. Farooque M. Holtz A. Olsson Y. Apoptosis of oligodendrocytes occurs for long distances away from the primary injury after compression trauma to rat spinal cord. Acta Neuropathol. (Berl.) 1999;98:473–480. doi: 10.1007/s004010051112. [DOI] [PubMed] [Google Scholar]
- Ling X. Liu D. Temporal and spatial profiles of cell loss after spinal cord injury: Reduction by a metalloporphyrin. J. Neurosci. Res. 2007;85:2175–2185. doi: 10.1002/jnr.21362. [DOI] [PubMed] [Google Scholar]
- Liu Q. Hendrickson W.A. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.Z. Xu X.M. Hu R. Du C. Zhang S.X. McDonald J.W. Dong H.X. Wu Y.J. Fan G.S. Jacquin M.F. Hsu C.Y. Choi D.W. Neuronal and glial apoptosis after traumatic spinal cord injury. J. Neurosci. 1997;17:5395–5406. doi: 10.1523/JNEUROSCI.17-14-05395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy D.N. Sroufe A.E. Pelt J.L. Burke D.A. Cao Q.L. Talbott J.F. Whittemore S.R. Serum biomarkers for experimental acute spinal cord injury: rapid elevation of neuron-specific enolase and S-100beta. Neurosurg. 2005;56:391–397. doi: 10.1227/01.neu.0000148906.83616.d2. [DOI] [PubMed] [Google Scholar]
- Lucas J.H. Wheeler D.G. Emery D.G. Mallery S.R. The endogenous antioxidant glutathione as a factor in the survival of physically injured mammalian spinal cord neurons. J. Neuropathol. Exp. Neurol. 1998;57:937–954. doi: 10.1097/00005072-199810000-00006. [DOI] [PubMed] [Google Scholar]
- Maikos J.T. Shreiber D.I. Immediate damage to the blood–spinal cord barrier due to mechanical trauma. J. Neurotrauma. 2007;24:492–507. doi: 10.1089/neu.2006.0149. [DOI] [PubMed] [Google Scholar]
- Matsuoka S. Ballif B.A. Smogorzewska A. McDonald E. R., III Hurov K.E. Luo J. Bakalarski C.E. Zhao Z. Solimini N. Lerenthal Y. Shiloh Y. Gygi S.P. Elledge S.J. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McClellan A.J. Xia Y. Deutschbauer A.M. Davis R.W. Gerstein M. Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- McEwen M.L. Springer J.E. A mapping study of caspases-3 activation following acute spinal cord contusion in rats. J. Histochem. Cytochem. 2005;53:809–819. doi: 10.1369/jhc.4A6467.2005. [DOI] [PubMed] [Google Scholar]
- McEwen M.L. Sullivan P.G. Springer J.E. Pretreatment with the cyclosporin derivative, NIM811, improves the function of synaptic mitochondria following spinal cord contusion in rats. J. Neurotrauma. 2007;24:613–624. doi: 10.1089/neu.2006.9969. [DOI] [PubMed] [Google Scholar]
- Nishimura R.N. Sharp F.R. Heat shock proteins and neuromuscular disease. Muscle Nerve. 2005;32:693–709. doi: 10.1002/mus.20373. [DOI] [PubMed] [Google Scholar]
- Noble L.J. Wrathall J.R. The blood–spinal cord barrier after injury: Pattern of vascular events proximal and distal to a transection in the rat. Brain Res. 1987;424:177–188. doi: 10.1016/0006-8993(87)91208-x. [DOI] [PubMed] [Google Scholar]
- Noble L.J. Wrathall J.R. Blood–spinal cord barrier disruption proximal to a spinal cord transection in the rat: time course and pathways associated with protein leakage. Exp. Neurol. 1988;99:567–578. doi: 10.1016/0014-4886(88)90173-2. [DOI] [PubMed] [Google Scholar]
- Noble L.J. Wrathall J.R. Distribution and time course of protein extravasation in the rat spinal cord after contusive injury. Brain Res. 1989;482:57–66. doi: 10.1016/0006-8993(89)90542-8. [DOI] [PubMed] [Google Scholar]
- Nordby H.K. Urdal P. The diagnostic value of measuring creatine kinase BB activity in cerebrospinal fluid following acute head injury. Acta Neurochir. (Wien) 1982;65:93–101. doi: 10.1007/BF01405445. [DOI] [PubMed] [Google Scholar]
- Ottens A.K. Kobeissy F.H. Fuller B.F. Liu M.C. Oli M.W. Hayes R.L. Wang K.K. Novel neuroproteomic approaches to studying traumatic brain injury. Prog. Brain Res. 2007;161:401–418. doi: 10.1016/S0079-6123(06)61029-7. [DOI] [PubMed] [Google Scholar]
- Pawson T. Scott J.D. Protein phosphorylation in signaling – 50 years and counting. Trends Biochem. Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Power J.H. Asad S. Chataway T.K. Chegini F. Manavis J. Temlett J.A. Jensen P.H. Blumbergs P.C. Gai W.P. Peroxiredoxin 6 in human brain: Molecular forms, cellular distribution and association with Alzheimer's disease pathology. Acta Neuropathol. 2008;115:611–622. doi: 10.1007/s00401-008-0373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar R. McEwen M.L. Springer J.E. Post-treatment with the cyclosporin derivative, NIM811, reduced indices of cell death and increased the volume of spared tissue in the acute period following spinal cord contusion. J. Neurotrauma. 2007;24:1618–1630. doi: 10.1089/neu.2007.0329. [DOI] [PubMed] [Google Scholar]
- Robinson M.B. Tidwell J.L. Gould T. Taylor A.R. Newbern J.M. Graves J. Tytell M. Milligan C.E. Extracellular heat shock protein 70: A critical component for motoneuron survival. J. Neurosci. 2005;25:9735–9745. doi: 10.1523/JNEUROSCI.1912-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheff S.W. Rabchevsky A.G. Fugaccia I. Main J.A. Lumpp J.E., Jr Experimental modeling of spinal cord injury: characterization of a force-defined injury device. J. Neurotrauma. 2003;20:179–193. doi: 10.1089/08977150360547099. [DOI] [PubMed] [Google Scholar]
- Schoonheim P.J. Veiga H. Pereira Dda C. Friso G. van Wijk K.J. de Boer A.H. A comprehensive analysis of the 14-3-3 interactome in barley leaves using a complementary proteomics and two-hybrid approach. Plant Physiol. 2007;143:670–683. doi: 10.1104/pp.106.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S.L. Bresnahan J.C. Beattie M.S. Apoptosis of microglia and oligodendrocytes after spinal cord contusion in rats. J. Neurosci. Res. 1997;50:798–808. doi: 10.1002/(SICI)1097-4547(19971201)50:5<798::AID-JNR16>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Siman R. McIntosh T.K. Soltesz K.M. Chen Z. Neumar R.W. Roberts V.L. Proteins released from degenerating neurons are surrogate markers for acute brain damage. Neurobiol. Dis. 2004;16:311–320. doi: 10.1016/j.nbd.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Siman R. Zhang C. Roberts V.L. Pitts-Kiefer A. Neumar R.W. Novel surrogate markers for acute brain damage: Cerebrospinal fluid levels corrrelate with severity of ischemic neurodegeneration in the rat. J. Cereb. Blood Flow Metab. 2005;25:1433–1444. doi: 10.1038/sj.jcbfm.9600138. [DOI] [PubMed] [Google Scholar]
- Springer J.E. Apoptotic cell death following traumatic injury to the central nervous system. J. Biochem. Mol. Biol. 2002;35:94–105. doi: 10.5483/bmbrep.2002.35.1.094. [DOI] [PubMed] [Google Scholar]
- Springer J.E. Azbill R.D. Knapp P. Activation of caspases-3 apoptotic cascade in traumatic spinal cord injury. Nat. Med. 1999;5:943–946. doi: 10.1038/11387. [DOI] [PubMed] [Google Scholar]
- Springer J.E. Azbill R.D. Mark R.J. Begley J.G. Waeq G. Mattson M.P. 4-Hydroxynonenal, a lipid peroxidation product, rapidly accumulates following traumatic spinal cord injury and inhibits glutamate uptake. J. Neurochem. 1997;68:2469–2476. doi: 10.1046/j.1471-4159.1997.68062469.x. [DOI] [PubMed] [Google Scholar]
- Springer J.E. Azbill R.D. Nottingham S.A. Kennedy S.E. Calcineurin-mediated BAD dephosphorylation activates the caspase-3 apoptotic cascade in traumatic spinal cord injury. J. Neurosci. 2000;20:7246–7251. doi: 10.1523/JNEUROSCI.20-19-07246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan P.G. Krishnamurthy S. Patel S.P. Pandya J.D. Rabchevsky A.G. Temporal characterization of mitochondrial bioenergetics after spinal cord injury. J. Neurotrauma. 2007;24:991–999. doi: 10.1089/neu.2006.0242. [DOI] [PubMed] [Google Scholar]
- Taylor S.W. Fahy E. Ghosh S.S. Global organellar proteomics. Trends Biotechnol. 2003;21:82–88. doi: 10.1016/S0167-7799(02)00037-9. [DOI] [PubMed] [Google Scholar]
- Tsai M.C. Shen L.F. Kuo H.S. Cheng H. Chak K.F. Involvement of acidic fibroblast growth factor in spinal cord injury repair processes revealed by a proteomics approach. Mol. Cell. Proteomics. 2008;7:1668–1687. doi: 10.1074/mcp.M800076-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.K. Ottens A. Haskins W. Liu M.C. Kobeissy F. Denslow N. Chen S. Hayes R.L. Proteomics studies of traumatic brain injury. Int. Rev. Neurobiol. 2004;61:215–240. doi: 10.1016/S0074-7742(04)61009-9. [DOI] [PubMed] [Google Scholar]
- Wang L.H. Strittmatter S.M. A family of rat CRMP genes is differentially expressed in the nervous system. J. Neurosci. 1996;16:6197–6207. doi: 10.1523/JNEUROSCI.16-19-06197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuermser L.A. Ho C.H. Chiodo A.E. Priebe M.M. Kirshblum S.C. Scelza W.M. Spinal cord injury medicine. 2. Acute care management of traumatic and nontraumatic injury. Arch. Phys. Med. Rehabil. 2007;88:S55–S61. doi: 10.1016/j.apmr.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Wu X. Yoo S. Wrathall J.R. Real-time quantitative PCR analysis of temporal-spatial alterations in gene expression after spinal cord contusion. J. Neurochem. 2005;93:943–952. doi: 10.1111/j.1471-4159.2005.03078.x. [DOI] [PubMed] [Google Scholar]
- Yan X. Liu T. Yang S. Ding Q. Liu Y. Zhang X. Que H. Wei K. Luo Z. Liu S. Proteomic profiling of the insoluble pellets of the transected rat spinal cord. J. Neurotrauma. 2009;26:179–193. doi: 10.1089/neu.2008.0533. [DOI] [PubMed] [Google Scholar]
- Yates D.M. Manser C. DeVos K.J. Shaw C.E. McLoughlin D.M. Miller C.C. Neurofilament subunit (NFL) head domain phosphorylation regulates axonal transport of neurofilaments. Eur. J. Cell. 2009;88:193–202. doi: 10.1016/j.ejcb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Yates J.R., 3rd Gilchrist A. Howell K.E. Bergeron J.J. Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell. Biol. 2005;6:702–714. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- Yong C. Arnold P.M. Zoubine M.N. Citron B.A. Watanabe I. Berman N.E. Festoff B.W. Apoptosis in cellular compartments of rat spinal cord after severe contusion injury. J. Neurotrauma. 1998;15:459–472. doi: 10.1089/neu.1998.15.459. [DOI] [PubMed] [Google Scholar]