Abstract
This paper presents a method for detection of cerebral white matter hyperintensities (WMH) based on run-time PD-, T1-, and T2- weighted structural magnetic resonance (MR) images of the brain along with labeled training examples. Unlike most prior approaches, the method is able to reliably detect WMHs in elderly brains in the absence of fluid-attenuated (FLAIR) images. Its success is due to the learning of probabilistic models of WMH spatial distribution and neighborhood dependencies from ground-truth examples of FLAIR-based WMH detections. These models are combined with a probabilistic model of the PD, T1, and T2 intensities of WMHs in a Markov Random Field (MRF) framework that provides the machinery for inferring the positions of WMHs in novel test images. The method is shown to accurately detect WMHs in a set of 114 elderly subjects from an academic dementia clinic. Experiments show that standard off-the-shelf MRF training and inference methods provide robust results, and that increasing the complexity of neighborhood dependency models does not necessarily help performance. The method is also shown to perform well when training and test data are drawn from distinct scanners and subject pools.
1 Introduction
Relevance of WMHs
White matter foci that are hyperintense on FLAIR images of the human brain are indicative of focal dysfunction of underlying axonal tracts. Common in a variety of clinical conditions, including multiple sclerosis, cerebrovascular disease, and depression, WMHs are important clinical measures in the elderly because their prevalence is strongly associated with cognitive function, longevity, disease progression, and the effects of disease-modifying treatments [1][2][3]. Because semi-quantitative manual grading of WMH severity is time-consuming and variable due to human subjectivity [4], a variety of fully automated methods have been developed to detect WMHs on FLAIR images in a robust, efficient, and objective manner [5][6].
Need for detecting WMH without FLAIR
However, while FLAIR images provide optimal contrast between WMHs and all other tissues, the detection of WMHs when no FLAIR is available is an increasingly important problem. Large-scale imaging studies are under pressure to collect a wide range of MR imaging sequences, including T1, T2, proton density (PD), diffusion tensor, functional, and perfusion MR, to capture the broadest possible range of biological phenomena in the brains of participants. Simultaneously, the studies are under pressure to scan each subject for the shortest amount of time possible due to scanner resource costs and the increases in head motion and subject discomfort that occur over the course of the scan session. Therefore, a growing list of large-scale imaging studies that have a strong interest in white matter dysfunction have nonetheless chosen to forgo FLAIR acquisition [3][7][8].
WMH detection without FLAIR using spatial and contextual priors
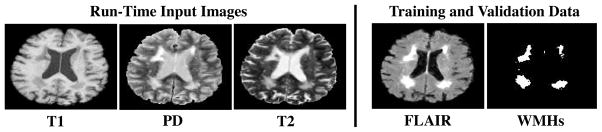
Because T1-weighted and double echo PD/T2-weighted acquisitions are nearly ubiquitous in large-scale imaging studies, we focus on WMH detection based solely on T1, T2, and PD input images. We use FLAIR exclusively for training data and the validation of automated methods (Fig. 1). WMHs are hyperintense on PD and T2, and hypointense on T1, but none of these modalities provide sufficient contrast between normal white matter (WM) and WMHs (Fig. 1). Therefore, we combine image intensity information with prior anatomical knowledge about where WMHs are known to occur in the brain and how they progress over time from one part of the brain to another. In particular, we employ a spatial prior– the prior probability of a WMH occurring at a given pixel, irrespective of imaging data– and a contextual prior– the conditional probability of a WMH occurring at a given pixel, given that WMHs have occurred at neighboring pixels. In elderly subjects, the spatial and contextual priors are highly structured and capture a characteristic spatial distribution of WMH occurrence and progression; specifically, WMHs in Alzheimer’s Disease and healthy aging tend to begin in periventricular zones and spread upward and outward (see Fig. 2 and [9]). The prior models that capture this progression are learned from FLAIR-based ground-truth WMH detections in a training phase, and are combined with intensity information at run-time in an MRF framework to detect WMHs in novel sets of coregistered (PD, T1, T2) test image sets.
Fig. 1.
A representative axial slice from the input images used for detecting WMHs at run time (left) and ground-truth data used for training the WMH detection method and validating the results (right).
Fig. 2.
Left: ADC subjects were divided into quintiles based on total WMH volume; voxels that had WMHs in more than 5% of subjects in the quintile are shown in red. Note that WMHs appear to progress systematically upwards and outwards from periventricular zones. Right: The contextual prior captures the characteristic inferior-to-superior progression of WMHs in elderly subjects. Each pixel is colored according to the probability that it is WMH, given that the pixel below it, vs. above it, is WMH. P(WMH|WMHBelow) is moderate at most pixels because if a downward neighbor is WMH, the upward propagation of WMHs may have arrived there and stopped; or it may have continued upward to include the pixel in question. Meanwhile P(WMH|WMHAbove) is generally high because if the upward progression of WMHs has already reached a particular pixel, it is likely to have already passed through the pixels below it. The WMH detection method uses this known spatial progression of WMH to help determine which pixels are WMH, based on the absolute position of the pixel and the presence of neighboring WMHs.
1.1 Prior Work
WMH detection without FLAIR
Few papers to date have dealt with the problem of automated WMH detection in the absence of FLAIR images, each using a comparatively simple model of WMH spatial distribution. One such method detected WMHs using a MRF system with a 2D and spatially invariant isotropic smoothing prior [10]. In another, the authors detected WMHs as outliers to models of other tissue classes instead of modelling them explicitly [11]. One method used boosted classifiers and Support Vector Machines to perform detection from PD and T1 images using spatially invariant isotropic smoothing and radial distance from center as a spatial prior. It also required separate training sets for mild, moderate, and severe WMH cases [12]. Finally, another used several run-time steps, including segmentations of grey matter, white matter, and CSF; segmentation of the thalamic nuclei; morphological post-processing to fix segmentation problems; and separation of WMHs into sub-classes based on image contrast [13]. The key difference between these methods and the current one is that the current method uses training data to directly capture the anatomical distribution and progression of WMHs in a model that allows spatial dependencies in WMH occurrence to vary arbitrarily across the image. Our method leverages this additional prior knowledge to directly model WMHs using a relatively straightforward run-time procedure that requires few steps or arbitrary parameter settings since it only fits parameters to a 3D intensity distribution and runs an existing, widely available MRF solver. Additionally, we focus on the elderly brain, whose morphological characteristics can be highly heterogeneous across a population due to diverse aging-related biological phenomena; the heterogeneity provides challenges to WMH detection that may differ from those associated with multiple sclerosis [10] and [11].
Use of contextual cues in WMH detection
While little attention has been paid to WMH detection in the absence of FLAIR, several methods have used neighborhood information during FLAIR-based WMH detection (e.g., [6][5]). Usually the use of contextual information amounts to fully-isotropic smoothing– that is, WMHs are considered more likely at a given pixel if they occur at neighboring pixels, regardless of their absolute positions or the directions in which neighboring WMHs do or do not occur. We extend these prior contextual methods by allowing the associations between neighboring WMH detections to vary with pixel position and direction of neighbors. As suggested above, the spatially-and directionally-variable nature of associations between neighboring WMHs in our contextual model allows us to more accurately capture the neurobiological course of spreading WMHs over the course of brain aging.
2 Methods
Data
We tested our method on a diverse pool of 114 elderly individuals who received a full clinical workup and structural MR scans including T1-weighted, double-echo PD/T2 weighted, and FLAIR scans at their times of enrollment into the University of California, Davis Alzheimer’s Disease Center (ADC). Subjects were 70–90 years of age; the subject pool included individuals with normal cognition, mild cognitive impairment, and dementia.
Pre-processing
All scans were pre-processed through a standardized pipeline. T1, T2, PD, and FLAIR were rigidly coregistered using cross-correlation as a similarity measure and previously-presented optimization methods [14]. Nonbrain tissues were manually separated from the brain on all scans. A strongly-validated, semi-automated method was used to detect WMHs based solely on the FLAIR scans and human input [15]. The skull-stripped T1-weighted image was then nonlinearly aligned to a minimum deformation template (MDT) based on moving control points in a multi-scale grid and using cubic spline interpolation to move image pixels between the control points [16][17]. The warp is constrained such that no region is permitted to collapse entirely. The T1, T2, PD, FLAIR, and map of ground-truth FLAIR-based WMH pixels were then warped to the space of the MDT image using the nonlinear alignment.
MRF Approach
We take a Bayesian MRF approach to WMH detection. Let yi denote a vector of three image intensities– PD, T1, and T2– associated with image pixel i. Our goal is to determine a binary label xi for each image pixel i: xi = 1 denotes the presence of a WMH at pixel i and xi = 0 denote the absence of WMH there, i.e. to find a set of labels X = {x1, x2, ··· xk} corresponding to image intensity vectors Y = {y1, y2 ··· yk} that maximizes the posterior probability of the labels given the image data, P(X|Y). By Bayes’ theorem, P(X|Y) ∝ Π(X) * L(Y|X), where Π(X) is the prior probability of a particular set of labels X irrespective of imaging data and L(Y|X) is the likelihood of observing image intensities Y given that the underlying labels are X. The prior probability of a specific label xi depends on a spatial prior– the prior probability that WMHs occur at pixel i– as well as a contextual prior– the conditional probability of xi given the labels at neighbors of pixel i. The likelihood depends on the statistical distribution of the (PD, T1, T2) image intensities Y relative to the underlying labels X.
MRF Prior: Π(x)
The MRF label prior involves spatial and contextual prior models whose parameters are learned from training data. We write the MRF prior as a Gibbs field:
where Z, the partition function, is the sum of exp(−H(X)) over all possible labelings, and H(X) is an energy function that takes on lower values when the label field X is more probable a priori. This Gibbs prior is equivalent to an MRF prior under straightforward technical restrictions [18]. The energy function is a sum of terms that represent energies from the spatial and contextual priors: H(X) = Hs(X) + Hc(X). The spatial prior penalizes pixel i if it is labeled as WMH but WMHs are deemed unlikely there according to a prior probability, αi (see sec. 2), of a WMH occurring at pixel i:
The contextual prior Hc(X) penalizes a label xi when it differs from the labels of its neighbors. Recall that the MRF formulation utilizes a graph in which all pixels in the image are attached to some arbitrary set of their immediate spatial neighbors (see Sec. 3 for more information); there is one term in Hc for each clique in this graph. Let δ be one such clique of nodes, Δ be the set of all such cliques, and Xδ be the assignment of labels (i.e., WMH or non-WMH) that X provides to the nodes of δ. Then, Hc is given by:
This is a Potts model in which neighboring labels within a group δ incur a fixed penalty of βXδ [19]. Generally, these β parameters encourage neighboring pixels to have the same label, but in some locations of the brain, they may actually be encouraged to be different. These β parameters are calculated from the training data (Sec. 2).
MRF Likelihood
The likelihood of a given set of image intensity vectors, given the underlying labels, comes from a tissue mixture model with one lognormal distribution for WM and one for WMH:
where π0 and π1 are mixture coefficients for non-WMH and WMH respectively, with π0 + π1 = 1 and log(y) is the component-wise log of vector y. We estimate π1, by taking the proportion of pixels in YH that are inliers to the distribution found for the pixels in YL.
A lognormal mixture model was chosen because the distributions of 3D intensity vectors for WM and WMH empirically followed asymmetric, “comet-like” patterns (Fig. 3). A Gaussian mixture model was initially tried without success, which led to adoption of this choice. As we explain below, the μ and Σ parameters are estimated at run time by an unsupervised method that fits the two lognormal distributions to (PD, T1, T2) triples sampled from a large number of pixels.
Fig. 3.
The intensity distributions of WM and WMH intensities empirically follow “comet-like” patterns. Pictured: WMH intensities and the fit distribution for them in one ADC subject.
Combining the equations for Π and L and taking the log, we have
In the following sections, we describe the Training phase that determines the values of α and β, followed by the Inference phase where the best set of labels X is determined for an input image Y.
Training
In the training phase the parameters αi and βXδ governing Hs and Hc respectively are estimated from the ground-truth FLAIR-based WMH detection. The αi values are the empirical probabilities of WMHs at each pixel in labeled training examples, i.e. sets of (X, Y) pairs gathered from ground-truth FLAIR-based WMH detection. That is, αi is the proportion of training examples that have a WMH at pixel i.
The βXδ values are calculated using the same training data as the αi values using Iterative Proportional Fitting (IPF) [20]. For each δ and for each possible label assignment to Xδ, IPF iteratively computes an estimate for βXδ using the following fixed point equation:
where is the value of βXδ at the nth iteration of IPF, is the empirical marginal probability of δ = Xδ calculated as the proportion of the training data in which that label configuration occurred in δ, and denotes the model marginal probability of Xδ: the integral of Π(X) over all X in which the assignment Xδ occurs. The model marginal is calculated through Sum-Product BP (Sec. 2). R(x) is a sigmoid regularization function that prevents divergence of the fixed point iteration.
Run-time inference Fitting the MRF Likelihood Distributions
Run-time processing of a novel image set begins by using an MLESAC-based procedure to robustly estimate the means and covariances of the lognormal distributions associated with the WM and WMH classes [21]. Specifically, we generate k random samples of 10 pixels each from among those pixels that are most likely to contain WMHs a priori, i.e. from among the 5% of pixels i with the highest αi. Similarly we generate k 10-pixel samples from among the 5% of pixels with the lowest αi. From each high-αi sample we estimate a candidate μ1 and Σ1 from the corresponding yi, and similarly a candidate μ0 and Σ0 is estimated from each low-αi sample. Let YL and YH be the yi corresponding to the low-αi pixels and high-αi pixels respectively. Let XL contain a WM label for each low-αi pixel and XH contain a WMH label for each high-αi pixel. Each candidate (μ0, μ1, Σ0, Σ1) is assigned a numerical score that summarizes how well it fits the high-αi and low-αi yi, as well as how many of the yi ∈ {YL, YH} are outliers. The score is
where ν is a fixed penalty for outliers and δ(i) indicates whether yi is an outlier, i.e. it is 1 when f(yi; μxi, Σxi) > T and 0 when f(yi; μxi, Σxi) < T. In our experiments, we set k,T, and ν to 100, 10−6, and −0.1 respectively. The highest-scoring (μ0, μ1, Σ0, Σ1) are our parameter estimates for the distributions. Given the parameters needed to calculate the likelihood and contextual prior, we then use Belief Propagation to infer labels X that maximize log(P(X|Y)) [22].
MRF Inference
In Belief Propagation (BP), inference is performed by propagating local evidence (beliefs) as messages. Here, we use the Factor Graph formulation of BP in order to simplify notation. Factor Graphs represent undirected graphs in a bipartite fashion with two types of nodes: factor nodes and variable nodes. In our method, variable nodes directly correspond with pixel labels xi and factor nodes each correspond to a δ ∈ Δ. In each BP iteration, each variable node sends a message to each factor node that represents a clique it is a member of, and each factor node sends a message to the variable nodes of the clique member nodes. These messages are called variable messages xi → δ(x) and factor messages δ → xi(x) respectively. For Max-Product BP, the version used to compute a set of maximum a posteriori labels, the messages are:
where x is a candidate label for xi, Δi denotes the set of δ containing i, the observation term
and the compatibility term C(Xδ) = S(βXδ) where S(u) is a regularization function that smoothes across values of K to avoid numerical implementation issues introduced by extreme-valued weightings. When computing the β terms using Sum-Product BP as referenced in Sec. 2, the sums in the above terms are replaced with products, the max is replaced with a sum, and O(x) = 1. The model marginals are then computed by:
for each possible configuration of labels Xδ for the given δ to form . [23]
3 Experiments
In this section, we test the method’s performance under varying training/inference conditions, training set sizes, neighborhood connectivity, and training data sources.
Training and Inference Methods
In these tests, we use leave-one-out cross-validation to evaluate MRF-based WMH detection on the ADC data set; for each subject, we estimate the α and β parameters from the remainder of the subjects and use them to detect WMHs on the left-out subject. Agreement between the ground-truth WMH volumes and our computed volumes is evaluated using the intraclass correlation coefficient (ICC). We compute these ICC values for our method under each of these conditions: In the No MRF method, we do not use an MRF-based system and instead simply threshold the Posterior probabilities deduced from the Hs and HL terms alone. In the 6-MRF Without Training method, we use the empirical marginals for the βXδ terms instead of performing a proper training method. Finally, the 6-MRF With Training method uses our complete system with its designed proper IPF-based training. The results of these experiments are available in Table 1 and an example is given in Fig. 5.
Table 1.
Intraclass correlation coefficients (ICCs) between ground-truth WMH volume and WMH volume estimated by our method on the ADC data set with several variations.
| No MRF | 6-MRF Without Training | 6-MRF with Training | |
|---|---|---|---|
| ICC | 0.909 | 0.872 | 0.916 |
Fig. 5.
Comparison of WMH detection results for a selected brain region (see green box, left, and ground-truth). Detected WMHs are shown in yellow.
In our experiments, our MRF-based method outperforms the No-MRF and untrained MRF versions.
Contextual Prior Connectivity
One variable parameter of the method is the connectivity of its Contextual Prior information, ie. what size groupings of neighboring pixels influence each other in the MRF system. Higher values allow the system to model more complex spatial patterns. In 2D images, this choice is generally whether or not diagonal pixels are considered neighbors. In 3D, neighborhoods are described in values between 6, ie. a pixel’s 4 nearest neighbors within the plane and 2 nearest in the Z direction; and 26, ie. all of a pixel’s neighbors in a 3×3×3 pixel box around it. Results of testing our method under varying connectivities are presented in Table 2. For these tests, as in the previous, we used the ADC dataset and leave-one-out cross validation.
Table 2.
Intraclass correlation coefficients (ICCs) between ground-truth WMH volume and WMH volume estimated by our method using various degrees of spatial prior directional connectivity for the ADC data set.
| 6-MRF | 10-MRF | 18-MRF | 24-MRF | |
|---|---|---|---|---|
| ICC | 0.916 | 0.909 | 0.898 | 0.862 |
In these experiments, we found that our method performs best using 6-connected neighborhoods, the smallest logical size within 3D space.
Training Set Size
One important property of any training-based classification method is the amount of training data it requires to give good results on test data. To test this property, we trained upon three different randomly selected subsets of the ADC dataset for each size: 10, 20, 40, 60, 80, and 100 subjects. We then ran the method to classify the dataset using these subsets as training data (Fig. 4).
Fig. 4.

Plotted mean μ and μ ± σ of ICC values between ground-truth WMH volume and WMH volume estimated by our method using differently-sized random subsets of the training set in cross validation. Note that these values are absolute, not percents, and are out of a maximum of 114 (training with all data). *Each of the size-10 ICC measures is without 1–2 test subjects for whom IPF did not converge.
For this dataset our method performs better when using more training data up until about 60 images, after which there is little improvement.
Training and Test Sets from Different Populations and Scanners
To test our method’s performance using a completely different dataset from that upon which it was trained, we employed ground-truth WMH map data of 51 subjects from the Chicago Health and Aging Project (CHAP), a longitudinal Epidemiological study of individuals with risk factors for Alzheimer’s Disease [24]. These images were preprocessed in the same fashion as the ADC data (Sec. 2) and used for training. We then tested (using 6-connected neighborhoods and standard training/inference) our dataset of 114 ADC subjects using this training data and obtained results with an ICC of 0.841, demonstrating our method’s ability to perform reasonably when classifying images from a dataset from an entirely different MRI scanner, study type (epidemiological vs. clinic-based cohorts), and population.
4 Discussion and Future Work
Summary of Results
Our method performs robust WMH detection with no FLAIR when using at least 60 training images and standard MRF training/inference, including when the sources of training and testing data differ significantly. While our method performs strongly in these experiments, there exist several routes through which it can be improved in the future. Specifically, we discuss why the method performed worse using higher connectivities and possible new applications such as longitudinal WMH detection and multi-class segmentation.
Higher Degrees of Neighborhood Connectivity
Increased complexity can model more complex spatial dependencies among WMHs, but did not perform well in our experiments (Sec. 3). This drop in performance can be explained by a combination of factors. Higher connectivities subdivide the training data into a larger set of parameters, requiring a larger amount of training data. Additionally, it is possible that higher connectivities result in overfitting to the training data. Finally, BP, used here in both training and inference, is technically not guaranteed to perform well in loopy graphs but empirically does for 4-connected 2D latices. As the connectivity of our model increases, so does the proportion of loops in the graph, which may decrease performance. Future work should determine which combination of factors causes the decrease.
Other Applications
In addition to improving the method itself, future work will test and extend it for use in other applications. Simply by using appropriate training data, it could be applied to other diseases and modalities. It could also be extended to classify multiple tissue types at once to create an overall brain tissue segmentation system. Another possibility would be to detect WMHs on longitudinal series of MRIs. With this change our method could not only improve the results of each detection by the additional information (eg. encouraging pixels with WMH at time 1 to remain WMH at time 2) but also generate models of disease progression.
Fig. 6.
Comparison of WMH detection results for a selected brain region (see green box, left, and ground-truth). Detected WMHs are shown in yellow.
References
- 1.Taylor WD, Steffens DC, et al. White Matter Hyperintensity progression and late-life depression outcomes. Arch Gen Psychiatry. 2003 November;60(11):1090–1096. doi: 10.1001/archpsyc.60.11.1090. [DOI] [PubMed] [Google Scholar]
- 2.Au R, Massaro JM, et al. Association of White Matter Hyperintensity volume with decreased cognitive functioning: the Framingham Heart Study. Arch Neurol. doi: 10.1001/archneur.63.2.246. (In Press) [DOI] [PubMed] [Google Scholar]
- 3.Dufouil C, de Kersaint-Gilly A, et al. Longitudinal study of blood pressure and White Matter Hyperintensities: The EVA MRI cohort. Neurology. 2001 April;56(7) doi: 10.1212/wnl.56.7.921. [DOI] [PubMed] [Google Scholar]
- 4.van Straaten E, Fazekas F, et al. Impact of White Matter Hyperintensities scoring method on correlations with clinical data: The LADIS study. Stroke. 2006;37(3) doi: 10.1161/01.STR.0000202585.26325.74. [DOI] [PubMed] [Google Scholar]
- 5.Admiraal-Behloul F, van den Heuvel D, et al. Fully automatic segmentation of White Matter Hyperintensities in MR images of the elderly. Neuroimage. 2005 November;28(3):607–617. doi: 10.1016/j.neuroimage.2005.06.061. [DOI] [PubMed] [Google Scholar]
- 6.Wu M, Rosano C, et al. A fully automated method for quantifying and localizing White Matter Hyperintensities on MR images. Psychiatry Research: Neuroimag. 2006 December;148(2–3):133–142. doi: 10.1016/j.pscychresns.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longstreth W, Manolio TA, et al. Clinical correlates of White Matter findings on cranial magnetic resonance imaging of 3301 elderly people: The Cardiovascular Health Study. Stroke. 1996 August;27:1274–1282. doi: 10.1161/01.str.27.8.1274. [DOI] [PubMed] [Google Scholar]
- 8.Mueller S, Weiner M, et al. Ways toward an early diagnosis in Alzheimer’s disease: The Alzheimer’s disease neuroimaging initiative (ADNI) Alzheimers Dement. 2005 July;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshita M, Fletcher E, et al. Extent and distribution of White Matter Hyperintensities in normal aging, mci, and ad. Neurology. 2006 December;67(12):2192–8. doi: 10.1212/01.wnl.0000249119.95747.1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leemput KV, Maes F, Bello F, Vandermeulen D, Colchester ACF, Suetens P. Automated segmentation of MS lesions from multi-channel MR images. 1999:11–21. [Google Scholar]
- 11.Van Leemput K, Maes F, Vandermeulen D, Colchester A, Suetens P. Automated segmentation of Multiple Sclerosis lesions by model outlier detection. Medical Imaging, IEEE Transactions. 2001;20(8):677–688. doi: 10.1109/42.938237. [DOI] [PubMed] [Google Scholar]
- 12.Azhar Quddus PF, Basir O. Adaboost and support vector machines for White Matter Lesion segmentation in MR images. Engineering in Medicine and Biology Society. 2005 doi: 10.1109/IEMBS.2005.1616447. [DOI] [PubMed] [Google Scholar]
- 13.Maillard P, Delcroix N, et al. An automated procedure for the assessment of White Matter Hyperintensities by multispectral (T1, T2, PD) MRI and an evaluation of its between-centre reproducibility based on two large community databases. Neuroradiology. 2008 January;50(1):31–42. doi: 10.1007/s00234-007-0312-3. [DOI] [PubMed] [Google Scholar]
- 14.Maes F, Collignon A, et al. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 15.Yoshita M, Fletcher E, et al. Current concepts of analysis of cerebral White Matter Hyperintensities on Magnetic Resonance Imaging. Top Magn Reson Imaging. 2005 December;16(6):399–407. doi: 10.1097/01.rmr.0000245456.98029.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kochunov P, Lancaster J, et al. Regional spatial normalization: toward an optimal target. J Comp Assist Tomog. 2001 Sep–Oct;25(5):805–16. doi: 10.1097/00004728-200109000-00023. [DOI] [PubMed] [Google Scholar]
- 17.Otte M. Elastic registration of fMRI data using bezier-spline transformations. IEEE Trans Med Imaging. 2001;20:193–206. doi: 10.1109/42.918470. [DOI] [PubMed] [Google Scholar]
- 18.Hammersley J, Clifford P. Markov fields on finite graphs and lattices. 1971. [Google Scholar]
- 19.Besag J. On the statistical analysis of dirty pictures. Journal of the Royal Statistical Society Series B. 1986;48(3):259–302. [Google Scholar]
- 20.Jirousek R, Preucil S. On the effective implementation of the iterative proportional fitting procedure. Computational Statistics & Data Analysis. 1995 February;19(2):177–189. [Google Scholar]
- 21.Torr P, Zisserman A. MLESAC: A new robust estimator with application to estimating image geometry. CVIU. 2000 April;78(1):138–156. [Google Scholar]
- 22.Freeman WT, Pasztor EC, YOTC Learning low-level vision. International Journal of Computer Vision. 2000;40 [Google Scholar]
- 23.Bishop CM. Pattern Recognition and Machine Learning. Springer Science+ Business Media; 2006. [Google Scholar]
- 24.Bienias JL, Beckett LA, Bennett DA, Wilson RS, Evans DA. Design of the chicago health and aging project (CHAP) J Alzheimers Dis. 2003 Oct;5:349–355. doi: 10.3233/jad-2003-5501. [DOI] [PubMed] [Google Scholar]