Summary
This work is motivated by a quantitative Magnetic Resonance Imaging study of the relative change in tumor vascular permeability during the course of radiation therapy. The differences in tumor and healthy brain tissue physiology and pathology constitute a notable feature of the image data—spatial heterogeneity with respect to its contrast uptake profile (a surrogate for permeability) and radiation induced changes in this profile. To account for these spatial aspects of the data, we employ a Gaussian hidden Markov random field (MRF) model. The model incorporates a latent set of discrete labels from the MRF governed by a spatial regularization parameter. We estimate the MRF regularization parameter and treat the number of MRF states as a random variable and estimate it via a reversible jump Markov chain Monte Carlo algorithm. We conduct simulation studies to examine the performance of the model and compare it with a recently proposed method using the Expectation-Maximization (EM) algorithm. Simulation results show that the Bayesian algorithm performs as well, if not slightly better than the EM based algorithm. Results on real data suggest that the tumor “core” vascular permeability increases relative to healthy tissue three weeks after starting radiotherapy, which may be an opportune time to initiate chemotherapy and warrants further investigation.
Keywords: hidden Markov random fields, Mann Whitney U statistic, Quantitative Magnetic Resonance Imaging, reversible jump MCMC, Swendsen-Wang algorithm, Image Analysis, quantitative MRI
1. Introduction
As a non-invasive visualization tool, quantitative Magnetic Resonance Imaging (qMRI) enables researchers to assess pathological and physiological changes of in vivo tissue that cannot be evaluated with conventional anatomic MRI. Recent applications of qMRI include Cao et al. (2005), Moffat et al. (2005), and Hamstra et al. (2005). These applications share a common goal: to use qMRI to predict (local) therapeutic efficacy early during treatment with the aim of individualizing treatment regimens.
Despite recent advances in cancer treatments, the median survival time of patients with high-grade gliomas (a type of brain cancer) has not substantially increased from approximately one year after diagnosis. This is largely attributable to the tight endothelial junctions in the tumor vascular structure (also known as the blood-tumor barrier, BTB) that blocks large molecules, such as traditional chemotherapeutic agents, from entering these tumors. This mechanism also protects healthy tissue in the brain from the cytotoxicity of chemotherapeutic agents (also known as the blood-brain barrier, BBB). Although it is known that radiation can increase vascular permeability via damage to the vascular structure, standard of care sequential radiotherapy followed by chemotherapy has had limited success in treating brain cancer (Medical Research Council, 2001).
Researchers at the University of Michigan, School of Medicine recently conducted a study aimed at determining if the increase in tumor vascular permeability induced by radiation is transient and whether there is a time at which this increase is maximum (Cao et al. 2005). If this hypothesis holds, it would suggest that chemotherapy should begin during the course of radiation therapy instead of after its completion. This was the first study to use quantitative, high-resolution MRI to assess radiation induced increases in tumor vascular permeability among glioma patients (Cao et al. 2005). Eleven subjects with primary, high-grade gliomas participated in the study. T1-weighted MRI1 was performed on each patient, with and without contrast enhancement, prior to the beginning of radiotherapy. The same imaging protocol was subsequently performed after the first and third week of radiotherapy, and at one, three and six months after the completion of radiotherapy. The contrast agent used, Gadolinium diethylenetriaminepentaacetic acid (Gd-DTPA), has approximately the same molecule weight as many chemotherapeutic agents used to treat gliomas. Hence, the contrast uptake (quantified as the difference, on the log scale, of the contrast enhanced and pre-enhancement MRI images) was used as a surrogate of vascular permeability to these drugs (Cao et al. 2005). In this paper, we focus on the change in contrast uptake from baseline to week three (the difference between contrast uptake at week 3 and baseline, e.g. Figures 1a and 2a), which is of special interest to the researchers. For simplicity, we hereafter refer to the change in contrast uptake image as the observed image.
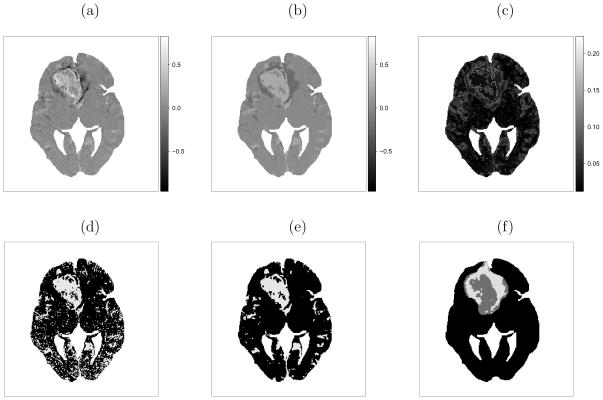
Figure 1.
Subject 1 results: (a) observed change in contrast uptake (black to white indicating larger increases in contrast uptake); (b) marginal posterior mean of change in contrast uptake μ̃zi and (c) standard deviation σ̃zi; thresholded image assuming (d) spatial independence (e) spatial dependence; (f) tumor “core/annulus”.
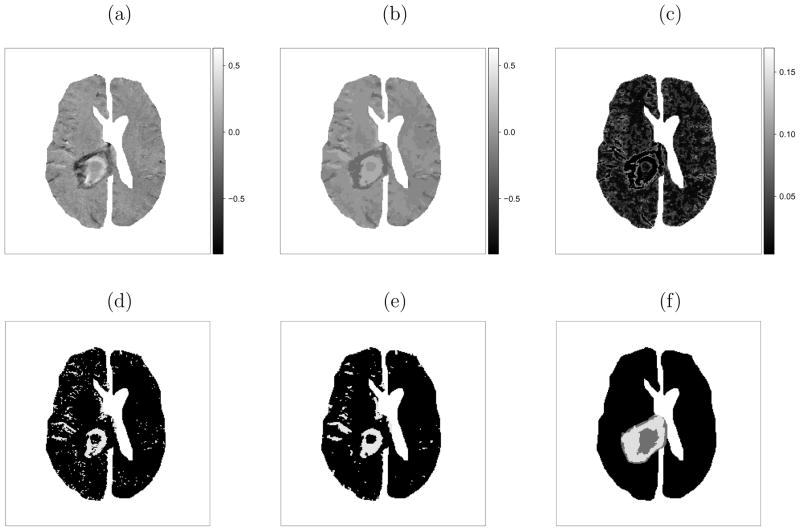
Figure 2.
Subject 2 results: (a) observed change in contrast uptake; (b) marginal posterior mean of change in contrast uptake μ̃zi and (c) standard deviation σ̃zi; thresholded image assuming (d) spatial independence and (e) spatial dependence; (f) tumor “core/annulus”.
It is known that solid tumors are physiologically different from surrounding healthy tissue, and that the contrast uptake, as well as its change, is heterogeneous. Many qMRI analyses ignore the inherent spatial correlation (at the pixel level) in the data, and treat the observed change in contrast uptake at each pixel as independent observations (e.g. Cao et al. 2005; Moffat et al. 2005; Hamstra et al. 2005), which results in biased variance estimates. To model the change in tumor/brain contrast uptake induced by radiation at the pixel-level, we use a model that accounts for the spatial correlation in the data and respects the distinct boundaries between tumor and healthy tissue. We introduce a layer of discrete hidden labels from a Markov random field (MRF, Besag 1974) which accounts for the spatial correlation in the data and avoids over-smoothing. A priori, the MRF encourages spatial continuity but allows for spatial heterogeneity. Like many similar models, we assume the observed data conditional on the hidden labels are independent and normally distributed (e.g. Lei and Udupa, 2002; Liang and Lauterber, 1999, Ch. 8).
The hidden MRF model we employ, known as the Potts model in statistical physics (Potts, 1952), has been applied in diverse areas such as disease mapping (Green and Richardson, 2002), agriculture (Dryden, Scarr, and Taylor, 2003), and landscape genetics (Francois, Ancelet, and Guillot, 2006; Guillot, Estoup, Mortier, and Cosson, 2005). All the above applications share the same feature as ours—spatial heterogeneity. However, our work builds on previous methods by integrating many of the individual ideas that have been previously published.
Firstly, we estimate the spatial regularization parameter, which is often fixed in the MRF models (e.g. Green and Richardson, 2002). Francois, Ancelet, and Guillot (2006) show that results can be sensitive to the choice of this parameter. However, estimation of the spatial regularization parameter requires a corresponding normalizing constant, which is computationally intractable. Some authors use pseudo-likelihood (Besag, 1974), which avoids the need to estimate the normalizing constant. However, this approach tends to overestimate the regularization parameter and over-smooth the data (Melas and Wilson, 2002). Instead, we use thermodynamic integration proposed by Ogata (1989) to estimate the ratio of these constants. This method was subsequently generalized by Gelman and Meng (1998). In 1997, Potamianos and Goutsias conducted a simulation study comparing several methods that approximate the normalizing constant in the Ising model (a two state Potts model) and recommend that Ogata’s thermodynamic integration method be used.
Secondly, we treat the number of states of the MRF as a model parameter and employ reversible jump Markov chain Monte Carlo (RJMCMC, Green, 1995; Richardson and Green, 1997) on a large scale dataset. The number of states in the MRF is traditionally assumed known (Khalkighi, Soltanian-Zadeh, and Lucas, 2002; Marroquin, Arce, and Botello, 2003). This is reasonable when there is substantial scientific justification, e.g. segmenting the brain into white matter, gray matter and cerebrospinal fluid. However, the segmentation labels lack a biologically meaningful interpretation in our application. Therefore, it is more appropriate to treat the number of labels as a model parameter. In this manuscript, we implement a complete Bayesian approach using RJMCMC and we propose a novel implementation of the reversible jump proposal inspired by the Swendsen-Wang algorithm (Swendsen and Wang, 1987). We focus on the marginal posterior distribution of change in contrast uptake, rather than prediction of the hidden labels. The labels are used to model spatial correlation and have no intrinsic scientific interpretation of interest.
The rest of this manuscript is organized as follows. In the next section we introduce notation and specify the model. We then discuss its implementation in Section 3. In Section 4, we present results from simulation studies and compare the results with the EM algorithm of Zhang et al. (2008). We also investigate the performance under violations to model assumptions and present results from the motivating example. We conclude by summarizing the strengths and weaknesses of our algorithm, and discuss future work.
2. Model Specification
Following convention, we use i = 1, 2, ···, N to index pixels (short for picture element). If pixel i and i′ share a common edge, we call them neighbors, denoted by i ~ i′. The set of neighbors of pixel i is denoted by ∂i = {i′: i′ ~ i}. Let y = (y1, y2 ···, yN)T denote the observed image (Figures 1a and 2a). In the proposed hidden MRF model, we introduce a latent discrete label Zi from a discrete state space 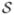 = {1, 2, ···, M} for each pixel i. The collection of latent labels, z = (Z1 = z1, Z2 = z2, ···, ZN = zN)T, is called a configuration. The set of pixels that share the same label is referred to as a component, and may consist of disjoint clusters of pixels.
= {1, 2, ···, M} for each pixel i. The collection of latent labels, z = (Z1 = z1, Z2 = z2, ···, ZN = zN)T, is called a configuration. The set of pixels that share the same label is referred to as a component, and may consist of disjoint clusters of pixels.
2.1 Distribution of the Data
We assume the observed data are conditionally independent given the hidden labels,
The hidden labels follow a Gibbs distribution with joint probability mass function
| (1) |
where I[·] is the indicator function (I[zi = zj] = 1 if zi = zj and 0 otherwise). The regularization parameter, β, controls the spatial smoothness of the MRF. When β = 0 the pixels are independent. When β is large, the correlation between pixels is strong (neighboring pixels have high tendency to assume the same label) and the configuration tends to be smooth. In principle, the MRF encourages neighboring pixels to share the same label. We note that the spatial correlation decreases as the distance between pixels increases. The normalizing constant in Equation (1), g(β, M) = Σz∈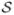 exp{βΣi~i′ I[zi = zi′]}, has MN outer summands, and therefore is not computationally tractable. Some authors (e.g., Chalmond, 1989; Won and Derin, 1992) avoid computing the normalizing constant by using the pseudo-likelihood,
exp{βΣi~i′ I[zi = zi′]}, has MN outer summands, and therefore is not computationally tractable. Some authors (e.g., Chalmond, 1989; Won and Derin, 1992) avoid computing the normalizing constant by using the pseudo-likelihood,
as an approximation to Pr(Z = z |β, M). However, Barker and Rayner (1997) show that, under certain circumstances, this pseudo-likelihood may lead to an improper posterior distribution. Others completely avoid estimating it by assuming fixed values for both β and M, the results of which may depend on the choice of β (Francois, Ancelet, and Guillot, 2006). We use thermodynamic integration2 (see Appendix 1 for details) as proposed by Ogata (1989), brought to the attention of the statistical community by Gelman and Meng (1998), and subsequently used by Green and Richardson (2002) and Higdon (1998), among others.
It follows that the joint distribution of the data and hidden labels is
Although we assume conditional independence of the observed data given the hidden labels, one can show that the observed data are marginally correlated because of the correlation introduced by the MRF labels.
2.2 Prior Distributions
A priori, regularization parameter, β, follows a uniform distribution on the interval [0, βmax] with βmax = 3. The number of components, M, is given a uniform distribution on the set of integers {Mmin, …, Mmax}, where Mmin = 1 and Mmax = 20. The component means, μk are independently and identically distributed (i.i.d.) Unif[ymin, ymax], where ymin and ymax are the minimum and maximum of the observed data, respectively. The inverse of the component variances are, a priori, i.i.d. gamma random variates: with ασ = 2.1 and βσ ~ Gamma(2.5, 5). The prior mode of βσ is 0.3, which favors smaller variances. We place a hyperprior distribution on βσ to reduce the dependence of the posterior of on its prior. In Section 4.1.1, we investigate other values of ασ in a sensitivity analysis.
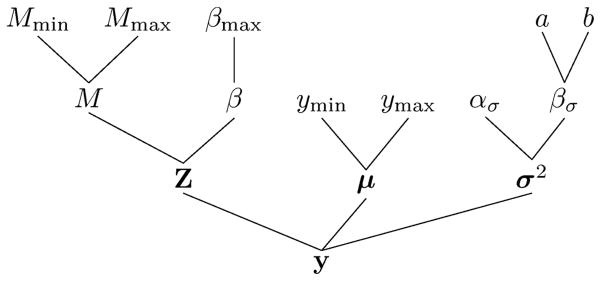
We illustrate the model structure in a Directed Acyclic Graph (DAG) in Figure 3 and write in vector form μ = (μ1, ···, μM), , and θ = (μ, σ2, βσ, β, M). Assuming (conditionally) independent prior distributions, the joint prior distribution is π(θ) = π(β)π(M)π(μ)π(σ2|βσ)π(βσ).
Figure 3.
Directed Acyclic Graph (DAG) of the proposed model
2.3 Joint Distribution and Conditional Posterior Distributions
By Bayes’ Theorem, the joint posterior distribution is proportional to
| (2) |
Note that the above joint distribution is invariant to permutations of the component labels conditional on M, and therefore the component means and variances are not identifiable. In many fixed-dimension problems with segmentation as the main goal, a constraint such as μ1 < μ2 < ··· < μk is imposed to resolve the identifiability issue. However, when M is treated as a parameter, the components may still be unidentifiable even with the ordering constraint. We discuss this further in Section 3.3.2.
The conditional posterior distributions of the model parameters are
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
for k = 1, 2, …, M, where Dk = {i: zi = k} denotes the set of pixel indices in component k, Nk is the number of pixels in component k, and ‘·’ denotes all other parameters and data.
2.4 Marginal Posterior Distribution of Change in Contrast Uptake
Our goal is to establish the underlying change in contrast uptake, μzi, and characterize it with its posterior mean, , and variance, . Both of these quantities are estimated via Markov chain Monte Carlo (MCMC). Let t denote the tth draw (1 ≤ t ≤ T). Then and , where when . Zhang et al. (2008) propose a similar measure: the “expected change in contrast uptake” defined by . However, this estimate depends only on the maximum likelihood estimate (MLE) of θ, and neglects the uncertainty in its estimation.
3. Algorithm Details
We initialize the Monte Carlo chain with a relatively large number of components, M = 15. The initial values of μ are evenly spaced on the range of observed data ([ymin, ymax]), while the components of σ are set to the interval width. The hidden labels are drawn independently from neighboring pixels, i.e. β = 0.
The parameters μ, σ2, and βσ can be updated via standard Gibbs sampling steps due to conjugacy. The spatial regularization parameter β requires a Metropolis-Hastings step. We update the hidden labels via the Swendsen-Wang algorithm (Swendsen and Wang, 1987), an efficient sampler for the Potts model. The most difficult part of the sampler is updating the number of components. Since the dimension of the parameter space is determined by the number of components, we employ RJMCMC to update M.
3.1 Updating the Spatial Regularization Parameter
When updating the spatial regularization parameter at the tth iteration, β(t), we use a Gaussian proposal distribution centered at β(t−1), , where the variance of the proposal, , is adaptively tuned during burn-in to give an acceptance rate of approximately 35%. Since we use a symmetric proposal distribution, the acceptance probability only depends on the ratio of the conditional posterior distributions in Equation (6). The ratios of normalizing constants are estimated before-hand on a grid of values for β and M (β = 0, 0.05, ···, 3.00 and M = 1, 2, ···, 20, see Appendix 1 for details), interpolating between values when necessary.
3.2 Updating the Hidden Labels via the Swendsen-Wang Algorithm
The kernel of the conditional posterior distribution of Z, Equation (7), consists of two parts—an interaction term (between neighboring pixels) and a likelihood term (sometimes referred to as the external field, Higdon, 1998). A pixel-wise updating scheme may not mix well in the presence of the interaction term (Higdon, 1998). Hence, we use the Swendsen-Wang algorithm, an efficient sampling scheme designed to speed up the mixing of the Potts model (see Appendix 2 for details).
3.3 Trans-dimensional moves
Applications of RJMCMC in Potts models include Green and Richardson (2002), who analyze disease mapping data using a trans-dimensional proposal that emulates the spatial dependence structure of the Potts model; and Dryden, Scarr, and Taylor (2003), who apply a similar scheme to analyze weed and crop images. However, we found that these proposals do not work well for our application: with tens of thousands of pixels per image, the acceptance rate using these proposals is practically zero.
Therefore, we propose a novel implementation of the trans-dimensional move inspired by the Swendsen-Wang algorithm. We first randomly choose between a split and a merge proposal with
3.3.1 Split Proposal
If a split proposal is chosen, we randomly pick a component k (1 ≤ k ≤ M) to split, i.e. . The dimension of the parameter space increases by two accordingly. To match the increase in dimension, we introduce two independent random variables, u1, u2 ~ Beta(2, 2), and define a bijective transformation that matches the first two moments,
We denote the proposed set of parameters with a superscript*: and . When there exists a component h (1 ≤ h ≤ M + 1) satisfying , the proposal is not likely to be accepted. We therefore reject such proposals. The common hidden labels define contiguous equivalence classes on the lattice. We base the proposed labeling on the equivalence classes. In contrast to the Swendsen-Wang update, we do not further break contiguous same-labeled regions with stochastic “bonds” (Appendix 2). Rather, we partition the configuration deterministically. This is equivalent to a Swendsen-Wang update with an infinite regularization parameter (β = ∞) where all same-labeled neighbors are “bonded”. Suppose there are L such equivalence classes ξ1, ξ2, ···, ξL with current labels E1, E2, ···, EL (by definition, L ≥ M, and when i ∈ ξl, zi = El). We randomly draw a label for these equivalence classes with probability proportional to the likelihood. Specifically, the probability that class ξl assumes a new label k is
| (8) |
where the are normalized so that for all l. Let denote the proposed label for class ξl from Equation (8), i.e. . The allocation probability in the current state conditional on the equivalence classes is .
We also compute the allocation probability for the proposed configuration given the same equivalence classes, , where , for k = 1, 2, ···, M.
It follows that the acceptance probability for the proposed split move is min(1, A) with
| (9) |
Here b(·) is the density function of the Beta(2, 2) distribution. The first line is the product of the posterior ratio and the proposal ratio, while the second line is the Jacobian of the bijective transformation.
3.3.2 Merge Proposal
We first randomly pick a pair of components k1 and k2 with adjacent means, . The parameters for the new component are determined as the inverse of the split proposal, i.e.
We denote the proposed set of parameters by and .
We propose a new configuration z* with M − 1 components based on the same-labeled contiguous equivalence classes. The new label of class ξl is drawn from , for k = 1, 2, ···, M − 1. The associated allocation probability is . Similarly, the allocation probability for the new configuration is , where , for k = 1, 2, ···, M.
Hence, the acceptance probability of the merge proposal is min(1, B), where
We note that identifiability issues may still arise under the constraints μ1 < μ2 < ··· < μM, when M is not fixed. For example, suppose there are three components labeled 1, 2, and 3, after the merge of components 1 and 2. The component with original label 3 is automatically relabeled as 2 under the constraints. It is clear that the component with new label 2 (originally labeled 3) is not the component with original label 2. Nevertheless, we are mainly interested in the “true” change in contrast uptake and are not interested in the labels. Thus, we marginalize over M and the labels (Section 2.4), and thus avoid the issue of non-identifiability.
4. Results
To evaluate the performance of the proposed method, we start with simulation studies under the model assumption of conditionally independent noise, and then investigate model performance under violations of this assumption. We define the image Mean Squared Error (MSE) of pixel intensities by , where is the simulated true value. We generate multiple datasets and compute the average MSE across the datasets, i.e. , where is the image MSE of the lth simulated dataset (1 ≤ l ≤ L). The results are compared with an EM algorithm (Zhang et al. 2008) as well as a fixed dimension MCMC. We also present results from the motivating example.
4.1 Simulation Study with Conditionally Independent Noise
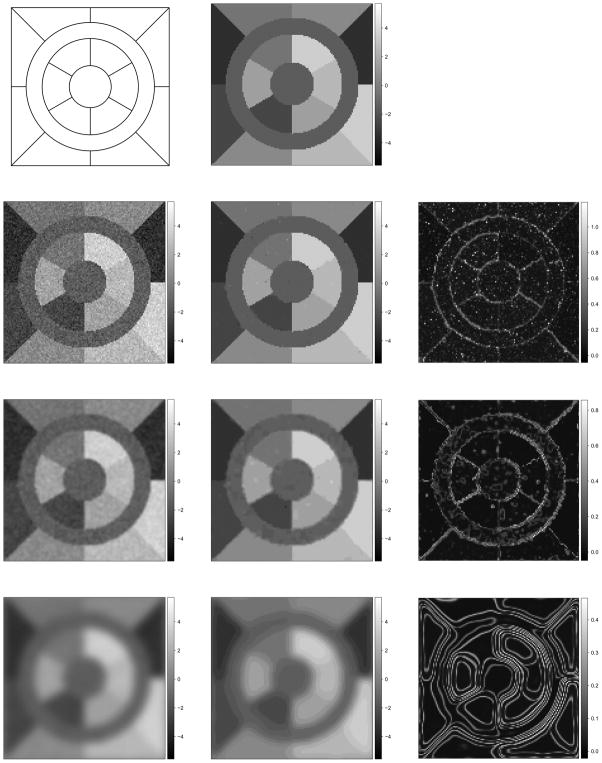
We divide a 128 × 128 lattice into 16 regions of various shapes (Figure 4). We assume the observed pixel values within the same region follow the same distribution, drawn randomly from eight candidate Gaussian distributions with means μ = (−3.5, −2.5, −1.5, −0.5, 0.5, 1.5, 2.5, 3.5)T and standard deviation 0.625. We generate 100 sets of true and observed images to compute the MSEs (L = 100, see Figure 4 for one example).
Figure 4.
One set of simulations: the skeleton (top left) the “true” scene ( , top middle), the observed images (y, left), the posterior mean (η̂i, middle) and standard deviation (ψ̂i, right) of pixel mean intensity when σ = 0, 0.85, and 3.40 from the second to fourth row.
Zhang et al. (2008) report biased parameter estimates when the pixels are treated as independent observations, which is equivalent to a Gaussian mixture model (GMM) with equal component weights. We find similar results with M = 8 and β = 0: the average MSEs are (mean ± standard deviation).
The EM algorithm (Zhang et al. 2008) yields smaller MSEs than the GMM by modeling the spatial correlation, i.e. (Table 1). However, our results also show that the number of components has a sizable impact on the average MSE. When the number of components is under-specified, e.g. M = 6, the performance of the EM algorithm is not satisfactory with . On the other hand, when M is over-specified, e.g. M = 12, the model fit improves with extra degrees of freedom, i.e. . These results present a potential problem with the EM algorithm—if both small MSE and correct estimation of M are important, the EM algorithm does not yield a “best” model under both criteria.
Table 1.
of the proposed method vs. fixed dimension MCMC and the EM algorithm.
| 0 | 0.42 | 0.85 | 1.70 | 3.40 | ||
|---|---|---|---|---|---|---|
| RJ | 0.012 ± 0.001 | 0.014 ± 0.001 | 0.157 ± 0.040 | 0.338 ± 0.084 | 0.659 ± 0.147 | |
| MCMC | 6 | 0.103 ± 0.005 | 0.105 ± 0.004 | 0.242 ± 0.049 | 0.430 ± 0.096 | 0.715 ± 0.158 |
| 8 | 0.020 ± 0.017 | 0.016 ± 0.010 | 0.145 ± 0.038 | 0.333 ± 0.070 | 0.666 ± 0.145 | |
| 12 | 0.013 ± 0.002 | 0.015 ± 0.001 | 0.147 ± 0.035 | 0.357 ± 0.083 | 0.656 ± 0.148 | |
| EM | 6 | 0.107 ± 0.010 | 0.111 ± 0.009 | 0.256 ± 0.037 | 0.413 ± 0.085 | 0.713 ± 0.151 |
| 8 | 0.032 ± 0.024 | 0.021 ± 0.016 | 0.142 ± 0.030 | 0.334 ± 0.070 | 0.666 ± 0.145 | |
| 12 | 0.014 ± 0.002 | 0.015 ± 0.001 | 0.151 ± 0.038 | 0.362 ± 0.086 | 0.657 ± 0.147 | |
In 99 out of the 100 simulated datasets, RJMCMC produces smaller MSEs than both EM and fixed dimension MCMC. The inter-quartile range of the decrease in image MSE is [0.002, 0.003] compared to EM and [0.001, 0.002] compared to the fixed dimension MCMC.
As an alternative, parallel to the EM algorithm, we also implement a fixed dimension MCMC algorithm (see results in Table 1). As with the EM algorithm, the choice of M impacts the average MSE. When M = 8, the average MSE is smaller than that of the EM algorithm, i.e. . The same holds true when M = 6 with . When the data are over-fitted with M = 12, the average MSE decreases to . In summary, the MCMC algorithm has smaller average MSE than the EM under all three choices for M. Furthermore, the MCMC algorithm appears to be less variable as evidenced by the smaller standard deviations of the .
We then run our proposed RJMCMC algorithm for 50000 iterations and discard the first half as burn-ins. Figure 4 displays the posterior mean (η̂i) and standard deviation (ψ̂i) for one simulated dataset. The posterior mode is M = 8 for all L datasets and the posterior probability that M = 8 ranges from 50% to 88%. The average MSE, (Table 1), is much smaller than both EM and fixed dimension MCMC under the true M = 8 with less variability. Although the decrease in average MSE compared to the other two algorithms when M = 12 is marginal, a detailed examination of all 100 simulations reveal that the image MSE of our RJMCMC algorithm is smaller than the other two algorithms, conditional on M = 12, in 99 out of the 100 simulated datasets. The inter-quartile range of the decrease in image MSE is [0.002, 0.003] compared to EM and [0.001, 0.002] compared to the fixed dimension MCMC.
We reason that the Bayesian algorithms explore the posterior distribution more efficiently and are less prone to getting stuck in local modes, whereas the EM algorithm only guarantees convergence to a local mode. Furthermore, we reason that the additional flexibility of changing between models (M) afforded by the RJMCMC algorithm, can further aid in exploring the posterior. We argue that one significant benefit of the proposed algorithm is that it produces satisfactory results (and in many cases better results) without the risk of over- or under-specifying the number of components.
4.1.1 Sensitivity Analysis
In Section 2.2, we have chosen diffused prior distributions that cover reasonable ranges for most parameters. We now investigate alternative values for ασ: ασ = 0.5 and ασ = 5.0. The impact on the MSEs is negligible, i.e. the average MSE is 0.012 ± 0.001 when ασ = 0.5 and 0.012 ± 0.003 when ασ = 5.0 (compared to 0.012 ± 0.001 when ασ = 2.1).
We also examine our proposed algorithm under various signal to noise ratios (SNR). The simulation set in the above section has signal level Δμ = μk+1 − μk = 1 and noise level σ = 0.625. We increase the noise level to σ = 0.65, 0.7, 0.75, and 0.8, while maintaining the same signal level. As the SNR decreases, the average MSE increases from 0.014 ± 0.005, 0.023 ± 0.014, 0.059 ± 0.024 to 0.083 ± 0.027.
4.2 Simulation Studies with Correlated Noise
We evaluate the robustness of the method to violations of the conditional independence assumption by applying Gaussian smoothing kernels on the simulated datasets. We follow the same Gaussian kernel parameters as Zhang et al. (2008), i.e. σ = 0.42, 0.85, 1.70, and 3.40. Larger values of σ indicate smoother images.
The “edge preservation” of the proposed algorithm is more evident with lighter smoothing (the bottom two rows of Figure 4). The average MSEs for all degrees of smoothing investigated are displayed in Table 1. Within each algorithm, the average MSE increases as smoothing gets heavier. When smoothing is light (σ = 0.42), the fixed dimension MCMC achieves smaller MSEs compared to the EM algorithm using the same value for M. Our algorithm results in the smallest average MSE regardless of the choice of M in the other two algorithms. However, when the smoothing is heavy, e.g. σ ≥ 0.85, none of the algorithms has satisfactory performance. Thus we suggest that no smoothing of the data be performed prior to running any of these algorithms.
4.3 Application
In the motivating study, eleven subjects received standard of care radiotherapy and MRI images were obtained before, during, and after treatment. All images were registered to anatomical Computed Tomography (CT) images obtained for treatment planning purposes. Due to space limitations, we only display the images on the same two subjects as Zhang et al. (2008).
In the original analysis, Cao et al. (2005) divided the tumor into two regions, one with relatively high pre-treatment contrast uptake and the other with relatively low initial contrast uptake. This more-or-less divides the tumor into a “core” (low initial contrast uptake) surrounded by an “annulus” (with high initial contrast uptake). Tumor cores are typically hypoxic (with low oxygen content) due to a lack of blood supply, which is known to have a protective effect against damage due to both radiation and chemotherapy and may be a source of tumor re-growth. Hence, the focus was on demonstrating that radiation therapy has a transient effect on the tumor “core” with respect to increasing the contrast uptake. The time when this increase is greatest may suggest an optimal time for initiating chemotherapy, allowing for more effective delivery of chemotherapeutic agents. Following Zhang et al. (2008), we use the 95th percentile of healthy tissue contrast uptake at baseline as the threshold to divide the tumor into core and annulus.
We then run the proposed algorithm for 50000 iterations after 25000 burn-in iterations. The acceptance rate of the trans-dimensional proposals is 12.5% for subject 1. The posterior mean of change in contrast uptake and associated standard deviation (Figure 1b and 1c) delineates the heterogeneous response of the tumor. The results from subject 2 are similar (Figure 2b and 2c), as are the results from all subjects (not shown).
4.3.1 Interpretation of the Results
As discussed in the introduction, large increases in contrast uptake may suggest more effective delivery of chemotherapy drugs. Hence, an optimal time to deliver chemotherapy may exist when the contrast uptake increase in the tumor is large relative to healthy tissue. One way to quantify this is to define a threshold and compare the proportions of healthy and diseased tissue that exceed this threshold. Although, there is no established threshold, we apply an illustrative threshold, 0.06, used by Zhang et al. (2008). For subject 1, 8.2% of the normal tissue exceeds the threshold, while 59.5% of the tumor “core” has an increase in contrast uptake above the threshold, compared to 15.6% in the tumor “annulus” (Figure 1e). Similarly, for subject 2, 4.0% of healthy tissue exceeds the threshold. While 54.3% of the tumor “core” has a change in contrast uptake that exceeds the threshold, compared to 12.7% in the “annulus” (Figure 2e). The percentages are similar to those reported by Zhang et al. (2008).
Since the choice of the above threshold is arbitrary and is not supported by biological justifications, we propose to use the Mann Whitney U statistic to summarize the differential change in contrast uptake of the tumor and healthy tissue. It can be interpreted as the probability that a randomly selected pixel in the tumor has a larger mean change in contrast uptake than a randomly selected pixel in healthy tissue. A larger U statistic indicates a larger increase in contrast uptake of the tumor relative to healthy tissue which is desirable (i.e. more contrast is entering the tumor). There is a connection between thresholds and the U statistic. The U statistic is equivalent to the empirical area under the receiver operating characteristics curve, or AUC (Section 4.3.4 of Pepe, 2003).
We compute the U statistic at each iteration for the tumor “core” versus healthy tissue and the tumor “annulus” versus healthy tissue. The 95% posterior credible interval of the U statistic between the tumor “core” and healthy tissue is (0.74, 0.76) for subject 1, which suggests the “core” region has a substantial increase, on average, in contrast uptake compared to healthy tissue. The tumor “core” of subject 2 is also well separated from healthy tissue with 95% posterior credible interval (0.81, 0.82). The U statistic between the tumor “annulus” and healthy tissue is under 0.5 for both subjects: (0.23, 0.25) for subject 1 and (0.25, 0.26) for subject 2, suggesting the “annulus” on average has decreased contrast uptake compared to healthy tissue. The U statistic for all eleven subjects are listed in Table 2. Most subjects demonstrate significant increase in contrast uptake in the tumor “core”. We also investigate the sensitivity of the results to alternative cutoff of tumor core vs. annulus, i.e. using 90% and 97.5% percentile of the baseline healthy tissue contrast uptake instead of 95% percentile. The results are listed in Table 2.
Table 2.
Posterior 95% credible interval of Mann Whitney U statistic between the tumor “core/annulus” and healthy tissue for all subjects
| Subject ID | “Core” | “Annulus” | ||||
|---|---|---|---|---|---|---|
| 90th | 95th | 97.5th | 90th | 95th | 97.5th | |
| 1 | (0.75, 0.77) | (0.74, 0.76) | (0.71, 0.73) | (0.26, 0.28) | (0.23, 0.25) | (0.20, 0.22) |
| 2 | (0.83, 0.84) | (0.81, 0.82) | (0.81, 0.82) | (0.28, 0.29) | (0.25, 0.26) | (0.23, 0.24) |
| 3 | (0.42, 0.44) | (0.40, 0.42) | (0.36, 0.38) | (0.08, 0.09) | (0.05, 0.07) | (0.04, 0.05) |
| 4 | (0.55, 0.57) | (0.53, 0.55) | (0.52, 0.53) | (0.18, 0.20) | (0.15, 0.16) | (0.13, 0.15) |
| 5 | (0.49, 0.54) | (0.46, 0.52) | (0.42, 0.49) | (0.25, 0.31) | (0.24, 0.30) | (0.24, 0.29) |
| 6 | (0.91, 0.92) | (0.91, 0.91) | (0.89, 0.90) | (0.57, 0.68) | (0.55, 0.56) | (0.53, 0.54) |
| 7 | (0.92, 0.96) | (0.90, 0.94) | (0.89, 0.93) | (0.31, 0.33) | (0.29, 0.31) | (0.29, 0.31) |
| 8 | (0.23, 0.25) | (0.30, 0.31) | (0.35, 0.36) | (0.92, 0.94) | (0.95, 0.97) | (0.96, 0.97) |
| 9 | (0.62, 0.65) | (0.59, 0.62) | (0.58, 0.60) | (0.21, 0.35) | (0.21, 0.39) | (0.21, 0.42) |
| 10 | (0.87, 0.89) | (0.86, 0.88) | (0.85, 0.87) | (0.59, 0.60) | (0.59, 0.60) | (0.59, 0.60) |
| 11 | (0.69, 0.72) | (0.68, 0.71) | (0.66, 0.68) | (0.25, 0.27) | (0.23, 0.25) | (0.20, 0.23) |
5. Conclusion
In this article, we implement a statistical imaging model that respects the key feature of spatial heterogeneity in the qMRI data. Compared with previous work, we integrate many ideas that have been previously discussed individually. First, we estimate the normalizing constant via thermal integration, instead of using the pseudo-likelihood. Second, we estimate the spatial regularization parameter from the data, rather than holding it fixed. Third, we acknowledge that there is no clear substantive knowledge regarding the number of components, given the lack of biological interpretation of the hidden labels. We therefore marginalize over both M and the labels, and focus on the marginal posterior distribution of the pixel change in contrast uptake, which is of primary scientific interest. Furthermore, the split scheme proposed by Richardson and Green (1997) is inadequate for the scale of data, we therefore propose a non-trivial implementation inspired by the Swendsen-Wang algorithm. Finally, we propose using the U statistic to summarize the differential change in contrast uptake of the tumor “core/annulus” versus healthy tissue.
We find the performance of our RJMCMC algorithm improves on the EM algorithm and the fixed dimension MCMC algorithm regardless of the number of components specified in the latter two algorithms. Furthermore, our algorithm is preferable when there is no clear basis for the choice of M, since the loss associated with a miss-specified M can be severe with both the EM and the fixed dimension MCMC algorithms.
There are alternatives to the proposed algorithm. For example, birth-and-death MCMC (BDMCMC, Stephens 2000) is an alternative trans-dimensional MCMC algorithm. Another alternative could be a spatial infinite mixture model (Guillot, Estoup, Mortier, and Cosson, 2005, Francois, Ancelet, and Taylor, 2006), which extends infinite mixture models using the Dirichlet process prior to the spatial setting. However, implementation of the these alternatives may be challenging giving the large volume of data.
Finally, we would like to address that it is a complicated issue to determine the optimal timing of administering chemotherapy with respect to radiation. The image model discussed in this manuscript provides crucial information, but its role is not to be over-stated. It is more appropriately considered as a piece of a large puzzle. Future research in the image model itself as well as the underlying biological science are much needed. One future direction of our research is in extending the image model by incorporating the repeated images that captures the longitudinal profile of tumor/brain contrast uptake.
Acknowledgments
This work was partially funded by NIH grants PO1 CA087684-5 and PO1 CA59827-11A1.
Appendix
1. Estimating the Ratio of Normalizing Constant
The outline of the estimating procedure follows the general method found in Ogata (1989) and Gelman and Meng (1998) with appropriate notational changes. For a given M (number of components), we wish to estimate the log ratio of normalizing constants λM(a, b) = ln[g(b, M)/g(a, M)] for b > a ≥ 0. Now
Therefore,
| (10) |
In order to estimate λM(a, b), we have two integrals to evaluate. The inner integral, or expectation, is estimated via MCMC. We use the Swendsen-Wang algorithm under the assumption that there is no likelihood term. The use of the Swendsen-Wang algorithm is key to accurately estimate this ratio as the Swendsen-Wang algorithm moves quickly through the state space in a way that cannot be done with single site updates. The outer integral in Equation (10) is evaluated numerically using the trapezoidal rule. The ratio is calculated for M = 2, …, 20 and β = 0, 0.01, …, 3. For values of a and b not on the grid on which the (inner) expectation is evaluated, we linearly interpolate.
For the RJMCMC proposal, the ratio ln[g(β, M)/g(β, M + 1)] is required. This can be estimated using the fact that ln[g(0, M)] = ln Σz∈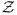 1 = N ln(M), where N is the number of pixels, and the following identity:
1 = N ln(M), where N is the number of pixels, and the following identity:
2. Details of the Swendsen-Wang Algorithm
The Swendsen-Wang algorithm stochastically partitions the configuration into same-labeled contiguous regions such that the label for these regions can be updated independently. This clever idea is implemented in the following three steps.
We first generate independently random variables, uii′ ~ Uniform(0, exp{βI[zi = zi′]}), for each pair of neighbors i ~ i′. Clearly, the joint distribution of all auxiliary variables u = {uii′}i~i′ is f(u) = Πi~i′ exp {−βI[zi = zi′]} I[0 ≤ uii′ ≤ exp{βI[zi = zi′]}].
-
By Bayes theorem, the posterior distribution of the labels conditional on the auxiliary variables is
(11) The second term in Equation (11) defines the range over which the posterior distribution is non-zero. More concretely, when 1 ≤ uii′ ≤ exp{βI[zi = zi′]} holds, it builds a virtual stochastic “bond” between i and i′ that requires both pixels to assume the same label. That is if pixel i and i′ assume the same label in the current configuration, they are “bonded” to assume a common label (could be different from the current one) with probability 1 − e−β. The bonds define an equivalence relation, and partition a configuration into contiguous regions of bonded pixels. We denote these equivalence classes by C1, C2, …, CJ. According to the definition, a component consists of one or more such equivalence classes.
- According to Equation (11), the labels of the equivalence classes can be updated independently. The new label of class j satisfies
Footnotes
A T1-weighted MRI uses short repetition time and echo time in which paramagnetic agents (e.g. Gadolinium) appear brighter in the image. Note that the qMRI images derived from T1-weighted MRI reflect the change in the intensities of the MRI images, and therefore reflect the change in the density of the paramagnetic agents, e.g. contrast uptake of Gd-DTPA.
Technically, we only need and compute the ratio of normalizing constants in this manuscript, as detailed in Appendix 1. However, the normalizing constant itself can be readily approximated from the ratios (Gelmen and Meng, 1998).
References
- Medical Research Council. Randomized Trial of Procarbazine, Lomustine, and Vincristine in the Adjuvant Treatment of High-Grade Astrocytoma: A Medical Research Council Trial. J Clin Oncol. 2001;19(2):509–518. doi: 10.1200/JCO.2001.19.2.509. [DOI] [PubMed] [Google Scholar]
- Barker SA, Rayner PJW. Unsupervised Image Segmentation Using Markov Random Field Models. 1997. [Google Scholar]
- Besag J. Spatial Interaction and the Statistical Analysis of Lattice Systems (with discussions) Journal of the Royal Statistical Society Series B. 1974;36:192–236. [Google Scholar]
- Cao Y, Tsien CI, Shen Z, Tatro DS, Ten Haken R, Kessler ML, Chenevert TL, Lawrence TS. Use of Magnetic Resonance Imaging to Assess Blood-Brain/Blood-Glioma Barrier Opening During Conformal Radiotherapy. Journal of Clinical Oncology. 2005;23(18):4127–4136. doi: 10.1200/JCO.2005.07.144. [DOI] [PubMed] [Google Scholar]
- Cappé O, Robert CP, Rydén T. Reversible Jump, Birth-and-Death and More General Continuous Time Markov Chain Monte Carlo Samplers. Journal of the Royal Statistical Society B. 2003;65:679–700. [Google Scholar]
- Chalmond B. An Iterative Gibbsian Technique for Reconstruction of Mary Images. Pattern Recognition. 1989;22:747–761. [Google Scholar]
- Dryden IL, Scarr MR, Taylor CC. Bayesian Texture Segmentation of Weed and Crop Images Using Reversible Jump Markov Chain Monte Carlo Methods. Journal of Royal Statistical Society, Series C. 2003;52:31–50. [Google Scholar]
- Francois O, Ancelet S, Guillot G. Bayesian Clustering Using Hidden Markov Random Fields in Spatial Population Genetics. Genetics. 2006;174:805–816. doi: 10.1534/genetics.106.059923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Meng X. Simulating Normalizing Constants: from IMportance Sampling to Bridge Sampling to Path Sampling. Statistical Science. 1998;13(2):163–185. [Google Scholar]
- Green PJ. Reversible Jump Markov Chain Monte Carlo Computation and Bayesian Model Determination. Biometrika. 1995;82(4):711–732. [Google Scholar]
- Green PJ, Richardson S. Hidden Markov Models and Disease Mapping. Journal of the American Statistical Association. 2002;97:1055–1070. [Google Scholar]
- Guillot G, Estoup A, Mortier F, Cosson JF. A Spatial Statistical Model for Landscape Genetics. Genetics. 2005;170(3):1261–80. doi: 10.1534/genetics.104.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamstra DA, Chenevert TL, Moffat BA, Johnson TD, Meyer CR, Mukherji SK, Quint DJ, Gebarski SS, Fan X, Tsien CI, Lawrence TS, Junck L, Rehemtulla A, Ross BD. Evaluation of the functional diffusion map as an early biomarker of time-to-progression and overall survival in high-grade glioma. PNSA. 2005;102(46):16759–16764. doi: 10.1073/pnas.0508347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon D. Auxiliary Variable Methods for Markov Chain Monte Carlo with Applications. Journal of the American Statistical Association. 1998:93. [Google Scholar]
- Khalighi MM, Soltanian-Zadeh HS, Lucas C. Unsupervised MRI Segmentation with Spatial Connectivity. Proceedings of SPIE, Medical Imaging; 2002. pp. 1742–1750. [Google Scholar]
- Liang ZP, Lauterbur PC. Principles of Magnetic Resonance Imaging: A Signal Processing Perspective. Section 2 Chapter 8. SPIE press book; 1999. [Google Scholar]
- Lei T, Udupa JK. Statistical Properties of X-ray CT and MRI from Imaging Physics to Image Statistics. Proceedings of the SPIE, Medical Imaging; 2002. pp. 82–93. [Google Scholar]
- Marroquin JL, Arce E, Botello S. Hidden Markov Measure Field Models for Image Segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2003;25:1380–1387. [Google Scholar]
- Melas DE, Wilson SP. Double Markov Random Fields and Bayesian Image Segmentation. Institute of Electrical and Electronics Engineers Transactions on Signal Processing. 2002;50:357–365. [Google Scholar]
- Moffat BA, Chenevert TL, Lawrence TS, Meyer CR, Johnson TD, Dong Q, Tsien C, Mukherji S, Quint DJ, Gebarski SS, Robertson PL, Junck LR, Rehemtulla A, Ross BD. Functional diffusion map: A noninvasive MRI biomarker for early stratification of clinical brain tumor response. PNSA. 2005;102(15):5524–5529. doi: 10.1073/pnas.0501532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata Y. A Monte Carlo Method for High Dimensional Integration. Numerische Mathematik. 1989;55:137–157. [Google Scholar]
- Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Chapter 4.3 Oxford Press; 2003. [Google Scholar]
- Potts RB. Some generalized order-disorder transformations. Proceedings of the Cambridge Philosophical Society. 1952;48:106–109. [Google Scholar]
- Richardson S, Green PJ. On Bayesian Analysis of Mixtures with an Unknown Number of Components. Journal of the Royal Statistical Society Series B. 1997;59(4):731–792. [Google Scholar]
- Stephens M. Bayesian Analysis of Mixture Models with an Unknown Number of Components - An Alternative to Reversible Jump Methods. The Annals of Statistics. 2000;28:40–74. [Google Scholar]
- Swendsen RH, Wang JS. Nonuniversal Critical Dynamics in Monte Carlo Simulations. Physical Review Letters. 1987;58:86–88. doi: 10.1103/PhysRevLett.58.86. [DOI] [PubMed] [Google Scholar]
- Won CS, Derin H. Unsupervised Segmentation of Noisy and Textured Images Using Markov Random Fields. Graphical Models and Image Processing. 1992;54:308–328. [Google Scholar]
- Zhang X, Johnson TD, Little RJA, Cao Y. Quantitative Magnetic Resonance Image Analysis via the EM Algorithm with Stochastic Variation. Annals of Applied Statistics. 2008;2(2):736–755. doi: 10.1214/07-AOAS157. [DOI] [PMC free article] [PubMed] [Google Scholar]