Abstract
Purpose
We identified molecular markers of erectile function, particularly those responding to erectile dysfunction treatment.
Materials and Methods
Sprague-Dawley retired breeder rats were intracorporeally injected with pVAX-hSlo, pSMAA-hSlo or the control plasmid pVAX. One week later the intracorporeal pressure-to-blood pressure ratio and gene expression were determined by microarray analysis and quantitative reverse transcriptase-polymerase chain reaction. Rat corporeal cells were transfected in vitro with pVAX-hSlo, pSMAA-hSlo or pVAX and the change in gene expression was determined. We also determined whether Vcsa1 expression was changed after pharmacotherapy using tadalafil.
Results
Animals treated with vectors expressing hSlo had significantly improved erectile function compared to that in controls, accompanied by changed expression of a subset of genes. Vcsa1 was one of the genes that was most changed in expression (the third of approximately 31,000 with greater than 10-fold up-regulation). Changes in gene expression were different than those observed in corporeal cells transfected in vitro, distinguishing gene expression changes that were a direct effect of hSlo over expression. When tadalafil was administered in retired breeder rats, the Vcsa1 transcript increased 4-fold in corporeal tissue compared to that in untreated controls.
Conclusions
Our study identifies a set of genes that are changed in response to improved erectile function, rather than as a direct effect of treatment. We noted Vcsa1 may act as marker of the restoration of erectile function after gene transfer and pharmacotherapy.
Keywords: penis, gene expression, tadalafil, muscle, smooth, microarray analysis
The development of ED is considered a multifactorial process, involving changes in the expression of several regulators of CSM tone.1 Recent microarray studies supports this hypothesis, demonstrating that the development of ED involves changes in the expression of multiple genes.2–6 However, few studies have focused on which genes are changed in expression following the restoration of erectile function after treatment. Published studies describe changes in gene expression in corporeal tissue after ED treatment with PDE5 inhibitors.5,7 These reports do not distinguish between the direct effect of the pharmacological agent on gene expression and effects that are secondary to improved erectile physiology.
Although the most prescribed drugs for ED are orally administered PDE5 inhibitors, potentially the next area of advancement for ED treatment will be a gene transfer approach.8 Intracorporeal injection of plasmids expressing the hSlo gene (encoding the MaxiK potassium channel) expressed from the cytomegalovirus promoter pVAX-hSlo or the smooth muscle α-actin promoter pSMAA-hSlo have been shown to restore erectile function in rat models of ED.9 Recently completed phase I clinical safety trials of the use of pVAX-hSlo to treat patients with ED show some evidence of efficacy.10 Despite evidence of the efficacy of intracorporeal injection of plasmids expressing hSlo to our knowledge there have been no published investigations of the impact of gene transfer on the expression of other genes or pathways.
We sought to identify molecular markers of erectile function, rather than the genes changed in direct response to treatment. One of the identified molecular markers was the Vcsa1 gene, which was up-regulated with the restoration of erectile function after gene transfer treatments. Subsequent experiments demonstrated that the Vcsa1 gene was also up-regulated following PDE5 mediated restoration of erectile function.
MATERIALS AND METHODS
Animals
Nine to 10-month-old male retired breeder Sprague-Dawley rats (Charles River Breeding Laboratories, Wilmington, Massachusetts) weighing greater than 500 gm were used in these experiments. They represent a commonly used model of age related ED.11,12 All animal protocols were approved by the Albert Einstein College of Medicine animal use committee.
Intracorporeal injection of 100 µg pVAX, pVAX-hSlo and pSMAA-hSlo into rats was previously described.9 One week after injection the ICP/BP response to electrostimulation of the cavernous nerve was determined.
In a second set of animals 1,000 µg pVAX-hSlo were administered to 10 animals 1 month before determining erectile function and gene expression. Five of these animals were treated with 2.5 mg/kg tadalafil orally 2 hours before ICP/BP measurement. A third group of 5 animals served as untreated controls and a fourth group of 5 was treated with 2.5 mg/kg tadalafil orally 2 hours before ICP/BP measurement. Mean ± SD ICP/BP and ANOVA were calculated in each treatment group.
Microarray Analysis
Following ICP/BP measurement the animals were sacrificed and corporeal tissue was isolated. Total RNA was extracted from frozen tissue with TRIzol®, as previously described.4,12,13 RNA was used to perform microarray analysis of global gene expression using the RGU-230A Affymetrix™ microarray. Quality control of RNA, labeling and hybridization was performed elsewhere according to Affymetrix protocols. Four chips each for control, pVAX-hSlo and pSMAA-hSlo treated animals were used for gene expression analysis using AffylmGUI software (http://www.bioconductor.org), as previously described.4
Rat CSM Cell Transfection
Rat CSM cells were isolated as described previously14 and grown in low glucose (1 gm/l) Dulbecco’s modified Eagle’s medium and 10% fetal bovine serum. Cells at passage 1 or 2 at 70% confluency were transfected with pVAX (control), pVAX-hSlo or pSMAA-hSlo using FuGENE® HD transfection reagent according to manufacturer instructions with a transfection reagent-to-DNA ratio of 3:2 in µl/µg. After 48 hours RNA was extracted with an RNeasy™ Mini Kit and used for quantitative RT-PCR.
Quantitative RT-PCR
Quantitative RT-PCR was performed as previously described.12,13,15 Appendix 1 lists the primers. Transcript expression was analyzed using the comparative crossing threshold (Ct) method (2−ΔΔCt), which was applicable because primer efficiency was found to be close to that of the housekeeping gene used to normalize samples.
APPENDIX 1.
Primers used to confirm microarray gene expression
| Gene | Primer |
|---|---|
| Vcsa1: | |
| Forward | 5′-GAGGGTGTCAGAGGCCC-3′ |
| Reverse | 5′-GAGCAGTTAGCTGCCACTGATA-3′ |
| Slo: | |
| Forward | 5′-TACTTCAATGACAATATCCTCACCCT-3′ |
| Reverse | 5′-ACCATAACAACCACCATCCCCTAAG-3′ |
| Expi: | |
| Forward | 5′-TGTTCCAATGGCTGTGGTCA-3′ |
| Reverse | 5′-GGCCATCAGTCGTGCTTATGA-3′; |
| Krt1-18: | |
| Forward | 5′-CAGACCTTGGAGATTGACCTGG-3′; |
| Reverse | 5′-TTGCTCCATCTGCACCCTGTA-3′; |
| Cav: | |
| Forward | 5′-ACCATCTTCGGCATCCCTATG-3′ |
| Reverse | 5′-AGGAAGCTCTTGATGCACGGT-3′; |
| Eef1a1: | |
| Forward | 5′-GTCAGAACGCAGGTGTTGTGAA-3′ |
| Reverse | 5′-GCCGGAATCTACGTGTCCAAT-3′; |
| Emp1: | |
| Forward | 5′-TCAAAGTGCATGCCCACCA-3′ |
| Reverse | 5′-GCGATGGAACATGTGCATCTC-3′ ‘; |
| RGD:1303126: | |
| Forward | 5′-TCTGACGGCAGGTCCTATGAGT-3′ |
| Reverse | 5′-TGGCCAGCATCTTTGCATC-3′. |
| Muc10: | |
| Forward | 5′-TCCCACCAAGGAGCAACATTAA-3′ |
| Reverse | 5′-GGATGTGGTTTTGGCTGGAAG-3′. |
| Alas2: | |
| Forward | 5′-ACCTCCCCTGCTGATTCAGAAT-3′ |
| Reverse | 5′-ACGGTATGTGTGGTCCTGCTTC-3′. |
| S100a9: | |
| Forward | 5′-ACCCTGAACAAGGCGGAATT-3′ |
| Reverse | 5′-TTTGTGTCCAGGTCCTCCATG-3′. |
| Pbsn: | |
| Forward | 5′-TGCTCACACTGGATGTGCTAGG-3′ |
| Reverse | 5′-TCCACGCTACTGGCAGCTAAGT-3′. |
| Rpl24: | |
| Forward | 5′-TCGAGCTGTGCAGTTTTAGTGG-3′ |
| Reverse | 5′-GCGGACTCACATTTGGCATTA-3′ |
| Glyceraldehyde-3-phosphate | |
| dehydrogenase | |
| Forward | 5′-GCCGCCTGCTTCACCACCTTCT-3′ |
| Reverse | 5′-GCATGGCCTTCCGTGTTCCTACC-3′ |
RESULTS
Treating Aging Rats With pVAX-hSlo or pSMAA-hSlo Restored Erectile Function
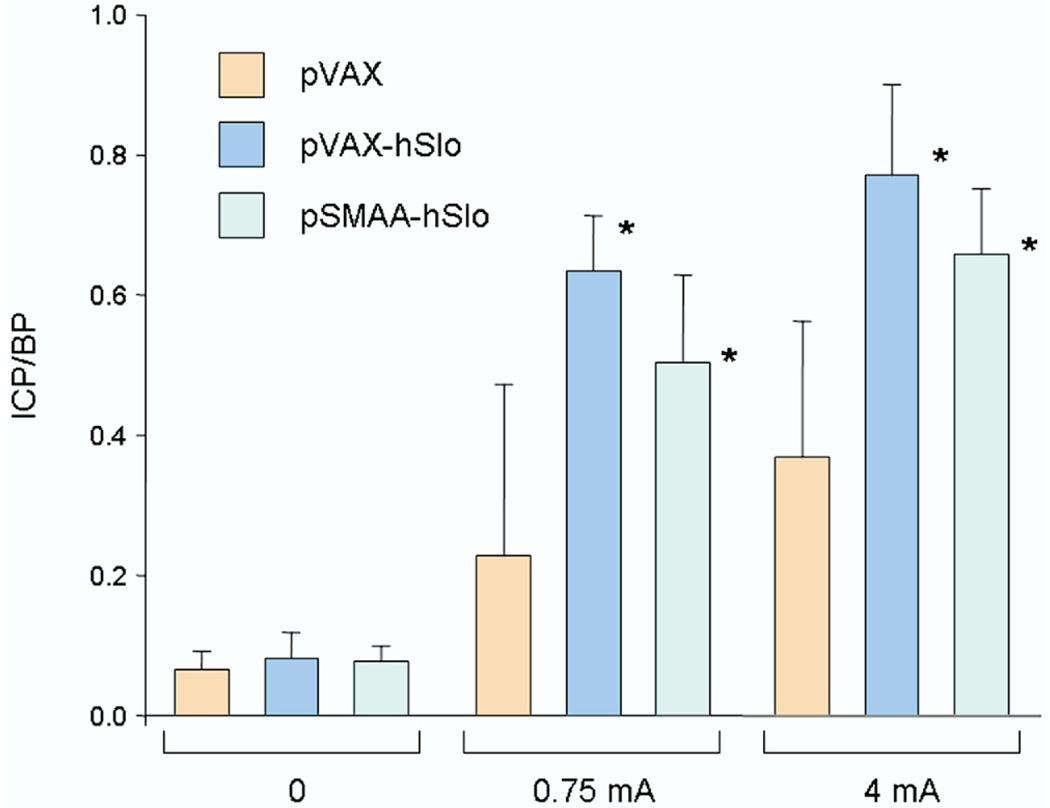
Three groups of 5 animals each received intracorporeal injections of 100 µg of plasmids expressing the hSlo gene (pVAX-hSlo and pSMAA-hSlo) or the empty backbone control (pVAX).9 Animals treated with either plasmid expressing hSlo showed significant improvement in erectile function compared to that in pVAX controls (fig. 1).
Figure 1.
Mean ICP/BP in 5 retired breeder rats treated with 100 µg pVAX, pVAX-hSlo and pSMAA-hSlo, respectively. Animals treated with pVAX-hSlo or pSMAA-hSlo showed significant ICP/BP increase in response to cavernous nerve stimulation with 0.75 or 4 mA compared to that in controls treated with empty plasmid vector pVAX. Asterisk indicates p <0.05.
Identification of Gene Expression Changes by Microarray Analysis
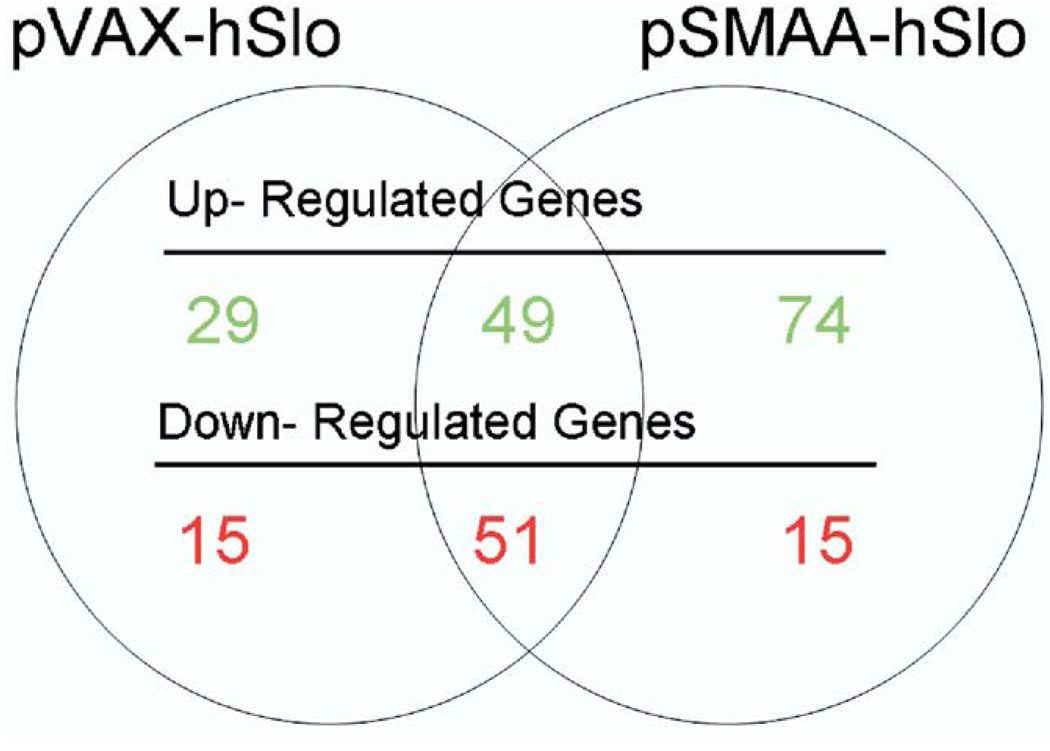
We compared gene expression in animals treated with pVAX-hSlo and pSMAA-hSlo to that in control animals treated with pVAX. In corpora treated with pVAX-hSlo 144 genes showed a statistically significant, greater than 1.5-fold change in expression compared to that in controls (B-statistic greater than 1), whereas animals treated with pSMAA-hSlo had a total of 189 genes with a greater than 1.5-fold level of change compared to that in controls. There was considerable overlap in genes that changed in expression (fig. 2), suggesting that treatment with either plasmid expressing hSlo triggered analogous physiological and molecular effects in vivo. Overall the changed genes represented less than 1% of the total genes (about 31,100) on the chip.
Figure 2.
Microarray analysis showed that approximately 144 and 189 genes were up-regulated and down-regulated in corpora of animals after treatment with pVAX-hSlo and pSMAA-hSlo, respectively. Intersection of circles indicates number of genes that 2 treatments shared in common.
We sorted the entire list of genes changed in expression by ontological themes using the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/home.jsp). Ontological analysis indicated that the keratins, which are the intermediate filament group of genes, were up-regulated after treatment (Appendix 2). Of the down-regulated genes those involved in transcription regulation were significantly overrepre-sented (Appendix 3).
APPENDIX 2.
Intermediate filament genes up-regulated after pVAX-hSlo or pSMAA-hSlo treatment
| Affymetrix Identification No. | Gene |
|---|---|
| 1371530_at | Keratin complex 2, basic, gene 8 |
| 1370868_at | Type II keratin kb1 |
| 1370863_at | Keratin complex 2, basic, gene 5 |
| 1388433_at | Keratin complex 1, acidic, gene 19 |
| 1373900_at | Keratin complex 2, basic, gene 7 (predicted) |
| 1372153_at | Type I keratin ka15 |
| 1388155_at | Keratin complex 1, acidic, gene 18 |
| 1373254_at | Keratin 10 |
Keratins belonging to the intermediate filament group of genes were up-regulated in corporeal tissue after treatment with pVAX-hSlo and pSMAA-hSlo (enrichment score 3.5).
APPENDIX 3.
Genes enriched in transcription regulation
| pVAX-hSlo |
pSMAA-hSlo |
||
|---|---|---|---|
| Affymetrix Identification No. |
Gene | Affymetrix Identification No. |
Gene |
| 1368813_at | ccaat/Enhancer binding protein (c/ebp), Δ | 1368813_at | ccaat/Enhancer binding protein (c/ebp), Δ |
| 1388088_a_at | Upstream transcription factor 2 | 1388088_a_at | Upstream transcription factor 2 |
| 1380552_at | Lymphoblastic leukemia derived sequence 1 | 1390399_at | Camp responsive element binding protein-like 2 |
| 1395695_at | ae Binding protein 1 (predicted) | 1380552_at | Lymphoblastic leukemia derived sequence 1 |
| 1367468_at | Scan domain-containing 1 (predicted) | 1367468_at | Scan domain-containing 1 (predicted) |
| 1372097_at | Interferon consensus sequence binding protein 1 | 1374974_at | Cell division cycle associated 7 |
| 1374974_at | Cell division cycle associated 7 | 1374085_at | Max dimerization protein 4 (predicted) |
Genes related to transcription regulation were identified as down-regulated in corporeal tissues after treatment with pVAX-hSlo (enrichment score 0.63) or pSMAA-hSlo (enrichment score 1.01)
Since genes with unknown function would not be included in an ontological grouping, we also focused on the 20 most up-regulated and down-regulated genes. Vcsa1 was up-regulated with each treatment (tables 1 and 2). This is of particular interest since Vcsa1 has been suggested as a marker of ED because it is down-regulated in several animal models of ED.3,12 Neither microarray analysis nor quantitative RT-PCR revealed significant changes in Slo gene expression (table 3). It is known that only a small number of cells uptake intracorporeally injected plasmid,11 so that it might be expected that hSlo expression would not be greatly affected in the whole corpora. This confirms past observations that despite the positive physiological effect of hSlo expressing plasmids on erectile function there is no effect on total Slo levels after 1 week.9
Table 1.
After pVAX-hSlo and pSMAA-hSlo treatment in rats 20 most up-regulated genes, respectively
| Gene Expression Change |
|||
|---|---|---|---|
| Gene Symbol | Gene | Log-Fold | Fold |
| pVAX-hSlo: | |||
| LOC362442* | Not available | 6.68 | 44.59 |
| Muc10* | Mucin 10, submandibular gland salivary mucin | 6.40 | 40.99 |
| RGD:708577* | Common salivary protein 1 | 6.06 | 36.70 |
| Vcsa1* | Variable coding sequence A1 | 5.56 | 30.86 |
| Not available* | Not available | 3.55 | 12.61 |
| Oit1_predicted* | Oncoprotein induced transcript 1 (predicted) | 2.99 | 8.92 |
| Igha* | Ig heavy chain (α polypeptide) | 2.84 | 8.06 |
| Cldn10_predicted* | Claudin 10 (predicted) | 2.77 | 7.69 |
| Tacstd1* | Tumor-associated calcium signal transducer 1 | 2.75 | 7.57 |
| Alas2† | Aminolevulinic acid synthase 2 | 2.70 | 7.29 |
| S100a9† | S100 calcium binding protein A9 (calgranulin B) | 2.50 | 6.25 |
| RGD1310209_predicted† | Similar to KIAA1324 protein (predicted) | 2.35 | 5.54 |
| LOC498228† | Not available | 2.27 | 5.16 |
| Hemgn | Hemogen | 2.21 | 4.87 |
| Krt1-18† | Keratin complex 1, acidic, gene 18 | 2.18 | 4.75 |
| Not available† | Not available | 2.17 | 4.73 |
| Not available | Not available | 2.10 | 4.39 |
| Hbb† | Hemoglobin β chain complex | 2.08 | 4.33 |
| Myo5c_predicted | Myosin VC (predicted) | 2.05 | 4.20 |
| RGD:1359209 | Globin, α | 2.03 | 4.12 |
| Acdc | Adipocyte complement related protein of 30 kDa | 2.03 | 4.11 |
| pSMAA-hSlo: | |||
| Muc10* | Mucin 10, submandibular gland salivary mucin | 7.07 | 50.05 |
| RGD:708577* | Common salivary protein 1 | 6.70 | 44.90 |
| LOC362442* | Not available | 6.34 | 40.16 |
| Vcsa1* | Variable coding sequence A1 | 6.06 | 36.70 |
| Pip† | Prolactin induced protein | 4.04 | 16.28 |
| Stfa3_predicted | Stefin A3 (predicted) | 3.96 | 15.66 |
| Not available* | Not available | 3.81 | 14.51 |
| Krtdap | Keratinocyte differentiation associated protein | 3.69 | 13.59 |
| Vcsa2 | Variable coding sequence A2 | 3.60 | 12.98 |
| Not available† | Not available | 3.44 | 11.83 |
| Oit1_predicted* | Oncoprotein induced transcript 1 (predicted) | 3.20 | 10.22 |
| Tacstd1* | Tumor-associated calcium signal transducer 1 | 3.19 | 10.18 |
| RGD:1359664 | Type II keratin Kb1 | 3.18 | 10.13 |
| Not available | Not available | 3.16 | 10.01 |
| Not available | Not available | 3.11 | 9.65 |
| Expi† | Extracellular peptidase inhibitor | 3.03 | 9.18 |
| Not available | Not available | 3.00 | 9.03 |
| Igha* | Ig heavy chain (a polypeptide) | 2.93 | 8.61 |
| Cldn10_predicted* | Claudin 10 (predicted) | 2.82 | 7.97 |
| Not available | Not available | 2.80 | 7.86 |
| RGD:1303044† | Type I keratin KA15 | 2.79 | 7.77 |
Top 20 changed genes per treatment group.
Genes changed in 2 treatment groups.
Table 2.
After pVAX-hSlo and pSMAA-hSlo treatment in rats 20 most down-regulated genes, respectively
| Gene Expression Change |
|||
|---|---|---|---|
| Gene Symbol | Gene | Log-Fold | Fold |
| pVAX-hSlo: | |||
| RGD:1303126* | SPARC-related modular calcium binding protein 1 | −2.40 | 5.77 |
| Col6a3_predicted | Procollagen, type VI, α3 (predicted) | −2.07 | 4.28 |
| Ptrf_predicted* | Polymerase I and transcript release factor (predicted) | −2.06 | 4.23 |
| Nedd4a† | Neural precursor cell gene 4A | −1.99 | 3.98 |
| Lcn2 | Lipocalin 2 | −1.97 | 3.89 |
| App† | Amyloid β (A4) precursor protein | −1.82 | 3.30 |
| Cav† | Caveolin | −1.74 | 3.04 |
| Prnd_predicted* | Prion protein dublet (predicted) | −1.72 | 2.95 |
| Not available* | Not available | −1.72 | 2.94 |
| Mfap2_predicted† | Microfibrillar-associated protein 2 (predicted) | −1.71 | 2.91 |
| RGD:1359529 | Similar to hypothetical protein dJ465N24.2.1 | −1.68 | 2.83 |
| Mmp3* | Matrix metallopeptidase 3 | −1.66 | 2.75 |
| Not available | Not available | −1.64 | 2.70 |
| LOC294435 | Not available | −1.59 | 2.54 |
| Aebp1_predicted | AE binding protein 1 (predicted) | −1.59 | 2.53 |
| Nisch_predicted | Nischarin (predicted) | −1.57 | 2.46 |
| Mtmr1_predicted† | Myotubularin related protein 1 (predicted) | −1.51 | 2.27 |
| Col18a1 | Collagen, type XVIII, α 1 | −1.48 | 2.20 |
| Eef1a1 | Eukaryotic translation elongation factor 1 α 1 | −1.46 | 2.14 |
| Not available | Not available | −1.46 | 2.13 |
| RGD1309414_predicted | Similar to KIAA0913 protein (predicted) | −1.46 | 2.13 |
| pSMAA-hSlo: | |||
| RGD:1303126* | SPARC-related modular calcium binding protein 1 | −2.00 | 4.00 |
| Ptrf_predicted* | Polymerase I and transcript release factor | −1.96 | 3.86 |
| LOC499533 | Not available | −1.77 | 3.15 |
| Prnd_predicted* | Prion protein dublet (predicted) | −1.66 | 2.77 |
| Mfap2_predicted | Microfibrillar-associated protein 2 (predicted) | −1.66 | 2.75 |
| RT1-Bb | RT1 class II locus Bb | −1.65 | 2.73 |
| Not available* | Not available | −1.61 | 2.60 |
| Mmp3* | Matrix metallopeptidase 3 | −1.53 | 2.35 |
| Not available | Not available | −1.52 | 2.31 |
| LOC360840† | Not available | −1.48 | 2.20 |
| Scgb2a1 | Secretoglobin, family 2A, member 1 | −1.45 | 2.11 |
| Not available | Not available | −1.44 | 2.08 |
| LOC503252 | Not available | −1.44 | 2.08 |
| Not available | Not available | −1.44 | 2.07 |
| LOC498539 | Not available | −1.43 | 2.05 |
| Ap1s1_predicted | Adaptor protein complex AP-1, Σ 1 | −1.43 | 2.05 |
| Lamc1† | Laminin, γ 1 | −1.43 | 2.05 |
| LOC360596 | Not available | −1.42 | 2.03 |
| Gnb2† | Guanine nucleotide binding protein, β polypeptide 2 | −1.42 | 2.00 |
| Fgfr1 | Fibroblast growth factor receptor 1 | −1.40 | 1.97 |
| Tm9sf4_predicted | Transmembrane 9 superfamily protein member 4 | −1.40 | 1.97 |
Top 20 changed genes per treatment group.
Genes changed in 2 treatment groups.
Table 3.
Quantitative RT-PCR of gene expression changes after pVAX-hSlo or pSMAA-hSlo intracorporeal injection
| Mean ± SD Fold Change vs pVAX |
||
|---|---|---|
| pVAX-hSlo | pSMAA-hSlo | |
| Vcsa1 | 10.56 ± 2.43 | 320 ± 82 |
| Slo | 1.23 ± 0.51 | 1.33 ± 0.61 |
| Muc10 | 17.425 ± 2.3 | 210.5 ± 142.5 |
| Alas2 | 19 ± 3.23 | 5.84 ± 1.34 |
| Pbsn | 4.7 ± 1.16 | 2.16 ± 1.05 |
| S100a9 | 6.46 ± 1.62 | 4.6 ± 1.875 |
| Krt1-18 | 1.56 ± 0.06 | 2.03 ± 0.23 |
| Expi | 11.16 ± 2.62 | 42.53 ± 11.23 |
We performed quantitative RT-PCR to confirm changes in expression of several of the genes identified as changed by microarray analysis (table 3). The ribosomal protein 24 gene (RPL24) was selected as the housekeeping gene since its expression was unchanged following treatment according to microarray analysis. Overall quantitative RT-PCR data supported the changes in gene expression identified by microarray data (tables 1 to 3). Differences in the fold change were likely due to the different methods of normalization used for quantitative RT-PCR and microarray analysis.
Gene Expression After CSM Cell Transfection With pVAX-hSlo or pSMAA-hSlo
We hypothesized that the changes in gene expression after treatment with pSMAA-hSlo or pVAX-hSlo were the result of a physiological improvement in erectile function, rather than a direct consequence of gene transfer of the plasmids expressing hSlo. To test this hypothesis we transfected rat CSM cells in vitro with pVAX-hSlo or pSMAA-hSlo plasmids and compared gene expression with that in cells transfected with pVAX. In contrast to the Slo levels detected after gene transfer of pVAX-hSlo and pSMAA-hSlo in vivo, Slo expression following the transfection of CSM cells with pVAX-hSlo was increased greater than 14-fold and greater than 100,000-fold with pSMAA-hSlo. Greater expression of the Slo gene from pSMAA-hSlo may be facilitated by greater efficiency of the smooth muscle α-actin promoter, as we have previously observed.9
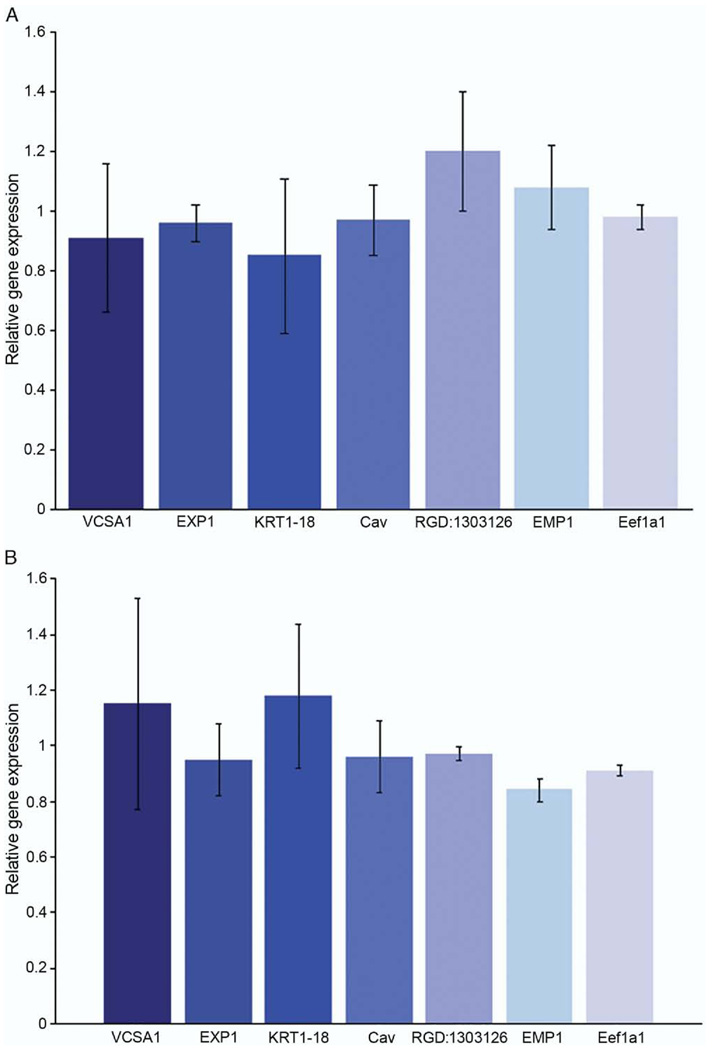
Using quantitative RT-PCR we analyzed genes that were up-regulated (Vcsa1, EXPI and KRT1–18) and down-regulated (Cav, RGD:1303126, EMP1 and Eef1a1) after gene transfer of pVAX-hSlo and pSMAA-hSlo administered in vivo (tables 1 and 2). None of these genes was significantly changed in expression in vitro (fig. 3). Therefore, the subset of genes identified as physiological markers for the recovery of erectile function following gene transfer are distinct from changes in gene expression that may occur in direct response to treatment, ie Slo gene over expression (tables 1 to 3).
Figure 3.
Transfection of rat CSM cells with pVAX-hSlo and pSMAA-hSlo did not affect expression of genes that were up-regulated or down-regulated after plasmid intracorporeal gene transfer. Change in gene expression was determined by quantitative RT-PCR, compared with that in control cells transfected with pVAX and averaged for 5 experiments. No significant change in gene expression was detected. A, transfection with pVAX-hSlo. B, transfection with pSMAA-hSlo. Error bars represent SE.
Tadalafil Administration Resulted in Vcsa1 Up-Regulation
Vcsa1, one of the genes changed as a physiological response to the recovery of erectile function, has previously been suggested to be a marker of ED in several animal models.3,12 We determined whether Vcsa1 expression correlated with the recovery of erectile function after the administration of the PDE5 inhibitor tadalafil alone or in association with pVAX-hSlo.
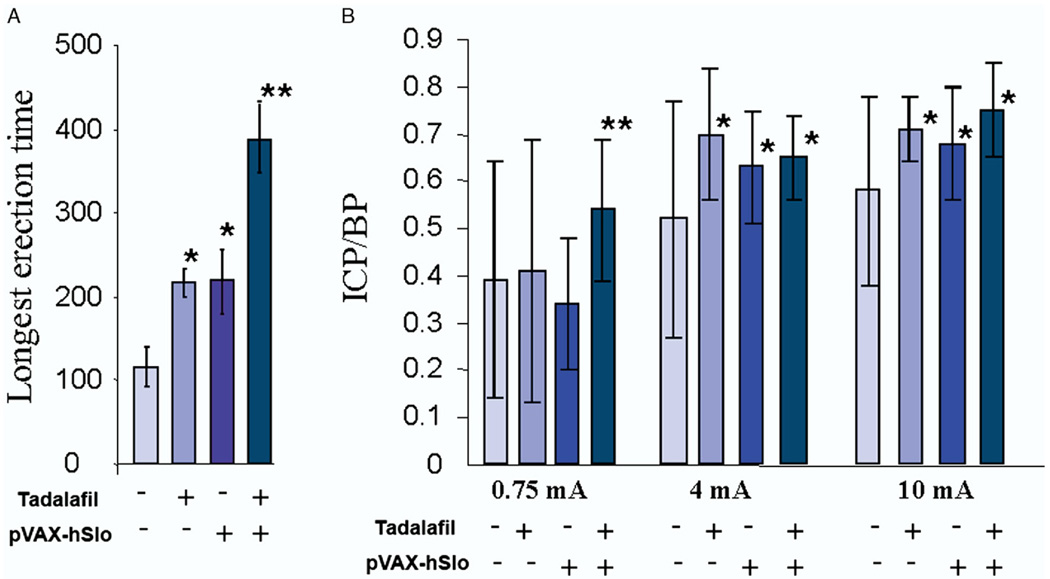
The longest visually observed erection and ICP/BP were determined (fig. 4). In rats treated with tadalafil or pVAX-hSlo the longest measured erection time was about 100 seconds longer than that in the untreated control groups, while a combination of treatments led to almost a 2-fold increase. Although all treatments produced a significant improvement in the ICP/BP response at 4 and 10 mA stimulation compared to that in control groups, combining the treatments showed a slight improvement in the ICP/BP ratio over that of the single treatment. At the lowest level of stimulation (0.75 mA) there was a significant increase in ICP/BP in animals receiving combined treatments compared to that in controls and animals receiving a single treatment. This suggests that combined treatments lower the level of stimulation needed to develop a significant increase in ICP/BP and, therefore, potentially an erection.
Figure 4.
Treatment with pVAX-hSlo and/or tadalafil improved erectile function. A, time of longest visually observed erection in seconds. B, ICP/BP response after electrostimulation at 0.75, 4 and 10 mA following various treatments to improve erectile function. Plus sign indicates 1,000 µg pVAX-hSlo intracorporeally injected 1 month before experiment and/or 2.5 mg/kg tadalafil administered orally 2 hours before erectile function evaluation. Error bars indicate animal with greatest ICP/BP difference from average in 5 animals. Single asterisk indicates significantly different vs control (p <0.05). Double asterisks indicate that tadalafil plus pVAX-hSlo was significantly different from pVAX-hSlo alone (p <0.05).
Improved Erection was Associated With Increased Vcsa1 Expression
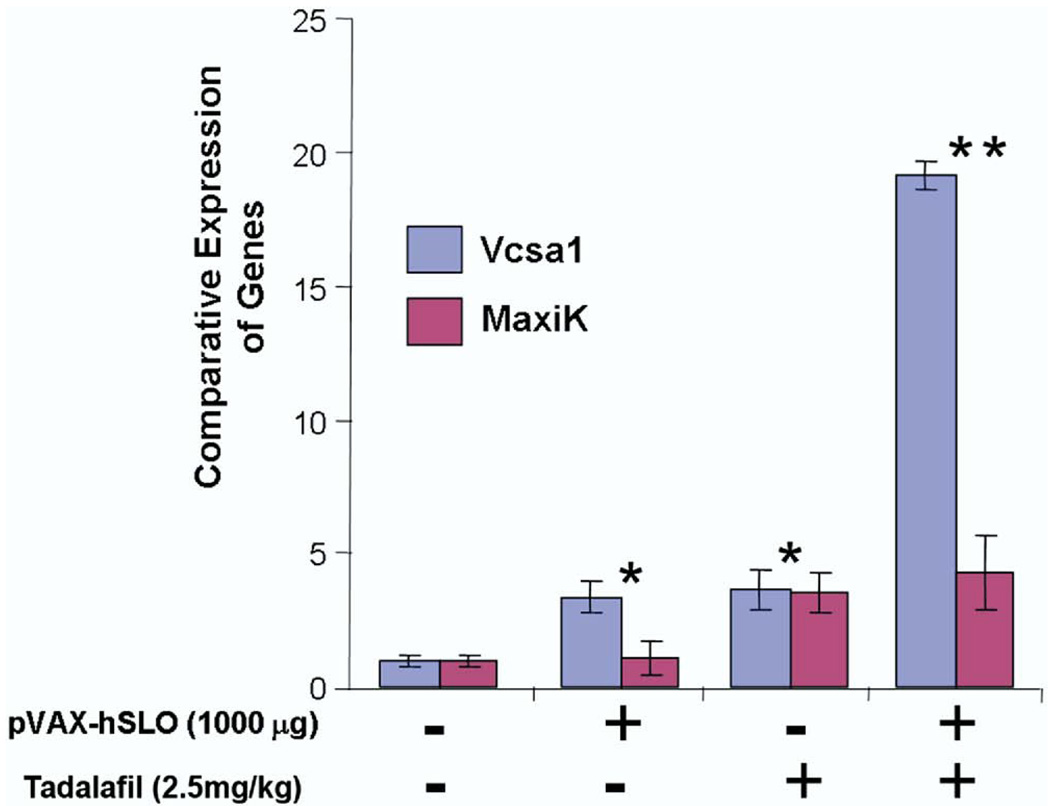
Quantitative RT-PCR was used to analyze expression of the Vcsa1 and hSlo transcripts after the administration of tadalafil, pVAX-hSlo or a combination of the 2 treatments. Intracorporeal gene transfer of pVAX-hSlo resulted in higher but not statistically higher levels of the hSlo transcript after 4 weeks (p >0.05, fig. 5). Quantitative RT-PCR was performed to verify whether the recovery of erection using different treatments still correlated with Vcsa1 up-regulation. Interestingly although tadalafil was administered only 2 hours before measuring erectile function, Vcsa1 expression was increased approximately 4-fold (figs. 4 and 5). Combination therapy seemed to have a synergistic effect on Vcsa1 expression. The detected level of the Vcsa1 transcript was approximately 20-fold greater than in untreated animals and 5-fold greater than after the individual treatments.
Figure 5.
Amount of Vcsa1 and MaxiK expression was measured by RT-PCR after erectile dysfunction treatment with pVAX-hSlo and/or tadalafil. After pVAX-hSlo or tadalafil Vcsa1 was up-regulated when erectile function was recovered, and combined treatment further enhanced Vcsa1 up-regulation. Single asterisk indicates significant increase in Vcsa1 expression in animals receiving pVAX-hSlo or tadalafil gene transfer vs that in untreated animals (p <0.05). Double asterisks indicate significant increase in Vcsa1 expression in animals with pVAX-hSlo plus tadalafil gene transfer vs that in animals receiving only 1 treatment type (p <0.05).
DISCUSSION
Our study shows that a subset of genes is changed in corporeal tissue after gene transfer of plasmids that express hSlo, resulting in improved erectile function in an aging rat model of ED. The genes that changed in these in vivo experiments were different than the genes that changed after over expression of hSlo in vitro. Therefore, we conclude that the subset of genes changed in vivo following intracorporeal injection of pVAX-hSlo or pSMAA-hSlo, which resulted in improved erectile function, represent molecular markers of erectile function. The change in expression may reflect the efficacy of ED treatment, rather than a direct response to treatment.
Our group is investigating a gene transfer approach to treat smooth muscle disorders of the urogenital system. The pVAX-hSlo vector has been shown to improve erectile and bladder function in animal models,11,16,17 and it has been evaluated in phase I clinical trials of ED treatment.10 Recently the gene transfer vector pSMAA-hSlo, in which hSlo is expressed from a smooth muscle specific promoter, was also shown to be effective for treating ED in aging rats.9 Gene transfer for ED treatment is useful for identifying markers of erectile function because 1) a single intracorporeal injection of pVAX-hSlo can improve erectile function in rats from 1 week to at least 6 months without a significant effect on the overall level of Slo in the penis and 2) only a small population of cells in the corpora actually take up the plasmid.11 Therefore, resulting gene changes are unlikely to be a direct effect of hSlo over expression, but rather a change in the physiology of corporeal tissue as a result of improved erectile function.
We identified a number of up-regulated and down-regulated genes on microarray analysis after the administration of pVAX-hSlo or pSMAA-hSlo. The 2 plasmids expressing hSlo each caused similar improvement in erectile function and there is considerable overlap between the lists of changed genes. Ontological analysis showed that intermediate filaments were up-regulated in animals treated with each plasmid. These proteins are involved in mechanically integrating the various components of the cytoplasmic space in eukaryotic cells. They are important regulators of smooth muscle tone and, therefore, over expression could be physiologically relevant, representing improved erectile function following gene transfer.18 Genes involved in transcriptional regulation are also overrepresented in the group of down-regulated genes, which may represent an adaptive response to improved erectile function.
The variable coding sequence protein A1, Vcsa1, was among the most up-regulated genes (tables 1 to 3). Vcsa1 has previously been suggested to be a marker of ED in several animal models.3,12 In addition, its human homologues ProL1 and hSMR3A/B are down-regulated in the corpora of patients with ED.13,19 Since we concluded that the set of genes that changed after treatment with plasmids expressing hSlo is indicative of restored erectile function, rather than a direct effect of treatment, we determined whether another ED treatment with a different mode of action would also change Vcsa1 expression. We treated retired breeder rats with the PDE5 inhibitor tadalafil. Although this treatment has a different mode of action to improve erectile function, it also resulted in a significant increase in Vcsa1 expression. Indeed, the change in Vcsa1 expression was impressively rapid, considering that tadalafil administration occurred 2 hours before the physiological/molecular determinations. In addition, our experiments showed a correlation between the degree of improvement of erection and the level of Vcsa1 up-regulation. Combined treatment with tadalafil and pVAX-hSlo in the animals demonstrated a significant increase in the longest observed erection time and greater sensitivity of the ICP/BP response to cavernous nerve stimulation with a corresponding 5-fold up-regulation of Vcsa1 when each treatment was used separately.
CONCLUSIONS
We identified a set of genes that act as molecular markers of erectile function. One of these markers, Vcsa1, is up-regulated in response to gene therapy and pharmacotherapy for ED. A combination of the 2 treatments causes a synergistic affect in improving erectile function, corresponding to a synergistic affect on Vcsa1 expression. Therefore, Vcsa1 might be useful for determining a quantitative measure of the efficacy of ED treatment. We are actively pursuing the development of immunoassays for the gene products of Vcsa1 in rats, and hSMR3 and ProL1 in humans. Indeed, to develop any practical assay for a marker of erectile function it will be necessary to confirm that changes at the gene expression level are mirrored at the protein level. Identifying an easily assayable objective marker for erectile function in humans would be of great advantage for evaluating the efficacy of ED treatments.
ACKNOWLEDGMENTS
Thomas Cao provided expertise on Excel®. RNA was labeled and hybridized to microarrays at the AECOM Affymetrix Facility, Albert Einstein College of Medicine.
Supported by National Institutes of Health/ National Institute of Diabetes and Digestive and Kidney Diseases Grants R21DK079594, R01DK077665 (KPD) and P01DK060037 (AM).
Abbreviations and Acronyms
- CSM
corporeal smooth muscle
- ED
erectile dysfunction
- ICP/BP
intracavernous pressure-to-systemic blood pressure ratio
- PDE5
phosphodiesterase type 5
- RT-PCR
reverse transcriptase-polymerase chain reaction
Footnotes
Study received Albert Einstein College of Medicine animal use committee approval.
REFERENCES
- 1.Andersson KE. Erectile physiological and patho-physiological pathways involved in erectile dysfunction. J Urol. 2003;170:S6. doi: 10.1097/01.ju.0000075362.08363.a4. [DOI] [PubMed] [Google Scholar]
- 2.Lin CS, Ho HC, Gholami S, Chen KC, Jad A, Lue TF. Gene expression profiling of an arteriogenic impotence model. Biochem Biophys Res Commun. 2001;285:565. doi: 10.1006/bbrc.2001.5191. [DOI] [PubMed] [Google Scholar]
- 3.User HM, Zelner DJ, McKenna KE, McVary KT. Microarray analysis and description of SMR1 gene in rat penis in a post-radical prostatectomy model of erectile dysfunction. J Urol. 2003;170:298. doi: 10.1097/01.ju.0000060882.75475.5a. [DOI] [PubMed] [Google Scholar]
- 4.Hipp JD, Davies KP, Tar M, Valcic M, Knoll A, Melman A, et al. Using gene chips to identify organ-specific, smooth muscle responses to experimental diabetes: potential applications to urological diseases. BJU Int. 2007;99:418. doi: 10.1111/j.1464-410X.2007.06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung HG, Shin JH, Kim KW, Yu JY, Kang KK, Ahn BO, et al. Microarray analysis of gene expression profile in the corpus cavernosum of hypercholes-terolemic rats after chronic treatment with PDE5 inhibitor. Life Sci. 2007;80:699. doi: 10.1016/j.lfs.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan CJ, Teal TH, Luttrell IP, Tran KB, Peters MA, Wessells H. Microarray analysis reveals novel gene expression changes associated with erectile dysfunction in diabetic rats. Physiol Genom. 2005;23:192. doi: 10.1152/physiolgenomics.00112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrade E, Andrade PM, Borra RC, Claro J, Srougi M. cDNA microarray analysis of differentially expressed genes in penile tissue after treatment with tadalafil. BJU Int. 2008;101:508. doi: 10.1111/j.1464-410X.2007.07285.x. [DOI] [PubMed] [Google Scholar]
- 8.Melman A, Feder M. Gene therapy for the treatment of erectile dysfunction. Nat Clin Pract Urol. 2008;5:60. doi: 10.1038/ncpuro1014. [DOI] [PubMed] [Google Scholar]
- 9.Melman A, Biggs G, Davies K, Zhao W, Tar MT, Christ GJ. Gene transfer with a vector expressing Maxi-K from a smooth muscle-specific promoter restores erectile function in the aging rat. Gene Ther. 2008;15:364. doi: 10.1038/sj.gt.3303093. [DOI] [PubMed] [Google Scholar]
- 10.Melman A, Bar-Chama N, McCullough A, Davies K, Christ G. The first human trial for gene transfer therapy for the treatment of erectile dysfunction: preliminary results. Eur Urol. 2005;48:314. doi: 10.1016/j.eururo.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Christ GJ, Rehman J, Day N, Salkoff L, Valcic M, Melman A, et al. Intracorporal injection of hSlo cDNA in rats produces physiologically relevant alterations in penile function. Am J Physiol. 1998;275:H600. doi: 10.1152/ajpheart.1998.275.2.H600. [DOI] [PubMed] [Google Scholar]
- 12.Tong Y, Tar M, Davelman F, Christ G, Melman A, Davies KP. Variable coding sequence protein A1 as a marker for erectile dysfunction. BJU Int. 2006;98:396. doi: 10.1111/j.1464-410X.2006.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong Y, Tar M, Melman A, Davies K. The opiorphin gene (ProL1) and its homologues function in erectile physiology. BJU Int. 2008;102:736. doi: 10.1111/j.1464-410X.2008.07631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson WF, Huebner JM, Rusch NJ. Enzymatic isolation and characterization of single vascular smooth muscle cells from cremasteric arte-rioles. Microcirculation. 1996;3:313. doi: 10.3109/10739689609148305. [DOI] [PubMed] [Google Scholar]
- 15.Tong Y, Tiplitsky SI, Tar M, Melman A, Davies KP. Transcription of G-protein coupled receptors in corporeal smooth muscle is regulated by the endogenous neutral endopeptidase inhibitor sialorphin. J Urol. 2008;180:760. doi: 10.1016/j.juro.2008.03.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christ GJ, Day NS, Day M, Santizo C, Zhao W, Sclafani T, et al. Bladder injection of “naked” hSlo/pcDNA3 ameliorates detrusor hyperactivity in obstructed rats in vivo. Am J Physiol Regul Integr Comp Physiol. 2001;281:R1699. doi: 10.1152/ajpregu.2001.281.5.R1699. [DOI] [PubMed] [Google Scholar]
- 17.Melman A, Zhao W, Davies KP, Bakal R, Christ GJ. The successful long-term treatment of age related erectile dysfunction with hSlo cDNA in rats in vivo. J Urol. 2003;170:285. doi: 10.1097/01.ju.0000063375.12512.6e. [DOI] [PubMed] [Google Scholar]
- 18.Tang DD. Intermediate filaments in smooth muscle. Am J Physiol Cell Physiol. 2008;294:C869. doi: 10.1152/ajpcell.00154.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong Y, Tar M, Monrose V, DiSanto M, Melman A, Davies KP. hSMR3A as a marker for patients with erectile dysfunction. J Urol. 2007;178:338. doi: 10.1016/j.juro.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]