Abstract
The purpose of this study was to develop ELISAs for key neural proteins, three synaptic and one glial, that exist in different intracellular compartments, which would be used as a measure of synaptic phenotype. These assays would be valuable to neurologically phenotype transgenic mouse models of human disease and also human disease itself using minimal amounts of post-mortem tissue. We showed that supernatant from crude brain tissue homogenates extracted in RIPA buffer containing 0.1% SDS bind to synaptophysin, synaptosome-associated protein of 25 kDa (SNAP-25), post-synaptic density-95 (PSD-95), and glial fibrillary acidic protein (GFAP) antibody pairs with high affinity and selectivity. Overall, RIPA + 0.1% SDS were more efficient than RIPA + 2% SDS or a buffer containing only 1% Triton-X-100. Diluting the brain extracts resulted in dose-dependent binding to the antibody pairs for each neural protein, with EC50s that varied from 8.6 µg protein for PSD-95 to 0.23 µg for GFAP. The assays were used to measure synaptic marker protein levels at various times during mouse development and GFAP in a model of disease accompanied by neuroinflammation. Comparison of ELISAs with Western blots by measuring marker levels in brain extract from developing mice showed a greater relative difference in values derived from ELISA. These ELISAs should be valuable to phenotype the synapse in neurological disease and their rodent models.
Keywords: Synapse, Synaptophysin, Synaptosome-associated protein of 25 kDa (SNAP-25), Post-synaptic density-95 (PSD-95), Glial fibrillary acidic protein (GFAP), Enzyme-linked immunoassay
Introduction
There has been a resurgence of interest in the abundance, structure and function of synapses and their relationship to neurological disorders. These disorders range from autism (Pfeiffer and Huber 2007; Tabuchi et al. 2007) to Alzheimer’s disease (AD) (Tanzi 2005) to traumatic brain injury (Scheff et al. 2005; Ansari et al. 2008). Sometimes, changes in function may be reflected by adjustments in the concentration of synaptic protein(s) present, which could modify the manner in which these proteins interact with other proteins in the pre-synaptic bouton or at the post-synaptic density. However, more often with constitutively expressed synaptic proteins, changes in synaptic number due to, for example, a loss of neurons, is reflected in changes in pre-synaptic and post-synaptic protein concentrations measured in extracted tissue. Dynamic changes in the density of synapses occur during development (Glantz et al. 2007), in age-related disease (Counts et al. 2006) and even in the mature adult (Knott et al. 2006). These changes may be quantified by measuring levels of synaptic proteins, or morphological structures such as dendritic spines that reflect a subset of synapses (Tsai et al. 2004). In AD, for example, convincing evidence supports the theory that synapses are lost as a prelude to overt neurodegeneration (Terry 2000). With increasing number of mouse models of neurological disease, it would be important to know whether changes in the expression of particular genes can alter local or regional synaptic expression of these proteins that might reflect changes in neural networks. An accurately quantifiable, sensitive and precise method that can detect alterations in these synaptic proteins in a discreet brain region would be of great benefit.
Synaptic number or density per region has been quantified using numerous methods, with each approach revealing particular advantages and disadvantages. A direct method is unbiased counting of the ultrastructure of synaptic densities in fixed brain sections using electron microscopy. This method is quantitative and robust, but it is highly labor intensive and usually a limited number of brain regions are counted (DeKosky and Scheff 1990; Scheff et al. 2006, 2007). Essentially all other methods used in the past have exploited the assumption that the quantitative expression of certain, constitutively expressed synaptic proteins is directly related to synaptic number. One method is immunohisto-chemical staining of synaptic boutons. Staining individual synaptic boutons with an antibody against a synaptic protein, such as anti-synaptophysin antibody, and counting individual puncta using light microscopy and unbiased, systematic stereological techniques is also quantitative, yet labor intensive and requires a great deal of confidence in a consistent immunological staining method (Calhoun et al. 1996; Mokin and Keifer 2006). A variation of this technique is to view immunohistologically stained brain sections at lower power magnification and estimate the overall intensity of synaptic marker protein staining using densitometry. Levels of numerous proteins have been measured with this technique to estimate synaptic abundance, with synaptophysin the most common (Calhoun et al. 1996). However, the results are highly variable, especially from laboratory to laboratory, even when employing an identical animal model (King and Arendash 2002; Fonseca et al. 2004; Jacobsen et al. 2006; Dong et al. 2007). Biochemical methods have been developed to measure levels of these proteins, but the disadvantage is that a large region of tissue is usually required for an assay and sometimes the assays are only semi-quantitative. Several groups have estimated synaptic density with measures of synaptic proteins on Western blot or other immunoblotting technique (Counts et al. 2006; Smith et al. 2007). The advantage of Western blot is that multiple samples can be measured at once; however, densitometric analysis of these data (especially if using chemiluminescence with film development) often do not quantitatively measure the true difference in protein amount between two samples due to technical issues (O’Callaghan et al. 1999). Lastly, ELISA is a frequently employed technique to quantitate synaptic protein levels. ELISA is highly quantitative (Schmidt et al. 1999). It is precise and accurate, with rather small coefficients of variation, but has the same limitation of Western blot in that it requires tissue extract for measurement and, therefore, synaptic changes cannot be localized to discreet brain subregions. However, if ELISAs are sensitive, small tissue punches may be sufficient tissue for assay (Mayer et al. 2005). Because of the difference in solubility of various synaptic proteins and their location in different cellular compartments, procedures for extraction often vary depending on the nature and localization of the protein.
Here, we have developed a panel of four ELISAs: one for the pre-synaptic vesicular protein, synaptophysin, the pre-synaptic SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein, SNAP-25 (synaptosome-associated protein of 25 kDa), the post-synaptic scaffolding protein, PSD-95 (post-synaptic density protein of 95 kDa) and the astrocyte intermediate filament protein, glial fibrillary acidic protein (GFAP). We have identified a single detergent-containing buffer system that will extract all four of these proteins from brain tissue and retain specific, antibody binding using small quantities of tissue. This panel of ELISAs may be particularly useful in phenotyping synaptic changes and astrocyte reactivity in various transgenic models of nervous system disease in addition to measuring synaptic changes and inflammation in post-mortem tissue of human nervous system disease.
Materials and methods
Animals
All animal procedures described here were approved by the Institutional Animal Use and Care Committee at the University of South Florida. Every effort was made to limit the number of animals used in these experiments. Adult Sprague–Dawley rats and C57Bl/6 mice were obtained from Harlan Industries (Indianapolis, IN) and JAX Laboratories (Bar Harbor, ME), respectively. Animals were housed 2–3 per cage on a 12 h light/dark cycle and had free access to food and water. Neonates were obtained from C57Bl/6 timed-pregnant dams (Harlan Industries) and mother and litter were maintained under standard conditions, and tissue obtained at various ages after the animals were euthanatized with the excess CO2. Brain tissue regions were also dissected from 15 to 16-month-old Tg2576 mice (n = 6) and non-transgenic controls (n = 6) (Hsiao et al. 1996) from a colony maintained at the University of South Florida. These mice express, hamster prion protein promoter-driven, human amyloid precursor protein bearing the double K670N, M671L mutations, the so-called Swedish mutation.
Tissue processing
Rats and mice were euthanatized by exposure to excess CO2, the animals decapitated, the skull removed, and the brain carefully removed from the cranium. Rat forebrain extract was used as the standard curve in all synaptic marker ELISAs. Whole brain was removed from a 3-month-old rat, and was extracted fresh. The brain was homogenized using a Wheaton Potter-Elvehjem glass tissue grinder with a Teflon pestle (Thermo Fisher, Waltham, MA) in ten volumes of RIPA buffer (25 mM Tris–HCl, 150 mM NaCl, 2 mM EDTA, 1% Triton-X-100, 1% sodium deoxycholate, 0.1% SDS, pH 7.4; all obtained from Sigma Chemical Co., St. Louis, MO, containing protease inhibitor set cocktail III, EMD Biosciences, Gibbstown, NJ). The extract was centrifuged at 40,000g for 30 min and the supernatant was recovered, diluted, aliquoted and frozen at −80°C. The protein concentration of the extract used for the standard was 4.6 µg/µl. Total protein in the standard and sample was measured with a bicinchoninic acid (BCA) procedure (Pierce, Thermo Scientific, Rockford, IL). Postnatal day 7 (P7) and P28 mice were euthanatized, frontal cortex was dissected and frozen at −80°C. This tissue was used to measure changes in synaptic protein expression over the period of early development in the mouse and for comparison of Western blot with ELISAs. In other experiments, efficiency of extraction was compared using rat or mouse brain that was extracted with 25 mM Tris–HCl, 150 mM NaCl containing 1% Triton-X-100, RIPA containing 0.1% SDS, or RIPA containing 2% SDS, all which contained protease inhibitor cocktail.
Antibodies
Sandwich ELISAs were developed and conducted for the following proteins: (1) pre-synaptic vesicular protein, synaptophysin, (2) the pre-synaptic membrane protein, SNAP-25, the post-synaptic scaffolding protein, PSD-95, and the astrocyte intermediate filament protein, GFAP. Each ELISA required a capture antibody and a “detection” antibody. For each ELISA, the capture antibodies were mouse monoclonals purchased from the following suppliers and used at the following dilutions for ELISA: (1) anti-synaptophysin, 1:250 (clone SY38, MAB368, Millipore, Temecula, CA), (2) anti-SNAP-25, 1:200 (clone SP14, MAB331, Millipore), (3) anti-PSD-95, 1:100 (clone 7E3-1B8, MAB1598, Millipore, or anti-PSD-95 from NeuroMab, clone K28/43, UC-Davis), (4) anti-GFAP, 1:250 (clone GA5, MAB360, Millipore). For each ELISA, the detection antibodies were various polyclonals purchased from the following suppliers and used at the following dilutions: (1) rabbit anti-synaptophysin, 1:2,000 (affinity purified, A0010, Dako North America, Carpinteria, CA); (2) rabbit anti-SNAP-25, 1:1,000 (IgG fraction, S9684, Sigma, St. Louis, MO); (3) sheep anti-PSD-95, 1:100 (51–6700 Invitrogen, Carlsbad, CA, formerly Zymed, or rabbit anti-PSD-95, ab18258, 1:400, Abcam, Cambridge, MA); (4) rabbit anti-GFAP, 1:1,000 (IgG fraction, Z0334, Dako).
Sandwich enzyme-linked immunosorbent assays
The method for these ELISAs (with the exception of antibodies) was identical regardless of the antigen. Unless otherwise mentioned, chemicals were purchased from Sigma. Capture antibodies were diluted in 10 mM phosphate buffer pH 8.0 and 50 µl was added to wells of a Costar hi-binding 96-well ELISA plate (3590, Lowell, MA). Importantly, the plate was tapped to ensure total coverage of the well. The plate was covered with parafilm to form tight seals at each well and left at room temperature overnight. The next morning, the wells were washed (diluent added to the top of the well and all liquid removed from the well) three times with buffer B (10 mM phosphate buffered saline, pH 7.5, 0.05% Tween 20). The wells were blocked by adding 300 µl of buffer B containing 5% dry milk (importantly, Saco Mix ‘n Drink) and the plate shaken (on an orbital shaker) for 1 h. The plates were washed with buffer B one time before the addition of samples or standard. The standard curve was constructed from rat brain extract. Standards were added to wells in 100 µl aliquots. The standard consisted of 1:1 dilutions of the extract in buffer B. 4.6 µg protein, in 1 µl RIPA buffer diluted to 100 µl in buffer B was arbitrarily designated as “100 U”. Thus, 2.3 µg protein was equal to 50 U, 1.15 µg protein, 25 U etc, and 9.2 µg protein was 200 U, and 18.4 µg protein was 400 U. Samples of rat or mouse brain were added in dilutions from 5 to 0.1 µl per 100 µl depending on the sensitivity of the standard and so that the sample absorbance values would fall near 50% binding (the linear range) in the standard curve. Samples and standards were pipetted in triplicate, or in some assays, in duplicate. The plate was covered with parafilm and then shaken at room temperature for 2 h. The wells were washed three times with buffer B and detection antibody, diluted in buffer B containing 5% dry milk, was added. The antibody was shaken at room temperature for 2 h and the wells were then washed three times. Developing antibody (horse radish peroxidase conjugated) anti-rabbit IgG (1:1,000) or anti-sheep IgG (1:1,000) (Chemicon) was diluted in buffer B plus 5% dry milk and added as 100 µl. The plates were shaken at room temperature for 45 min. Wells were washed three times with buffer B and 100 µl of tetramethylbenzidine substrate was added, the plate shaken by hand until the zero substrate blank becomes the most faint, pale blue (usually from 2 to 3 min depending on the assay). To stop development, 50 µl of 1 M H2SO4 was added and absorbance measured immediately at 450 nm on a Victor spectrophotometric microplate reader (Perkin-Elmer, Waltham, MA) or a BioTek Synergy HT (Winooski, VT). The sigmoidal standard was evaluated with a non-linear four-parameter fit using “Workout” software (or Gen 5 for the BioTek) and sample amounts were obtained using the fitted standard curve.
Western blots for specificity and comparison with ELISA
Mouse brain tissue extracts diluted with 2× reducing Laemmli sample buffer were loaded onto pre-cast 4–20% gradient SDS-PAGE gels (Invitrogen gels, Invitrogen, Carlsbad, CA) and electrophoresed using standard conditions. Protein was electrophoretically transferred to a polyvinylidene difluoride membrane (PVDF, Immobilon, Millipore, Billerica, MA) and the membranes probed with various antibodies. The primary antibodies described above were used at the following dilutions for Western blot: (1) anti-synaptophysin, MAB368 1:3,000; rabbit anti-syn-aptophysin, 1:3,000, (2) anti-SNAP-25 MAB331 1:2,000, rabbit anti-SNAP-25 1:5,000, (3) anti-PSD-95 MAB1598 1:2,000, sheep anti-PSD-95, 1:1,000, (4) anti-GFAP MAB360 1:1,000, rabbit anti-GFAP 1:5,000. Blots were developed using horseradish peroxidase conjugated secondary antibodies (Millipore) at the following dilutions: goat anti-mouse IgG 1:3,000, goat anti-rabbit IgG 1:5,000 and rabbit anti-sheep IgG 1:1,000 (Invitrogen) and Super-signal chemiluminescent substrate (Pierce Chemical, Thermo Scientific, Rockford, IL). Blots were exposed to film for various lengths of time and developed. Blocking and antibody incubation steps were conducted in buffer B containing 5% non-fat dry milk.
For quantitative comparison with ELISA, extracts from frontal cortex from P7 and P28 mice subjected to Western blot for synaptophysin, SNAP-25 and PSD-95 (10 µg protein loaded) or GFAP (1 µg protein loaded) and the chemiluminescent blots imaged on a Kodak Image Station 4000MM (Carestream Molecular Imaging) and mean integrated density determined with Kodak Molecular Imaging Software (version 5.0.1.27). For each blot, the density was normalized to the band density of the constitutively expressed gene, glyceraldehyde phosphate dehydrogenase.
Data analysis
Differences among sample means from the frontal cortex of different ages of developing rats or from APPsw mice and their non-transgenic littermates were evaluated by analysis of variance and specific differences between individual pairs of means was determined using Bonferonni’s test. A P < 0.05 was considered a significant difference.
Results
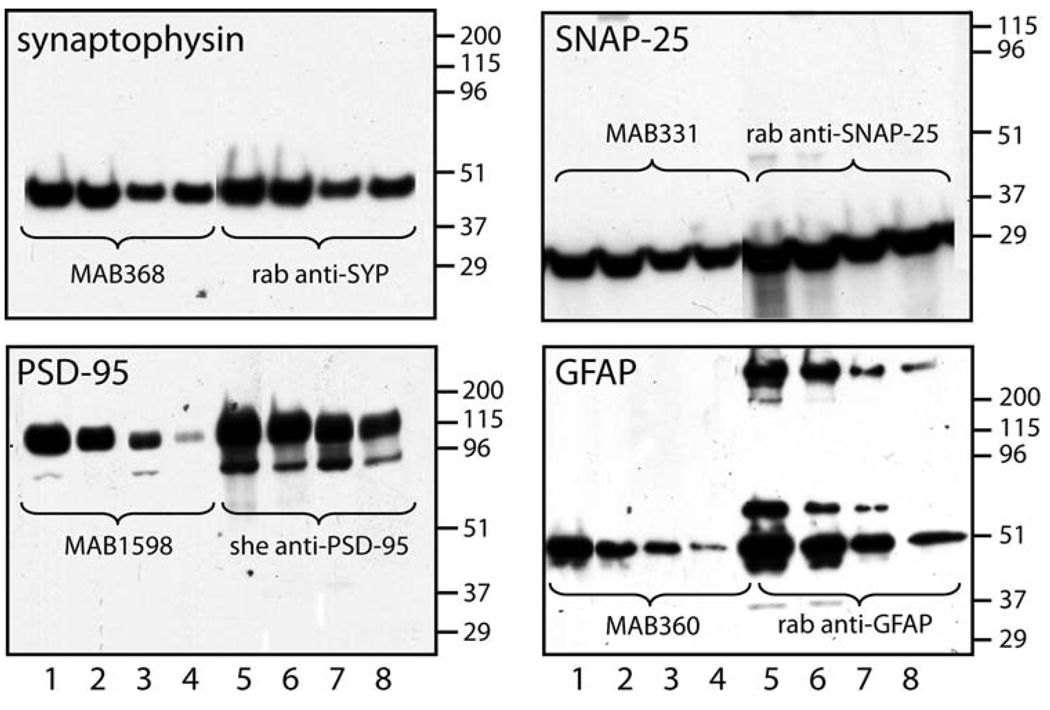
To determine the specificity and appropriate immunoreactivity for the antibodies used in the sandwich ELISA, polyclonal-monoclonal pairs for each of the four synaptic proteins were subjected to Western blot. Each polyclonal-monoclonal pair recognized the appropriate molecular weight and selective antigen on Western blot; and the signal was less intense when total protein loaded was reduced (Fig. 1). Identical molecular weight proteins were detected by the monoclonal and polyclonal antibody pairs. In the GFAP blot, signals from aggregates of the monomer were observed at high-molecular weights (Fig. 1). Thus, it appears that the selectivity and sensitivity of these antibodies were sufficient for ELISA, although efficient detection in Western blot does not necessarily mean the antibodies will recognize non-denatured protein (ie. protein not exposed to high concentrations of SDS).
Fig. 1.
Specificity of synaptic protein antibodies used for sandwich ELISAs. Supernatants from RIPA buffer extracted rat brain were subjected to SDS-PAGE, blotted to PVDF membrane, and probed with monoclonal (lanes 1–4) and polyclonal (lanes 5–8) antibodies. Different amounts of total protein were added to each gel. For synaptophysin lanes 1, 2, 5, 6 contain 10 µg and lanes 3, 4, 7, 8 contain 2 µg total protein. For SNAP-25 and PSD-95 lanes 1, 2, 5, 6 contain 15 µg and lanes 3, 4, 7, 8 contain 5 µg total protein. For GFAP lanes 1, 2, 5, 6 contain 1 µg and lanes 3, 4, 7, 8 contain 0.2 µg total protein. Blots were developed using horseradish peroxidase conjugated secondary antibodies with a chemiluminescent substrate. Blots were exposed to film for various lengths of time and developed using standard techniques. Representative data are shown from an experiment that was repeated twice with similar results
Because each of these proteins is either an integral membrane protein or is part of a protein scaffold or the cytoskeleton, the conditions of extraction are crucial for maximizing the efficiency of recovery. Thus, we compared the maximum total binding (absorbance) for each antibody pair when whole rat brain tissue was extracted in buffer containing 1% Triton-X-100, RIPA buffer containing 0.1% SDS, or RIPA buffer containing 2% SDS. The goals for optimizing the conditions of extraction are a balance of efficiency of extraction and retention of binding to the capture and detection antibodies. The results from a representative experiment are shown in Fig. 2. Rat brain extracted in 1% Triton-X-100 was as efficient at extracting synaptophysin and SNAP-25 as compared to either RIPA buffer. This was somewhat surprising because these proteins are a vesicular or bouton transmembrane protein. On the other hand, either variation of RIPA buffer was superior to Triton-X-100 in extracting PSD-95, but especially GFAP (Fig. 2). Surprisingly samples extracted with buffer containing 2% SDS maintained the ability to bind to all four antibody pairs. Nonetheless, because 0.1% SDS RIPA (standard RIPA buffer) was as effective as 2% SDS RIPA, this was the buffer chosen for developing each assay further. Similar results were obtained when mouse brain extract was used as the standard, and absorbance was not different from background when mouse hepatic or renal extracts were assayed up to 40 µg per well (data not shown).
Fig. 2.
Effect of extraction buffer on the magnitude of immunoreactivity for synaptophysin, SNAP-25, PSD-95 and GFAP in sandwich ELISA. Whole hemispheres of rat brain were extracted in the designated buffer and the indicated concentrations of total protein were subjected to ELISA. The average of duplicate samples is shown from a single experiment. This experiment was repeated twice with similar results
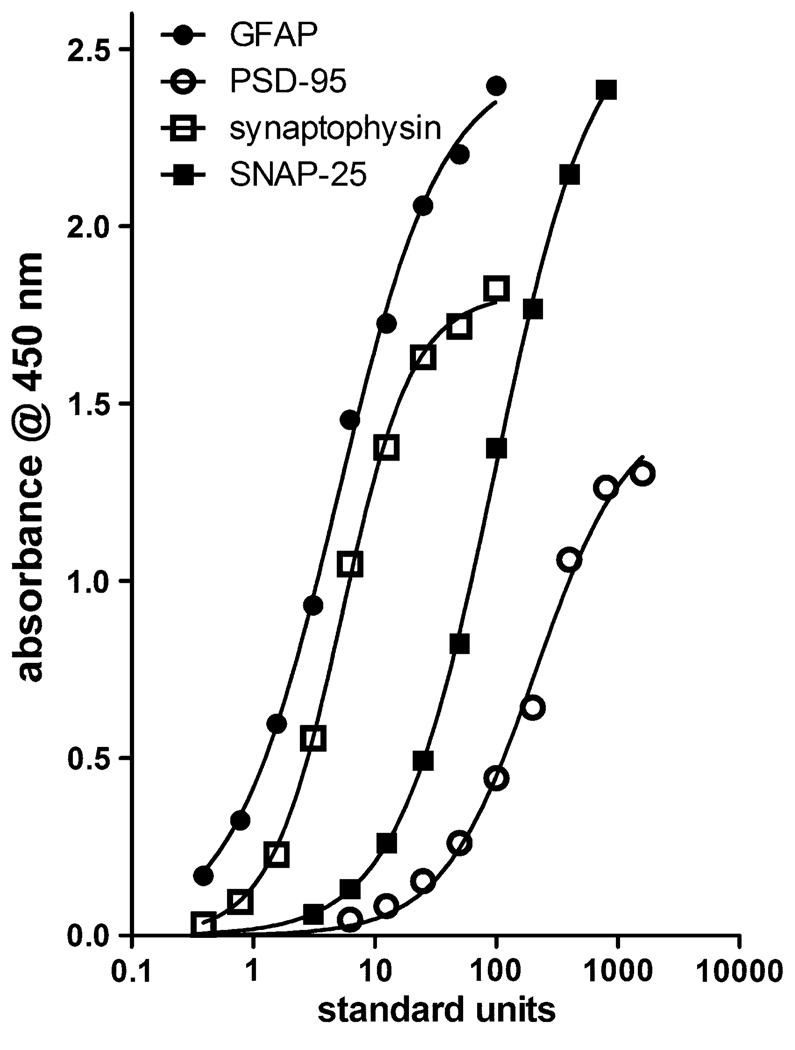
Thus, each antibody pair was examined for dose-dependent binding of antigen to the antibody pairs for the development of standard curves. These curves could then be used to extrapolate the amount of each synaptic antigen in an unknown sample. RIPA-extracted whole rat brain was aliquoted and used as the standard. A known amount of total protein standard (4.6 µg) was arbitrarily designated as 100 U, and equal, serial dilutions of this protein amount were considered 50, 25 U etc. Typical standard curves are shown in Fig. 3, with summary profiles of non-linear, four-parameter fits of replicates of these curves are found in Table 1. Each curve varied with respect to slope and maximum binding in the curve, however, all assays were highly sensitive requiring 8.6 µg protein (187.7 U) for half maximal binding in the PSD-95 curve and only 0.23 µg (5 U) protein for the GFAP assay. All samples from a single experiment were measured for the individual neural proteins within a single assay. Intra-assay coefficients of variation were somewhat larger than might be observed for a typical radioimmunoassay, but are more in line with typical ELISAs (Siew et al. 2004). Each antibody pair recognized antigen in human brain tissue extracts as effectively as rat and mouse extracts (data not shown).
Fig. 3.
Dose-dependent immunoreactivity of rat brain protein extract in sandwich ELISAs. Dilutions of rat brain protein extract were subjected to sandwich ELISA using the standard assay protocol and absorbance measured at 450 nm. The quantitative parameters for these curves are shown in Table 1
Table 1.
Synaptic marker ELISA parameters
| ELISA | Parameters in four parameter curve fit |
|||||
|---|---|---|---|---|---|---|
| n | Min (abs) | Max (abs) | EC (50) | Slope | Intra-assay CV (%) | |
| Synaptophism | 9 | 0.02 ± 0.00 | 1.46 ± 0.23 | 17.2 ± 2.9 | 1.49 ± 0.12 | 8.4 |
| SNAP-25 | 8 | 0.01 ± 0.00 | 2.53 ± 0.12 | 137.5 ± 22.9 | 1.08 ± 0.08 | 8.7 |
| PSD-95 | 8 | 0.01 ± 0.00 | 1.05 ± 0.09 | 187.7 ± 16.7 | 1.46 ± 0.14 | 7.6 |
| GFAP | 7 | 0.04 ± 0.01 | 2.46 ± 0.07 | 5.0 ± 0.7 | 1.05 ± 0.04 | 6.0 |
Data are shown as mean ± SEM
n number of statndard curve trials, MIN minimum absorbance in four-paramter fit, MAX maximum absorbance in four-parameter fit, EC50 concentration of extract where 50% of maximum binding (absorbance) is observed, slope the slope of the four-parameter fit, intra assay CV intra assay coefficient of variation
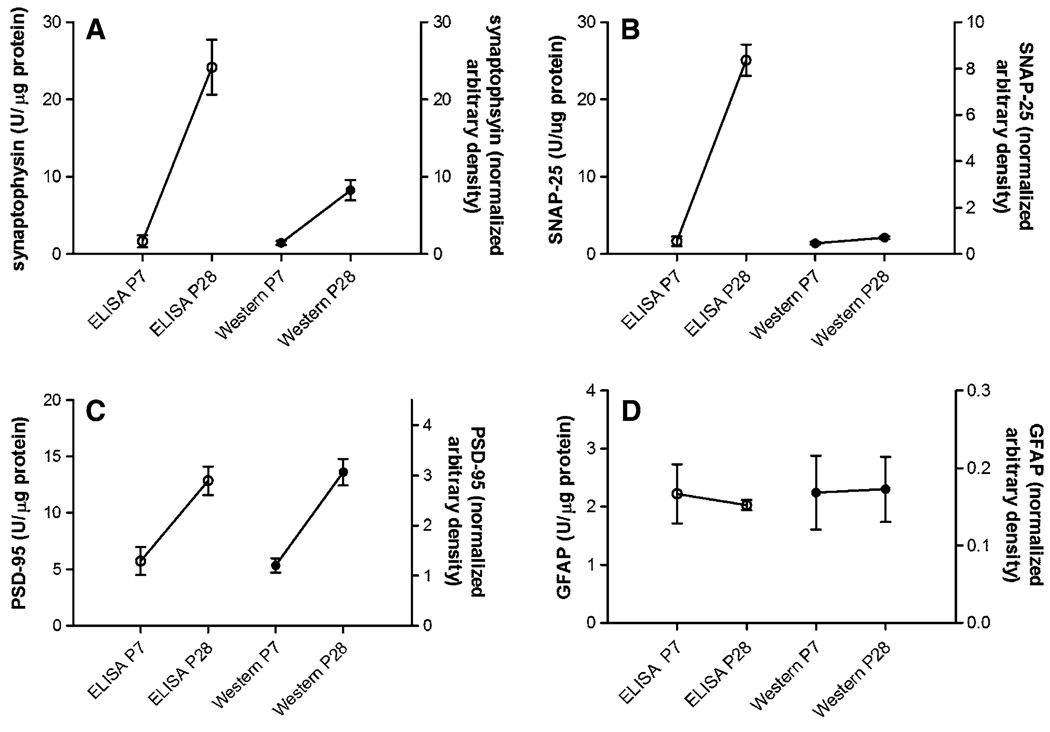
These assays were employed to measure levels of the synaptic markers and GFAP levels in brain tissues from various mouse models. As shown in Fig. 4, levels of synaptophysin, SNAP-25 and PSD-95 in frontal cortex from C57Bl/6 mice showed developmentally related increases from P7 to P28. The synaptophysin and SNAP-25 ELISAs were markedly more robust at detecting large differences between the P7 and P28 samples. Both the PSD-95 ELISA and Western blot showed a smaller 2–3-fold difference in levels between the two ages and there were no developmental differences in GFAP levels.
Fig. 4.
Relative levels of synaptophysin (a), SNAP-25 (b), PSD-95 (c) and GFAP (d) in frontal cortex extracts from P7 (n = 5) and P28 (n = 5) mouse pups and measured by ELISA and Western blot. ELISA units are on the left y axis and units for Western blot on the right y axis. Note that for synaptophysin and SNAP-25, the dynamic range measured by ELISA was greater than that measured by Western blot. The optical density of immunoreactive bands on Western blots was used as a measure of protein abundance. Blots were normalized using optical density of constitutively expressed GAPDH
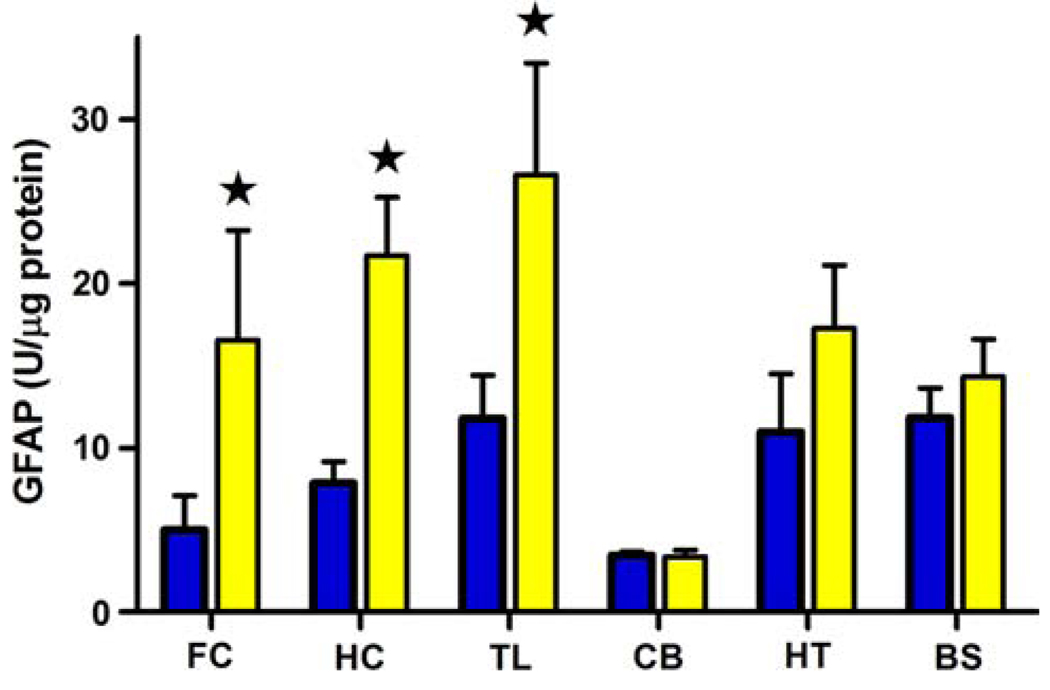
Finally, we measured GFAP levels in six brain regions dissected from 15-month-old APPsw (an age with distinct amyloid deposition) mice and their non-transgenic littermate controls. GFAP was significantly elevated in frontal cortex, temporal lobe and hippocampus of APPsw mice as compared to control regions (P < 0.05; Fig. 5). No differences in GFAP levels were found in cerebellum, hypothalamus, and brainstem between APPsw and non-transgenic mice.
Fig. 5.
GFAP immunoreactivity in frontal cortex (FC), hippocampus (HC), temporal lobe (TL), cerebellum (CB), hypothalamus (HT) and brain stem (BS) in 15–16-month-old non-transgenic (nt) (dark bars n = 6) or APPsw mice (light bars n = 6). All values are shown as the mean ± SEM and expressed as units/µg protein as described in “Methods”. *P ≤ 0.05 compared with the value of the non-transgenic animal
Discussion
The major advance made in this study was the identification of RIPA buffer as sufficient for extraction of four neural proteins that exist in different cellular compartments. Synaptophysin is a neuronal vesicular membrane protein that spans the membrane four times with both N- and C-termini in the cytoplasm (Sudhof et al. 1987). SNAP-25 is a “SNARE” protein localized to the active site of the synapse and is essential for fusion of a vesicle to the membrane for secretion (Sorensen 2005). PSD-95 is an abundant post-synaptic scaffolding protein vital for interfacing membrane proteins to the cytoskeleton and regulating glutaminergic receptor trafficking (Kim and Sheng 2004). GFAP is an intermediate filament protein directly associated with the growth of astrocyte cell bodies upon activation by injury or disease (Porchet et al. 2003). The sensitivity of these ELI-SAs is high, since each protein can be measured with a RIPA-extracted sample containing between 8.6 and 0.2 µg of total protein. Given this fact, all four proteins may be quantified from a single brain region or sub-region or even a micro tissue punch from brain. Because these antibodies cross-react with both human and rodent proteins, the assays provide a simple, yet, powerful means to phenotype a measure of synapses of human neurological disease and mouse models of human disease. ELISAs have been developed previously for each of these antigens, however, most were developed separately using soluble or detergent-containing extraction buffers (Schlaf et al. 1996; O’Callaghan et al. 1999; Honer et al. 2002; Nithianantharajah et al. 2004; Siew et al. 2004). Data here indicates that RIPA buffer containing 0.1% SDS was equally as effective as RIPA containing 2% SDS in extracting each of the four proteins, but both buffers were more effective than buffer containing 1.0% Triton-X-100 in solubilizing GFAP. In addition, these ELISAs are efficient in measuring both human and rodent proteins. Thus, extraction of rodent or human brain tissue in RIPA buffer containing 0.1% SDS results in an extract which can be used to measure all four proteins in ELISA.
Measuring synaptic markers by ELISA has several advantages over other methods. It is significantly more sensitive (Schlaf et al. 1996) and sometimes crude homogenates will contain proteins that quench the measurement of the protein of interest in other methods such as Western blot (O’Callaghan et al. 1999). Because of the sensitivity, small, sub-regional, tissue punch samples may be collected and used for measuring proteins in ELISA. Of course the major drawback, as with measuring any protein present at the synapse, is the assumption that the level of the protein reflects the abundance of functional synapses in that sample, yet technically, there is no measure of the abundance of functional synapses. In addition, it is not known whether protein levels reflect the number of densities counted ultra-structurally, but counting synapses reveals nothing about functionality either. Whether measurement of synaptic protein is related to the number of ultrastructural post-synaptic densities is not known, and in the end, may be less important than identifying changes in the levels of these synaptic proteins as a biomarkers of disease or injury.
To demonstrate the effectiveness of these assays, we measured the four proteins during post-natal development in mouse brain frontal cortex. The results showed that an age-related increase in the level of synaptophysin, SNAP-25 and PSD-95 from P7 through P28 in frontal cortex, similar to what was found previously in the rat (Aya-ay et al. 2005) (Sans et al. 2000) and human tissue (Glantz et al. 2007). The magnitude of change for each of these proteins was more robust with synaptophysin and SNAP-25 ELISA compared with quantification of the density of chemiluminescent signal of the immunoreactive bands on Western blot. The magnitude of change for PSD-95 was the same for both methods. This was not surprising since Western blot quantitation has inherent technical issues (O’Callaghan et al. 1999) for quantification, especially when using chemiluminescence detection by film. Lastly, we measured GFAP levels in various brain regions of 15-month-old APPsw mice which deposit amyloid in forebrain. Similar to AD (Ross et al. 2003) brain tissue and AD mouse model brain tissue measured by other methods (Holcomb et al. 1998), GFAP levels were significantly elevated in frontal cortex, hippocampus and temporal lobe as compared to regions from age-matched littermate controls. In human superior frontal cortex, (but not cerebellum) synaptophysin, SNAP-25 and PSD-95 levels were significantly reduced in extracts from AD subjects as compared to age-matched controls (not shown).
In conclusion, we have identified an extraction method for four proteins whose expression may change in neurological disease with alterations in the abundance of synapses. In addition, these assays should be useful to characterize and regionally identify synaptic changes that may be present in various genetic mouse models of neurological disease.
Acknowledgments
This work was supported in part by the National Institutes of Health (AG022101) and Alzheimer’s Association (Grant no. IIRG-02-3758). The authors would like to thank Christopher C. Leonardo and Yun Bai for their assistance.
Contributor Information
Paul E. Gottschall, Email: pegottschall@uams.edu, Department of Pharmacology and Toxicology, College of Medicine, University of Arkansas for Medical Sciences, Slot #611, 4301 West Markham Street, Little Rock, AR, USA.
Joanne M. Ajmo, Department of Molecular Pharmacology and Physiology, School of Basic Biomedical Sciences, University South Florida College of Medicine, Tampa, FL 33612, USA
Autumn K. Eakin, Department of Molecular Pharmacology and Physiology, School of Basic Biomedical Sciences, University South Florida College of Medicine, Tampa, FL 33612, USA
Matthew D. Howell, Department of Pharmacology and Toxicology, College of Medicine, University of Arkansas for Medical Sciences, Slot #611, 4301 West Markham Street, Little Rock, AR, USA
Hina Mehta, Department of Pharmacology and Toxicology, College of Medicine, University of Arkansas for Medical Sciences, Slot #611, 4301 West Markham Street, Little Rock, AR, USA.
Lauren A. Bailey, Department of Pharmacology and Toxicology, College of Medicine, University of Arkansas for Medical Sciences, Slot #611, 4301 West Markham Street, Little Rock, AR, USA
References
- Ansari MA, Roberts KN, Scheff SW. Oxidative stress and modification of synaptic proteins in hippocampus after traumatic brain injury. Free Radic Biol Med. 2008 doi: 10.1016/j.freeradbiomed.2008.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya-ay J, Mayer J, Eakin AK, Muffly BG, Anello M, Sandy JD, Gottschall PE. The effect of hypoxic-ischemic brain injury in perinatal rats on the abundance and proteolysis of brevican and NG2. Exp Neurol. 2005;193:149–162. doi: 10.1016/j.expneurol.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Calhoun ME, Jucker M, Martin LJ, Thinakaran G, Price DL, Mouton PR. Comparative evaluation of synaptophysin-based methods for quantification of synapses. J Neurocytol. 1996;25:821–828. doi: 10.1007/BF02284844. [DOI] [PubMed] [Google Scholar]
- Counts SE, Nadeem M, Lad SP, Wuu J, Mufson EJ. Differential expression of synaptic proteins in the frontal and temporal cortex of elderly subjects with mild cognitive impairment. J Neuropathol Exp Neurol. 2006;65:592–601. doi: 10.1097/00005072-200606000-00007. [DOI] [PubMed] [Google Scholar]
- DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol. 1990;27:457–464. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- Dong H, Martin MV, Chambers S, Csernansky JG. Spatial relationship between synapse loss and beta-amyloid deposition in Tg2576 mice. J Comp Neurol. 2007;500:311–321. doi: 10.1002/cne.21176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca MI, Zhou J, Botto M, Tenner AJ. Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer’s disease. J Neurosci. 2004;24:6457–6465. doi: 10.1523/JNEUROSCI.0901-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz LA, Gilmore JH, Hamer RM, Lieberman JA, Jarskog LF. Synaptophysin and postsynaptic density protein 95 in the human prefrontal cortex from mid-gestation into early adulthood. Neuroscience. 2007;149:582–591. doi: 10.1016/j.neuroscience.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcomb L, Gordon MN, McGowan E, Yu X, Benkovic S, Jantzen P, Wright K, Saad I, Mueller R, Morgan D, Sanders S, Zehr C, O’Campo K, Hardy J, Prada CM, Eckman C, Younkin S, Hsiao K, Duff K. Accelerated Alzheimer-type phenotype in trans-genic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat Med. 1998;4:97–100. doi: 10.1038/nm0198-097. [DOI] [PubMed] [Google Scholar]
- Honer WG, Falkai P, Bayer TA, Xie J, Hu L, Li HY, Arango V, Mann JJ, Dwork AJ, Trimble WS. Abnormalities of SNARE mechanism proteins in anterior frontal cortex in severe mental illness. Cereb Cortex. 2002;12:349–356. doi: 10.1093/cercor/12.4.349. [DOI] [PubMed] [Google Scholar]
- Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- Jacobsen JS, Wu CC, Redwine JM, Comery TA, Arias R, Bowlby M, Martone R, Morrison JH, Pangalos MN, Reinhart PH, Bloom FE. Early-onset behavioral and synaptic deficits in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2006;103:5161–5166. doi: 10.1073/pnas.0600948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- King DL, Arendash GW. Maintained synaptophysin immunore-activity in Tg2576 transgenic mice during aging: correlations with cognitive impairment. Brain Res. 2002;926:58–68. doi: 10.1016/s0006-8993(01)03294-2. [DOI] [PubMed] [Google Scholar]
- Knott GW, Holtmaat A, Wilbrecht L, Welker E, Svoboda K. Spine growth precedes synapse formation in the adult neocortex in vivo. Nat Neurosci. 2006;9:1117–1124. doi: 10.1038/nn1747. [DOI] [PubMed] [Google Scholar]
- Mayer J, Hamel MG, Gottschall PE. Evidence for proteolytic cleavage of brevican by the ADAMTSs in the dentate gyrus after excito-toxic lesion of the mouse entorhinal cortex. BMC Neurosci. 2005;6:52. doi: 10.1186/1471-2202-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokin M, Keifer J. Quantitative analysis of immunofluorescent punctate staining of synaptically localized proteins using confocal microscopy and stereology. J Neurosci Methods. 2006;157:218–224. doi: 10.1016/j.jneumeth.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Nithianantharajah J, Levis H, Murphy M. Environmental enrichment results in cortical and subcortical changes in levels of synaptophysin and PSD-95 proteins. Neurobiol Learn Mem. 2004;81:200–210. doi: 10.1016/j.nlm.2004.02.002. [DOI] [PubMed] [Google Scholar]
- O’Callaghan JP, Imai H, Miller DB, Minter A. Quantitative immunoblots of proteins resolved from brain homogenates: underestimation of specific protein concentration and of treatment effects. Anal Biochem. 1999;274:18–26. doi: 10.1006/abio.1999.4260. [DOI] [PubMed] [Google Scholar]
- Pfeifer BE, Huber KM. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J Neurosci. 2007;27:3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchet R, Probst A, Bouras C, Draberova E, Draber P, Riederer BM. Analysis of glial acidic fibrillary protein in the human entorhinal cortex during aging and in Alzheimer’s disease. Proteomics. 2003;3:1476–1485. doi: 10.1002/pmic.200300456. [DOI] [PubMed] [Google Scholar]
- Ross GW, O’Callaghan JP, Sharp DS, Petrovitch H, Miller DB, Abbott RD, Nelson J, Launer LJ, Foley DJ, Burchfiel CM, Hardman J, White LR. Quantification of regional glial fibrillary acidic protein levels in Alzheimer’s disease. Acta Neurol Scand. 2003;107:318–323. doi: 10.1034/j.1600-0404.2003.02098.x. [DOI] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, 2nd, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheff SW, Price DA, Hicks RR, Baldwin SA, Robinson S, Brackney C. Synaptogenesis in the hippocampal CA1 field following traumatic brain injury. J Neurotrauma. 2005;22:719–732. doi: 10.1089/neu.2005.22.719. [DOI] [PubMed] [Google Scholar]
- Scheff SW, Price DA, Schmitt FA, Mufson EJ. Hippocampal synaptic loss in early Alzheimer’s disease and mild cognitive impairment. Neurobiol Aging. 2006;27:1372–1384. doi: 10.1016/j.neurobiolaging.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Scheff SW, Price DA, Schmitt FA, DeKosky ST, Mufson EJ. Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology. 2007;68:1501–1508. doi: 10.1212/01.wnl.0000260698.46517.8f. [DOI] [PubMed] [Google Scholar]
- Schlaf G, Salje C, Poethke R, Felgenhauer K, Mader M. A novel enzyme-linked immunosorbent assay for determination of synap-tophysin as compared with other quantification procedures. J Neuroimmunol. 1996;67:59–65. doi: 10.1016/0165-5728(96)00049-5. [DOI] [PubMed] [Google Scholar]
- Schmidt GR, Hossner KL, Yemm RS, Gould DH, O’Callaghan JP. An enzyme-linked immunosorbent assay for glial fibrillary acidic protein as an indicator of the presence of brain or spinal cord in meat. J Food Prot. 1999;62:394–397. doi: 10.4315/0362-028x-62.4.394. [DOI] [PubMed] [Google Scholar]
- Siew LK, Love S, Dawbarn D, Wilcock GK, Allen SJ. Measurement of pre- and post-synaptic proteins in cerebral cortex: effects of post-mortem delay. J Neurosci Methods. 2004;139:153–159. doi: 10.1016/j.jneumeth.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Smith R, Klein P, Koc-Schmitz Y, Waldvogel HJ, Faull RL, Brundin P, Plomann M, Li JY. Loss of SNAP-25 and rabphilin 3a in sensory-motor cortex in Huntington’s disease. J Neurochem. 2007;103:115–123. doi: 10.1111/j.1471-4159.2007.04703.x. [DOI] [PubMed] [Google Scholar]
- Sorensen JB. SNARE complexes prepare for membrane fusion. Trends Neurosci. 2005;28:453–455. doi: 10.1016/j.tins.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Lottspeich F, Greengard P, Mehl E, Jahn R. A synaptic vesicle protein with a novel cytoplasmic domain and four transmembrane regions. Science. 1987;238:1142–1144. doi: 10.1126/science.3120313. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE. The synaptic Abeta hypothesis of Alzheimer disease. Nat Neurosci. 2005;8:977–979. doi: 10.1038/nn0805-977. [DOI] [PubMed] [Google Scholar]
- Terry RD. Cell death or synaptic loss in Alzheimer disease. J Neuropathol Exp Neurol. 2000;59:1118–1119. doi: 10.1093/jnen/59.12.1118. [DOI] [PubMed] [Google Scholar]
- Tsai J, Grutzendler J, Duff K, Gan WB. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat Neurosci. 2004;7:1181–1183. doi: 10.1038/nn1335. [DOI] [PubMed] [Google Scholar]