Abstract
Replicative senescence forms a major barrier to tumor progression. Cancer cells bypass this by using one of the two known telomere maintenance mechanisms: telomerase or the recombination-based alternative lengthening of telomeres (ALT) mechanism. The molecular details of ALT are currently poorly understood. We have previously shown that telomerase is actively repressed through complex networks of kinase, gene expression, and chromatin regulation. In this study, we aimed to gain further understanding of the role of kinases in the regulation of telomerase expression in ALT cells. Using a whole human kinome small interfering RNA (siRNA) screen, we highlighted 106 kinases whose expression is linked to human telomerase reverse transcriptase (hTERT) promoter activity. Network modeling of transcriptional regulation implicated c-Myc as a key regulator of the 106 kinase hits. Given our previous observations of lower c-Myc activity in ALT cells, we further explored its potential to regulate telomerase expression in ALT. We found increased c-Myc binding at the hTERT promoter in telomerase-positive compared with ALT cells, although no expression differences in c-Myc, Mad, or Max were observed between ALT and telomerase-positive cells that could explain decreased c-Myc activity in ALT. Instead, we found increased expression of the c-Myc competitive inhibitor TCEAL7 in ALT cells and tumors and that alteration of TCEAL7 expression levels in ALT and telomerase-positive cells affects hTERT expression. Lower c-Myc activity in ALT may therefore be obtained through TCEAL7 regulation. Thus, TCEAL7 may present an interesting novel target for cancer therapy, which warrants further investigation.
Introduction
The induction of the permanent cell cycle arrest known as cellular senescence is a major barrier to tumor progression [1–3]. Most human cancers bypass telomere-dependent replicative senescence through transcriptional up-regulation of the telomerase subunits human telomerase RNA (hTR) and human telomerase reverse transcriptase (hTERT) [4,5]. However, up to 77% of some mesenchymal tumor types [6,7] use the intertelomeric and intratelomeric recombination-based mechanism known as alternative lengthening of telomeres (ALT) [8]. The molecular details behind the decision to activate ALTor telomerase during the process of immortalization are still poorly understood. ALT has prognostic significance in a number of tumor types [9–13], and because ALT tumors do not express significant levels of telomerase, telomerase-centered therapeutics are unlikely to be of benefit to ALT tumor treatment. An improved understanding of regulatory processes underlying the ALT phenotype may yield potential targets for future intervention therapy.
Our previous studies have shown the existence of a complex network of interactions through kinases [14], gene expression alterations [15], and chromatin modification [16] that regulate telomerase expression in ALT and telomerase-positive cells. Network modeling of expression data also highlighted a potentially decreased level of c-Myc activity in ALT cells, which we confirmed using an ELISA assay for c-Myc DNA binding [15]. The Myc gene family is frequently overexpressed in cancer [17] and is involved in the regulation of the cell cycle, angiogenesis, and cell adhesion during neoplasia [18]. c-Myc overexpression has been seen to be essential for an effective senescence bypass in melanoma and has proved an interesting target for senescence induction therapy [19–21]. Furthermore, the expression of the catalytic subunit of telomerase, hTERT, is activated by c-Myc [22]. Given the strict regulatory networks surrounding telomerase and the decreased c-Myc activity in ALT cells, we have previously suggested that c-Myc may be an important factor in the decision between the use of ALT or telomerase during immortalization. Because we have previously observed no significant difference in expression of c-Myc between ALT and telomerase-positive cells, the regulatory mechanisms behind the lower c-Myc activity in ALT remain to be elucidated.
In this study, we aimed to gain a further understanding of the role of kinases in the complex regulatory network surrounding hTERT expression in ALT cells. Through the use of a small interfering RNA (siRNA) kinase screen, we elucidated 106 kinases with the ability to affect hTERT promoter activity by at least two-fold. Investigation of transcriptional regulators of the kinase hits revealed c-Myc to be a critical regulator of these kinases. In addition, through further investigation of c-Myc regulation, we show that alterations in expression of the c-Myc competitive inhibitor TCEAL7 may be one mechanism by which c-Myc activity and hTERT expression are differentially regulated in ALT cells.
Materials and Methods
Cell Culture
SKLU (lung adenocarcinoma), C33a (cervical carcinoma), and SUSM1 (liver fibroblasts) were cultured in Dulbecco's modified Eagle medium (10% FBS and 2 mM l-glutamine) at 10% CO2. HT1080 (fibrosarcoma), KMST6 (lung fibroblasts), WI38-SV40 (SV40 immortalized lung fibroblasts), WI38 (normal lung fibroblasts), and IMR90 (normal lung fibroblasts) were cultured in minimum essential medium (10% FBS and 2 mM L-glutamine) at 5% CO2. A2780 (ovarian carcinoma) and 5637 (bladder carcinoma) were cultured in RPMI 1640 (10% FBS and 2 mM l-glutamine) at 5% CO2. All culture media and reagents were purchased from Invitrogen (Paisley, UK).
c-Myc Transcription Factor Binding Assays
Nuclear extracts were obtained from each cell line using the nuclear extract kit from Active Motif (Active Motif, Rixensart, Belgium) as per the manufacturer's instructions. c-Myc activity was measured in duplicate samples for each cell line using TransAM transcription factor binding ELISAs (Active Motif) in triplicate as per the manufacturer's instructions. Briefly, nuclear extracts were incubated for 1 hour at room temperature with mild agitation in microwells coated with consensus c-Myc oligonucleotide. After incubation, wells were washed and probed for 1 hour with 1:1000 dilution of c-Myc antibody. Antibody binding was detected using HRP-conjugated secondary and the addition of developing and stop solutions. Optical density of wells was measured at 450 nm, and a standard curve of recombinant c-Myc protein was included in each assay along with blank negative control and positive control wells containing Jurkat cell extract.
siRNA and Overexpression Plasmid Transfections
Whole kinome siRNA library screen (Applied Biosystems, Inc, Foster City, CA) in SKLU cells was performed as previously published [14]. In brief, 50 nM siRNA or noncoding siRNA control (Applied Biosystems, Inc) were cotransfected with 250 ng of hTERT reporter plasmid in 96-well plates (Thermo-Fisher Scientific, Rochester, NY) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cell lysis was achieved using passive lysis buffer (Promega, Madison, WI). Luciferase assay system (Promega) was used to determine luciferase activities according to the manufacturer's instructions. TCEAL7 siRNA transfections were performed using Dharmacon Smartpools (Thermo-Fisher Scientific) in 75-cm2 flasks. TCEAL7 siRNA transfections for c-Myc activity were performed in 125-cm2 flasks. pCMV-XL4:TCEAL7 overexpression plasmid was purchased from Origene Technologies, Inc (Rockville, MD), and all transfections were performed in 75-cm2 flasks. All transfections were performed in triplicate.
Bioinformatic Analysis
Fully MIAME-compliant gene expression microarray data were previously submitted to GEO under the accession numbers GSE14487 and GSE17118. Data were normalized to the 75th percentile using Genespring GX (version 7.3; Agilent Technologies UK Ltd., South Queensferry, UK), and c-Myc, Mad, and Max probe-normalized intensities were exported to Excel (Microsoft Corporation, Redmond, WA) for visualization. The interval plot of mesenchymal tumor data was produced using Minitab (version 15; Minitab, Inc, State College, PA). Transcriptional regulation networks were generated using transcriptional regulation algorithms in MetaCore (Genego, St Joseph, MI; filters: positive and negative regulations, all mechanisms).
c-Myc Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as per the manufacturer's instructions (Millipore, Watford, UK) as previously published [14]. No antibody pull-downs were performed as negative controls for each immunoprecipitation. Data are presented as percentage of input after subtraction of any background detected in the no antibody control.
RNA Extraction and Quantitative Polymerase Chain Reaction
RNA was extracted using the Nucleospin II RNA extraction kit (Macherey-Nagel, Duren, Germany) according to the manufacturer's instructions. TCEAL7 and hTERT quantitative polymerase chain reaction (qPCR) and hTERT promoter ChIP qPCR was performed using the DyNAmo Hot star SYBR Green kit (Finnzymes, Espoo, Finland) and Opticon 2 DNA Engine from MJ Research. Primer sequences were as follows:
TCEAL7f: GAAGGAAGAGCTGGTTGAT
TCEAL7r: GACCCCTTATCTCTTCCAA
hTERTf: CTGCTGCGCACGTGGGAAGC
hTERTr: GGACACCTGGCGGAAGGAG
CHIPTERTf: TCCCCTTCACGTCCGGCATT
CHIPTERTr: AGCGGAGAG AGGTCGAATCG
All qPCRs were normalized to S15 expression before taking fold of control. S15 primers were as follows:
S15f: TCCGCAAGTTCACCTACC
S15r: CGGGCCGGCCATGCTTTACG
All qPCR experiments were performed in triplicate.
Telomerase Activity Assay
The TRAPeze XL kit was used for telomere repeat amplification protocol (TRAP) assay according to the manufacturer's instructions (Millipore) as previously published [14].
Statistical Analysis
Statistical analysis of ELISA, reverse transcription (RT)-PCR, and TRAP assays were performed by unpaired t-test, using GraphPad Prism software (GraphPad Software, San Diego, CA).
Results
siRNA Kinase Screen Highlights 106 hTERT Regulatory Kinases
To probe kinase regulation of the ALT phenotype, we undertook an siRNA kinase screen using an hTERT promoter luciferase reporter construct in the ALT cell line SKLU. Three separate siRNA for 719 kinases and kinase-related genes were investigated. As in our previous study [14], the cutoff we applied for hit assessment was a more-than two-fold change in hTERT promoter activity in at least two of three siRNAs. This resulted in 106 kinases that met the criteria (Figure 1 and Table W1). Examination of the regulatory effect of these hits shows that 89 activate the hTERT reporter (promoter activity downregulated by siRNA) and 17 are inhibitors.
Figure 1.
SiRNA kinase screen highlights 106 kinases with hTERT regulatory functions. Kinases with two siRNA that induce a two-fold or more change in hTERT promoter activity in the same direction over promoter-less control plasmid. Each point indicates the average change in promoter activity of two or more different siRNA for each kinase assayed in triplicate; error bars, SE of triplicate repeats.
This interesting result indicates that SKLU cells retain many active signaling components, which may be required for hTERT expression in telomerase-positive cells. In the context of the ALT phenotype, it is likely that their effects on telomerase are muted by additional regulatory systems including epigenetic changes at the hTERT promoter and the altered gene expression networks we have previously observed. It will be of interest to investigate the potential roles of the inhibitory kinase hits identified here in these processes in future studies.
Transcriptional Regulation Networks of 106 hTERT Regulatory Kinases Highlight the Importance of c-Myc
Given that we have previously shown that the ALT phenotype involves an altered transcriptional profile, we used network modeling to investigate which transcription factors might affect and/or be affected by the kinases identified as hits. Application of the transcriptional regulation network inference algorithm in MetaCore from Genego returned several networks centered on individual high-degree transcription factor neighbors of the hits, including a number with known hTERTregulatory functions (Table 1). Union of the networks surrounding the hTERT transcriptional activators STAT3, SP1, and c-Myc (Figure 2A) and repressors p53 and the Androgen receptor (Figure 2B) highlights the interplay between multiple candidate pathways that might influence the expression of hTERT in ALT cells.
Table 1.
Transcriptional Regulation Analysis of 106 hTERT Regulating Kinases in MetaCore from Genego Shows Known hTERT Regulating Transcription Factors and c-Myc as the Most Significant Regulator.
| No. | Network | % of Hits Regulated | P |
| 1 | c-Myc | 17 | 1.50e-48 |
| 2 | HNF4-α | 15 | 2.42e-40 |
| 3 | p53 | 14 | 1.28e-37 |
| 4 | SP1 | 13 | 6.67e-35 |
| 5 | STAT3 | 11 | 1.73e-29 |
Figure 2.
Transcriptional regulation networks highlight synergistic regulation of hTERT regulatory kinases by known hTERT transcriptional regulators. Merged networks of the five highest scoring transcriptional regulators of 106 siRNA kinase screen hits of known hTERT transcriptional activators (A) and repressors (B). Green lines between objects indicate an activating interaction, whereas red lines indicate an inhibitory reaction. Overlaid line colors (red, green, or blue) indicate transcriptional networks generated for each transcription factor before network merge. Symbols on the lines indicate the following: TR = transcriptional regulation, +P = phosphorylation, B = binding, and ? = unknown mechanism. Blue circles next to kinases indicate an hTERT activating kinase, and red circles indicate an hTERT inhibiting kinase.
We have previously shown that c-Myc activity is lower in ALT cells compared with telomerase-positive cells resulting in changes in the expression of c-Myc downstream targets. Interestingly, c-Myc regulates the largest percentage of kinase hits in this study (Table 1), showing not only that c-Myc itself is involved with telomerase regulation in ALT but also that its downstream kinases are regulators themselves of telomerase expression. Given the increasing evidence for the central role of c-Myc in this process, we further investigated the factors potentially regulating c-Myc activity in ALT cells.
Telomerase-Positive Cells Have Increased Binding of c-Myc at the hTERT Promoter but Show No Change in Binding Partner Expression
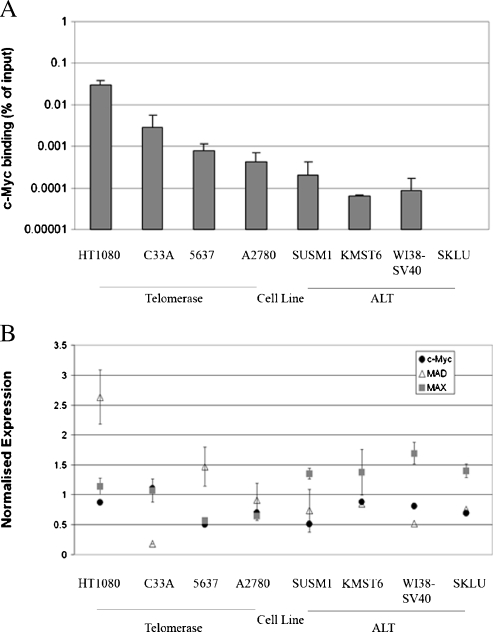
The role of epigenetic changes in hTERT promoter silencing is well established. However, the landscape of sequence-specific transcription factors associated with the hTERT promoter in ALT remains unclear, although it is predictable that many positively acting factors will be excluded or present in profoundly altered complexes. To compare the influence of c-Myc at the hTERT promoter between ALT and telomerase-positive cells, we performed ChIP using a c-Myc antibody and qPCR for the hTERT promoter in the resulting pull-down (Figure 3A). Within the telomerase-positive cell lines, HT1080 and C33a showed the highest levels of c-Myc immunoreactivity. A2780 cells had the lowest levels of c-Myc detected at the hTERT promoter; however, these were two-, five-, and seven-fold greater than SUSM-1, WI38-SV40, and KMST6, respectively (note the log scale). No signal above background was detected in SKLU cells. This is consistent with our previous findings of decreased c-Myc activity and inhibitory chromatin modifications at the hTERT promoter in ALT cells and might reflect either decreased site occupancy or epitope masking in ALT cells.
Figure 3.
c-Myc shows increased binding at the hTERT promoter in telomerase-positive cells, which cannot be explained by common c-Myc binding partner expression patterns in ALT and telomerasepositive cells. (A) ChIP analysis of the hTERT promoter was completed using an antibody against c-Myc performed in duplicate. Each immunoprecipitate was quantified in triplicate by qPCR analysis and related back to an input sample in each experiment to normalize the data. Error bars, SE of duplicate experiments. (B) Gene expression microarray data for c-Myc, Mad, and Max in ALT cell lines (SUSM1, KMST6, WI38-SV40, and SKLU) and telomerase-positive cell lines (HT1080, C33a, 5637, and A2780). Results are representative of one probe for c-Myc and themean and SE of duplicate probes for Mad and triplicate probes for Max. Data were previously submitted to GEO under accession number GSE14487.
Relative levels of Myc/Max versus Mad/Max binding have previously been observed to regulate the hTERT promoter chromatin environment, raising the interesting possibility that E-boxes might play a role in establishment of epigenetic silencing of telomerase in ALT. Using our gene expression microarray data on the ALT and telomerase cell lines, we investigated whether the difference in c-Myc binding at the hTERT promoter could be accounted for by expression differences in Mad and Max. We observed no significant difference in expression of c-Myc itself or Mad and Max between the ALT and telomerase-positive cell lines (Figure 3B).
Given the lack of differences in expression of c-Myc, Max, or Mad between ALTand telomerase cell lines that could explain the decrease of c-Myc activity observed in ALT, we decided to investigate other possible mechanisms for c-Myc regulation.
Differential Expression of c-Myc Inhibitor TCEAL7 in ALT and Telomerase-Positive Cell Lines
The transcription elongation factor A (SII)-like protein TCEAL7 has recently been shown to inhibit c-Myc activity through the competitive binding of E-boxes [23]. Such competitive inhibition would allow regulation of c-Myc transcriptional activity without the need for a down-regulation in expression. We examined TCEAL7 expression to assess whether it might regulate c-Myc activity in ALT and telomerase-positive cells. Figure 4A shows RT-qPCR for TCEAL7 in the ALT and telomerase-positive cell lines. Three of the four ALT cell lines have high levels of TCEAL7 expression, whereas the telomerase-positive cells have little or no expression of this gene. Coimmunoprecipitation experiments showed no direct interaction between TCEAL7 and c-Myc in any of the ALT or telomerase-positive cell lines (data not shown), suggesting that TCEAL7 regulation of c-Myc activity in ALT is through competitive inhibition.
Figure 4.
TCEAL7 expression is increased in ALT cell lines and mesenchymal tumors. (A) S15 normalized qPCR of TCEAL7 in telomerase-positive cell lines (HT1080, A2780, 5637, and C33A) and ALT cell lines (SKLU, KMST6, WI38-SV40, and SUSM1). Error bars, SE of triplicate experiments. (B) Interval plot of gene expression microarray data for TCEAL7 in threemesenchymal tumor types (liposarcoma, peritoneal mesothelioma, and malignant peripheral nerve sheath tumor) submitted to GEO under accession number GSE17118. Crosshairs show mean expression in telomerase-positive and ALT tumors; error bars, 95% confidence intervals drawn using Minitab (version 15). t = 1.99, P = 0.053, df = 42.
ALT is predominantly found in tumors of mesenchymal origin in vivo [6,7]; therefore, to investigate whether the increased expression of TCEAL7 is observed in ALT tumors, we used our gene expression microarray data on three mesenchymal tumor types: liposarcomas, peritoneal mesotheliomas, and malignant peripheral nerve sheath tumor [15]. Figure 4B shows that, in our mesenchymal tumor cohort, ALT tumors express higher levels of TCEAL7 than telomerasepositive tumors (t = 1.99, P = 0.053, df = 42). This suggests that TCEAL7 could regulate c-Myc activity both in vitro and in vivo.
TCEAL7 Affects c-Myc Activity, hTERT Expression, and Telomerase Activity
To investigate the effects of altering TCEAL7 in ALT cells, we performed siRNA knockdowns using a pool of three different siRNA. Figure 5A shows that, in all four ALT cell lines, TCEAL7 expression was reduced in comparison to nonspecific siRNA control. We therefore examined the effect of TCEAL7 knockdown on c-Myc activity in ALT cells using transcription factor binding ELISAs. Figure 5B shows that TCEAL7 knockdown caused strong induction of c-Myc activity in WI-38 and SUSM1 cells, suggesting suppression of c-Myc DNA binding activity in ALT cells which overexpress TCEAL7. In contrast, c-Myc DNA binding activity was not significantly affected in SKLU or KMST6 cells. As SKLU and KMST6 express less TCEAL7 than WI-38 and SUSM1, it is possible that other factors contribute to low c-Myc activity these cells.
Figure 5.
Knockdown of TCEAL7 expression leads to increased c-Myc activity and increased hTERT expression in some ALT cells. TCEAL7 knockdown using a pool of three independent TCEAL7 siRNA (Dharmacon) or noncoding siRNA control. Transfections were performed in duplicate, and two independent experiments were analyzed. (A) qPCR of TCEAL7 expression normalized to S15 expression before taking fold of control transfections. Error bars derived from two independent experiments with PCR done in triplicate. (B) hTERT expression was normalized to S15 expression before taking fold of control transfections. Error bars derived from two independent experiments with PCR done in triplicate. c-Myc activity was measured using transcription factor ELISA assays (Active Motif) performed in duplicate. Recombinant c-Myc (Active Motif) was used to create a standard curve for relative quantification. Results represent mean and SE of replicates derived from two independent transfections. P values were calculated using GraphPad Prism.
Because we have seen lower levels of c-Myc binding at the hTERT promoter in ALT cells, we investigated whether TCEAL7 expression knockdown and the resulting changes in c-Myc activity are sufficient to alter the expression of hTERT in ALT cells using RT-qPCR analysis of full-length hTERT (Figure 5B). TCEAL7 knockdown significantly induced hTERT expression in SUSM1 cells, confirming that TCEAL7 can play a role in the suppression of hTERT expression in some ALT cells. Interestingly, hTERT expression was decreased in SKLU and KMST6, also suggesting active E-box-dependent regulation of telomerase, although c-Myc activity was relatively unaffected in these cells. Therefore, additional mechanisms for c-Myc suppression may also be present. One possibility is that TCEAL7 knockdown preferentially relieves Mad rather than c-Myc targeting to E-boxes in these cells resulting in further reduction of hTERT expression. In WI38-SV40 cells, hTERT was not significantly affected as a result of TCEAL7 knockdown.
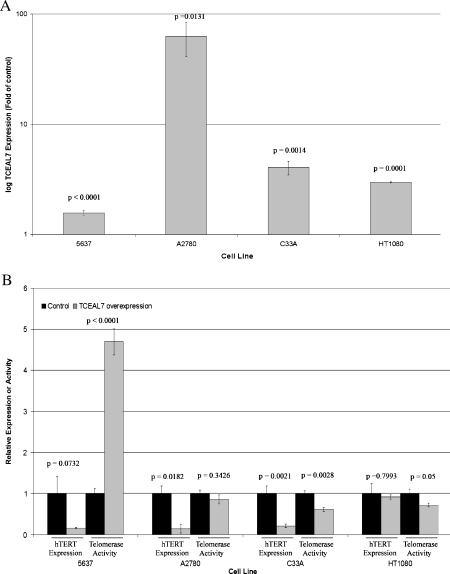
Because telomerase-positive cells express little or no TCEAL7, we investigated the effects of TCEAL7 overexpression in these cells. A complementary DNA expression construct was transfected into the four telomerase-positive cell lines, and the expression of TCEAL7 was analyzed by RT-qPCR compared with an empty vector control. In all four cell lines, TCEAL7 expression was significantly increased in comparison to empty vector control (Figure 6A). The effect of TCEAL7 overexpression on hTERT expression was then investigated using RTqPCR (Figure 6B). Expression of hTERT was decreased significantly after TCEAL7 overexpression in A2780 and C33A cells. A decrease of borderline significance was also observed in 5637 cells, although the expression was unaffected in HT1080 cells. Because levels of c-Myc binding at the hTERT promoter were highest in these cells (Figure 3), more TCEAL7 may be required for successful competition.
Figure 6.
Overexpression of TCEAL7 expression leads to decreased hTERT expression and telomerase activity in telomerase-positive cells. TCEAL7 complementary DNA overexpression plasmid (Origene) or empty vector control plasmid was transfected into telomerase-positive cells in duplicate, and two independent experiments were analyzed. (A) qPCR of TCEAL7 expression normalized to S15 expression before taking fold of control transfections. Error bars derived from two independent experiments with PCR done in triplicate. (B) hTERT expression was normalized to S15 expression before taking fold of control transfections. Error bars derived from two independent experiments with PCR done in triplicate. Telomerase activity was measured using TRAP assay. Error bars derived from duplicate assays for two independent transfections. P values were calculated using GraphPad Prism.
Telomerase activity was assessed by TRAP assay after TCEAL7 overexpression, revealing significantly decreased levels in C33A and HT1080 cells (Figure 6B). Interestingly, although 5637 cells showed decreased levels of hTERT gene expression, telomerase activity in fact increased with TCEAL7 overexpression. 5637 cells have relatively low levels of hTERT transcription, and the down-regulation of hTERT expression may trigger a regulatory feedback loop to maintain basal telomerase activity through, for example, posttranscriptional mechanisms such as alternative RNA splicing or hTERT phosphorylation. Regardless, taken together, the current results clearly indicate that TCEAL7 is a novel regulator of hTERT expression and telomerase activity in both ALTand telomerase-positive cell lines.
Discussion
ALT is predominantly found in tumors of mesenchymal origin [6,7] and has prognostic significance in multiple tumor types [9–13]. Improved understanding of the molecular mechanisms behind the decision to activate telomerase or ALT for immortalization may therefore provide novel opportunities for intervention in these tumors. We have previously shown that telomerase is strictly regulated at both the chromatin and gene expression levels in ALT cells [15,16]. Furthermore, we have also shown that kinases play a role in regulation of telomerase in telomerase-positive cells [14].
In this study, we initially probed the kinome for hTERT regulatory kinases in ALT cells using an siRNA kinase hTERT luciferase reporter screen. This highlighted 106 kinases through which hTERT promoter activity is regulated. A greater proportion of the hits observed were activators, such that their knockdown resulted in decreased hTERT promoter activity. Thus, our screening approach revealed that a substantial portion of the signaling machinery potentially required to support hTERT expression remains active in ALT cells. This observation may have future therapeutic implications because it has been suggested (although not experimentally observed) that the emergence of ALT could represent a mechanism of resistance to telomerase directed therapies. Furthermore, those hits identified as repressors of the hTERT promoter might be worthy of investigation as candidate ALT targets in their own right.
We previously adopted a network modeling approach to analyze a unique ALT-specific gene expression signature. In this study, we used our screening hits as a profile of active signaling pathways relevant to telomerase regulation in ALT and applied network building algorithms to investigate mechanisms of transcriptional control on these pathways (Table 1). The algorithm returned several hTERT regulatory transcription factors as highly significant neighbors of the kinase list, and we constructed network models around hTERT-activating and -repressing factors highlighting candidate pathways that may contribute to hTERT transcriptional repression in ALT. We have therefore shown that the complex network of interactions by which the expression of telomerase is regulated in ALT cells further extends to kinase regulation, consistent with our findings in telomerasepositive cells [14].
The most significant transcriptional regulator of the 106 kinase hits was c-Myc, a known hTERT transcriptional activator [22]. Overexpression of c-Myc is common in cancer, playing a regulatory role in the cell cycle [19], angiogenesis, and cell migration [18,24]. c-Myc therefore presents an attractive target for anticancer therapy. A recent study found that the potent anticancer drug butein causes a decrease in telomerase expression and inhibits cell proliferation through the suppression of c-Myc expression [25]. Furthermore, recent evidence that chromatin remodeling is required before c-Myc can access the hTERT promoter for activation [26] is consistent with our previous data showing that chromatin modification also plays a role in the lack of hTERT expression in ALT cell lines [16]. We have previously shown that c-Myc activity is significantly decreased in ALT cells compared with telomerase-positive cells [15], suggesting that c-Myc may have a role the decision between the two telomere maintenance mechanisms through telomerase regulation. To investigate this further, we used ChIP to assess the relative levels of c-Myc binding at the hTERT promoter in ALT and telomerase-positive cell lines. We found that telomerase-positive cells had more c-Myc present at the hTERT promoter when compared with ALT cells. c-Myc binds DNA at E-box sequences as a dimer with binding partner Max to activate target genes [27]. In turn, the activities of Max are antagonized by dimerization with Mad, and the resulting binding of Mad/Max to E-boxes within promoter regions leads to gene repression [28]. Furthermore, Mad has been found to dimerize with and recruit the histone demethylase RBP2 to E-boxes in the hTERT promoter and repress transcription through chromatin remodeling [29]. The expression levels of Mad and Max in ALT and telomerase-positive cells, therefore, may have an influence on the activity of c-Myc and telomerase expression. However, investigation of the expression patterns using our gene expression microarray data showed no significant difference in the expression of c-Myc, Mad, or Max between ALT and telomerase-positive cell lines.
The lack of binding partner interactions able to account for the decrease in c-Myc activity suggests a further layer of regulation of c-Myc activity in these cells. To this end, we investigated the potential for TCEAL7 regulation of c-Myc activity in ALT and telomerase-positive cells. TCEAL7 is a member of the transcription elongation factor A (SII)-like gene family and is relatively poorly studied. The expression of TCEAL7 is epigenetically silenced in epithelial ovarian cancer, and forced expression of TCEAL7 in ovarian tumor cell lines induced apoptosis [30]. Furthermore, TCEAL7 has been observed to have decreased expression in a variety of human tumors [23], and recently, polymorphisms in the gene have been associated with a reduced risk of invasive serous ovarian cancers [31]. The role of TCEAL7 in cancer risk may be due to its function as a regulator of cyclin D1 expression and c-Myc activity [23]. TCEAL7 binds to E-boxes of gene promoters and thereby competitively inhibits gene activation by Myc-Max complexes. Such competitive inhibition would effectively decrease c-Myc activity without the need for decreased c-Myc expression, which would be consistent with our findings. RT-qPCR showed that ALT cell lines, in general, express a high level of TCEAL7, whereas telomerase-positive cell lines express little or no TCEAL7. Because ALT is predominantly seen in mesenchymally derived tumors [6,7], we also investigated TCEAL7 expression in our gene expression microarray data for three mesenchymal tumor types: liposarcomas, peritoneal mesotheliomas, and malignant peripheral nerve sheath tumors. We found that TCEAL7 expression is increased in ALT tumors compared with telomerase-positive tumors (P = .053), suggesting that TCEAL7 overexpression may be a c-Myc regulatory mechanism both in vitro and in vivo.
To investigate the effect of perturbing TCEAL7 regulation in ALT cells, we performed siRNA knockdown of TCEAL7 with a pool of three separate siRNA oligonucleotides. TCEAL7 expression was successfully knocked down in all four ALT cell lines and was associated with significantly increased c-Myc activity in the two ALT cell lines expressing high levels of TCEAL7, WI38-SV40, and SUSM1, suggesting that TCEAL7 represents a potential c-Myc regulatory factor in these cells. Furthermore, examination of hTERT expression in these cell lines showed that the resulting c-Myc activity increase was sufficient to create an increase in hTERT expression in these cells, which reached significance in SUSM1.
The telomerase regulatory role of TCEAL7 was further investigated in telomerase-positive cells. These cells expressed little or no endogenous TCEAL7, however, overexpression resulted in a decrease in hTERT expression in all four cell lines tested, which reached significance in A2780 and C33A. We also found that in three of the telomerasepositive cell lines, TCEAL7 overexpression induced a decrease in telomerase activity. This decrease reached statistical significance in two of these cell lines. Consistent with the decreased expression of TCEAL7 in a wide variety of human tumors [23], these data combined suggest that TCEAL7 expression in ALT and telomerase-positive cells is a potentially important telomerase regulatory factor. The precise mechanism by which TCEAL7 affects telomerase expression remains to be investigated, although our ELISA results support inhibition of c-Myc DNA binding in at least some cases, and we ruled out direct protein-protein interactions by coimmunoprecipitation. Nevertheless, because most transcription factors including c-Myc regulate gene expression through complex mechanisms such as recruitment of chromatin remodeling activities, it is likely that direct competition with c-Myc is only part of the mechanism of action of TCEAL7; therefore, this warrants further investigation in future studies.
In this study, we have highlighted 106 kinases with the ability to regulate hTERT expression in ALT cells. Furthermore, we have shown that these kinases are regulated by known hTERT regulatory transcription factors and, in particular, the transcription factor c-Myc. Given that we have previously shown multiple gene expression changes in c-Myc downstream targets and decreased c-Myc activity in ALT cells [15], this study adds further weight to the importance of c-Myc in the regulation of telomere maintenance mechanisms. In addition, we also highlight competitive inhibition by TCEAL7 as a potential mechanism by which c-Myc activity could be regulated in ALT cells with the ability to regulate hTERT expression in both ALT and telomerase-positive cells. TCEAL7, therefore, presents an interesting novel therapeutic target. Further investigation of other regulatory targets of TCEAL7 may improve understanding of the complex network of interactions involved in the decision between telomere maintenance mechanisms in carcinogenesis and present us with novel targets for intervention.
Supplementary Material
Footnotes
This work was supported by Cancer Research UK, European Community grant Health-F2-2007-200950, Glasgow University, and Italian Association for Cancer Research.
This article refers to a supplementary material, which is designated by Table W1 and is available online at www.neoplasia.com.
References
- 1.Majumder PK, Grisanzio C, O'Connell F, Barry M, Brito JM, Xu Q, Guney I, Berger R, Herman P, Bikoff R, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14:146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–155. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Bilsland AE, Fletcher-Monaghan A, Keith WN. Properties of a telomerase-specific Cre/Lox switch for transcriptionally targeted cancer gene therapy. Neoplasia. 2005;7:1020–1029. doi: 10.1593/neo.05385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez JM, Grimm J, Josephson L, Weissleder R. Integrated nanosensors to determine levels and functional activity of human telomerase. Neoplasia. 2008;10:1066–1072. doi: 10.1593/neo.08350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, Wharton SB, Jellinek DA, Arbuckle SM, Yoo J, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11:217–225. [PubMed] [Google Scholar]
- 7.Ulaner GA, Hoffman AR, Otero J, Huang HY, Zhao Z, Mazumdar M, Gorlick R, Meyers P, Healey JH, Ladanyi M. Divergent patterns of telomere maintenance mechanisms among human sarcomas: sharply contrasting prevalence of the alternative lengthening of telomeres mechanism in Ewing's sarcomas and osteosarcomas. Genes Chromosomes Cancer. 2004;41:155–162. doi: 10.1002/gcc.20074. [DOI] [PubMed] [Google Scholar]
- 8.Royle NJ, Mendez-Bermudez A, Gravani A, Novo C, Foxon J, Williams J, Cotton V, Hidalgo A. The role of recombination in telomere length maintenance. Biochem Soc Trans. 2009;37:589–595. doi: 10.1042/BST0370589. [DOI] [PubMed] [Google Scholar]
- 9.Cairney CJ, Hoare SF, Daidone MG, Zaffaroni N, Keith WN. High level of telomerase RNA gene expression is associated with chromatin modification, the ALT phenotype and poor prognosis in liposarcoma. Br J Cancer. 2008;98:1467–1474. doi: 10.1038/sj.bjc.6604328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa A, Daidone MG, Daprai L, Villa R, Cantu S, Pilotti S, Mariani L, Gronchi A, Henson JD, Reddel RR, et al. Telomere maintenance mechanisms in liposarcomas: association with histologic subtypes and disease progression. Cancer Res. 2006;66:8918–8924. doi: 10.1158/0008-5472.CAN-06-0273. [DOI] [PubMed] [Google Scholar]
- 11.Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, McKay MJ, Reddel RR, Royds JA. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 12.Ulaner GA, Huang HY, Otero J, Zhao Z, Ben-Porat L, Satagopan JM, Gorlick R, Meyers P, Healey JH, Huvos AG, et al. Absence of a telomere maintenance mechanism as a favorable prognostic factor in patients with osteosarcoma. Cancer Res. 2003;63:1759–1763. [PubMed] [Google Scholar]
- 13.Villa R, Daidone MG, Motta R, Venturini L, DeMarco C, Vannelli A, Kusamura S, Baratti D, Deraco M, Costa A, et al. Multiple mechanisms of telomere maintenance exist and differentially affect clinical outcome in diffuse malignant peritoneal mesothelioma. Clin Cancer Res. 2008;14:4134–4140. doi: 10.1158/1078-0432.CCR-08-0099. [DOI] [PubMed] [Google Scholar]
- 14.Bilsland AE, Hoare S, Stevenson K, Plumb J, Gomez-Roman N, Cairney C, Burns S, Lafferty-Whyte K, Roffey J, Hammonds T, et al. Dynamic telomerase gene suppression via network effects of GSK3 inhibition. PLoS One. 2009;4:e6459. doi: 10.1371/journal.pone.0006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafferty-Whyte K, Cairney CJ, Will MB, Serakinci N, Daidone MG, Zaffaroni N, Bilsland A, Keith WN. A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT. Oncogene. 2009;28(43):3765–3774. doi: 10.1038/onc.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkinson SP, Hoare SF, Glasspool RM, Keith WN. Lack of telomerase gene expression in alternative lengthening of telomere cells is associated with chromatin remodeling of the hTR and hTERT gene promoters. Cancer Res. 2005;65:7585–7590. doi: 10.1158/0008-5472.CAN-05-1715. [DOI] [PubMed] [Google Scholar]
- 17.Lu X, Pearson A, Lunec J. The MYCN oncoprotein as a drug development target. Cancer Lett. 2003;197:125–130. doi: 10.1016/s0304-3835(03)00096-x. [DOI] [PubMed] [Google Scholar]
- 18.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, Gudkov AV, Prochownik EV, Nikiforov MA. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene. 2008;27:1905–1915. doi: 10.1038/sj.onc.1210823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang D, Mannava S, Grachtchouk V, Tang WH, Patil S, Wawrzyniak JA, Berman AE, Giordano TJ, Prochownik EV, Soengas MS, et al. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27:6623–6634. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C-H, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci USA. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chien J, Narita K, Rattan R, Giri S, Shridhar R, Staub J, Beleford D, Lai J, Roberts LR, Molina J, et al. A role for candidate tumor-suppressor gene TCEAL7 in the regulation of c-Myc activity, cyclin D1 levels and cellular transformation. Oncogene. 2008;27:7223–7234. doi: 10.1038/onc.2008.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC? Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- 25.Moon D-O, Kim M-O, Lee J-D, Choi YH, Kim G-Y. Butein suppresses c-Myc-dependent transcription and Akt-dependent phosphorylation of hTERT in human leukemia cells. Cancer Lett. 286(2):172–179. doi: 10.1016/j.canlet.2009.05.028. (in press) [DOI] [PubMed] [Google Scholar]
- 26.Bazarov AV, Hines WC, Mukhopadhyay R, Beliveau A, Melodyev S, Zaslavsky Y, Yaswen P. Telomerase activation by c-Myc in human mammary epithelial cells requires additional genomic changes. Cell Cycle. 2009;8(20):3373–3378. doi: 10.4161/cc.8.20.9856. [DOI] [PubMed] [Google Scholar]
- 27.Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 28.Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 29.Ge Z, Li W, Wang N, Liu C, Zhu Q, Bjorkholm M, Gruber A, Xu D. Chromatin remodeling: recruitment of histone demethylase RBP2 by Mad1 for transcriptional repression of a Myc target gene, telomerase reverse transcriptase. FASEB J. 2010;24(2):579–586. doi: 10.1096/fj.09-140087. [DOI] [PubMed] [Google Scholar]
- 30.Chien J, Staub J, Avula R, Zhang H, Liu W, Hartmann LC, Kaufmann SH, Smith DI, Shridhar V. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–5100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 31.Peedicayil A, Vierkant RA, Shridhar V, Schildkraut JM, Armasu S, Hartmann LC, Fridley BL, Cunningham JM, Phelan CM, Sellers TA, et al. Polymorphisms in TCEAL7 and risk of epithelial ovarian cancer. Gynecol Oncol. 2009;114:260–264. doi: 10.1016/j.ygyno.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.