Abstract
Bcl-6 and Blimp-1 have recently been identified as key transcriptional regulators of effector and memory differentiation in CD4+ T cells and CD8+ T cells. Bcl-6 and Blimp-1 were previously known to be critical regulators of effector and memory differentiation of B lymphocytes. The new findings unexpectedly point to the Bcl-6 and Blimp-1 regulatory axis as a ubiquitous mechanism for controlling effector and memory lymphocyte differentiation and function. Bcl-6 and Blimp-1 are antagonistic transcription factors and can function as a self-reinforcing genetic switch for cell-fate decisions. However, their influences in different lymphocytes are complex. Here we review and examine the commonalities and differences in the functions of these transcription factors in CD4+ follicular helper TFH lymphocytes, effector CD8+ T lymphocytes and B lymphocytes.
CD4+ T cells, CD8+ T cells and B cells are distinct lymphocyte populations with unique, critical roles in adaptive immunity. Each population has been thought to be controlled by quite different transcriptional regulation for its individual effector and memory differentiation programs1. Bcl-6 and Blimp-1 (encoded by Prdm1) are transcriptional repressors with the ability to block each other's expression (Fig. 1). The recent identification of central roles for Bcl-6 and Blimp-1 in the differentiation of follicular helper CD4+ T cells2–4 and CD8+ T cell effector and memory differentiation processes5–7, combined with their well documented roles in B cells, has revealed an unexpected common system used in key fate decisions made by mature CD4+ T cells, CD8+ T cells and B cells.
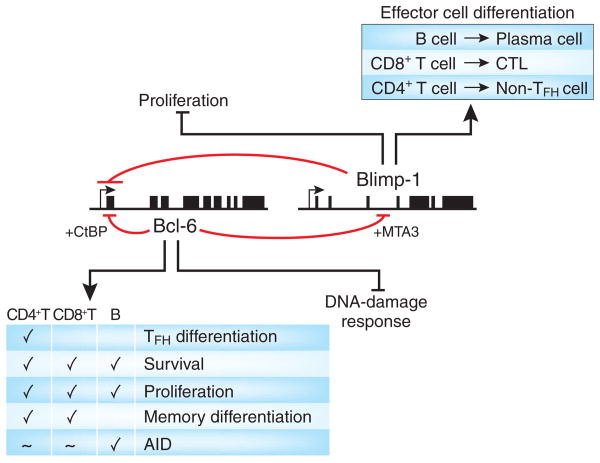
Figure 1.
Bcl-6 and Blimp-1 are reciprocally antagonistic transcription factors. Mechanisms of Bcl-6 and Blimp-1 antagonism and their broad influences on lymphocyte differentiation and function. CTL, cytotoxic T lymphocyte.
Bcl-6 and Blimp-1 in B cell differentiation
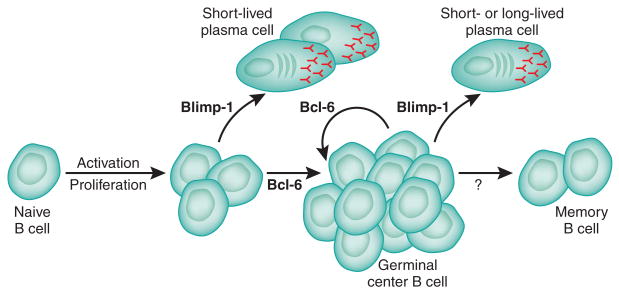
Bcl6 was first identified as a proto-oncogene frequently expressed in non-Hodgkin's lymphoma as a result of chromosomal translocations8–12. Bcl-6 protein was found to be highly expressed in germinal center B cells, which led to the conclusion that many B cell lymphomas begin as germinal center B cells that then acquire dysregulated Bcl-6 expression11. The germinal center reaction is the process by which antigen-specific B cells mature into the long-lived plasma cells (antibody-secreting cells) and memory B cells that constitute the cellular components of immunological memory for the B cell lineage1,13 (Fig. 2). After activation and costimulation, B cells differentiate into either short-lived plasma cells or germinal center B cells1,14 (Fig. 2). Short-lived plasma cells stop proliferating and elaborate a large secretory apparatus for producing antibodies to quickly combat infection1,14. In contrast, germinal center B cells have a minimal secretory apparatus and are instead specialized for proliferation and affinity maturation, the exquisitely complex process of rapidly evolving a B cell receptor of higher affinity for the production of high-affinity, isotype-switched antibody11,15. This process involves rapid proliferation and substantial amounts of DNA damage (both class-switch recombination and somatic hypermutation, controlled by activation-induced cytidine deaminase (AID)16). Bcl-6 is a crucial inducer of germinal center B cell proliferation and a crucial inhibitor of the DNA-damage response11 (Figs. 1 and 2). Bcl-6-deficient mice have an absence of germinal center B cells and affinity maturation17–19. Bcl-6-deficient B cells are nevertheless able to differentiate into plasma cells and secrete antibodies18,20, although long-term antibody responses in Bcl6−/− mice are considerably diminished, as the germinal center reaction is required to produce the majority of long-lived plasma cells19 (Fig. 2). Constitutive expression of Bcl-6 in B cells in vivo results in large germinal centers, which again confirms the central role of Bcl-6 in germinal center B cell differentiation21.
Figure 2.
Roles of Bcl-6 and Blimp-1 in B cell differentiation. Bcl-6 and Blimp-1 are required at various steps of B cell differentiation (as indicated by arrow labels).
Plasma cell differentiation, in contrast, critically depends on Blimp-1 (refs. 22,23) and the absence of Bcl-6 (refs. 11,23). The role of Blimp-1 in B cell development was first demonstrated by the ability of ectopic Blimp-1 expression to induce human B lymphoma cells to differentiate into cells with plasma cell features24. Blimp-1 is highly expressed in plasma cells and controls many genes important for plasma cell differentiation23,25–27, including Xbp1, which induces formation of the secretory apparatus necessary for the production of large amounts of antibody28,29. Blimp-1 also inhibits genes involved in cellular proliferation, such as Myc and Bcl6, which thereby allows the terminal differentiation of plasma cells into a post-mitotic state23,30.
Genetic ablation of Blimp-1 in B cells prevents mature B cells from differentiating into either short-lived or long-lived plasma cells, which results in dramatically reduced antibody titers22. Interestingly, germinal centers are enlarged in these mice, and memory B cells are generated22. Together with experiments using ectopic expression of Blimp-1, these data suggest that Blimp-1 is a master regulator of plasma cell differentiation with limited or no roles in germinal center B cell and memory B cell differentiation (Fig. 2).
Bcl-6 protein can bind to Prdm1 and directly repress Blimp-1 expression20,31,32 (Fig. 1) and thereby induce germinal center differentiation while inhibiting plasma cell differentiation11. Conversely, Blimp-1 protein can bind to Bcl6 and directly repress Bcl-6 production33 (Fig. 1) and thereby induce plasma cell differentiation while inhibiting germinal center B cell differentiation23,25. Although details about the kinetics of the expression of Bcl-6 and Blimp-1 during B cell activation and differentiation in vivo still remain to be fully elucidated25,34, the reciprocal antagonism between Bcl-6 and Blimp-1 clearly provides a powerful mechanism for inducing and then reinforcing bimodal fate ‘decisions’ in B cell effector and memory differentiation.
Bcl-6 and Blimp-1 in CD4+ T cell differentiation
Although Bcl-6 and Blimp-1 have been studied intensively in B cells for some 15 years, the functions of Bcl-6 and Blimp-1 in T lymphocytes remained relatively unexplored until recently25. A series of new data has now demonstrated that Bcl-6 is the master regulator of CD4+ follicular helper T cells (TFH cells)2–4 and that Blimp-1 is a critical antagonist of TFH differentiation2.
CD4+ T cells can differentiate into at least four different effector lineages (T helper type 1 (TH1) cells, TH2 cells, TH17 cells and regulatory T cells (Treg cells)), which allows them to powerfully tailor the larger immune response to best clear a given infection or control inflammation35. CD4+ T cell help to B cells is essential for the generation of germinal centers, memory B cells and long-lived plasma cells1. CD4+ TH2, TH1 and TH17 cells are not required for germinal center formation or B cell help36–38. Therefore, it had been proposed, mainly on the basis of gene-expression profiling and in vitro T cell help–B cell help assays, that TFH may represent a distinct CD4+ T cell effector lineage specialized for B cell help39–41. However, unlike the other effector lineages, until recently TFH cells had no lineage-specifying master regulator transcription factor (such as T-bet, GATA-3, RORγt or Foxp3)42.
Three new studies have now shown that Bcl-6 is a master regulator of TFH differentiation2–4. Constitutive expression of Bcl-6 in CD4+ T cells drives nearly absolute TFH differentiation in vivo2. These TFH cells induce larger germinal centers and higher antigen-specific antibody titers2. In contrast, Bcl6−/− CD4+ T cells are unable to differentiate into TFH cells in vivo, which demonstrates that Bcl-6 is necessary for TFH differentiation2–4. Bcl6−/− CD4+ T cells become activated and proliferate in response to protein immunization but are unable to drive germinal center formation2–4. These results confirm that TFH cells are a distinct subset of helper CD4+ T cells, reveal that Bcl-6 is a TFH master regulator both necessary and sufficient for TFH differentiation, and demonstrate that TFH cells are uniquely able to drive the germinal center reaction.
Interestingly, although TFH cells have high Bcl-6 expression42, the remaining antigen-specific CD4+ T cells (non-TFH cells) have high Blimp-1 expression 2,43,44, which indicates that Bcl-6 versus Blimp-1 expression may be a central cell fate decision of differentiating CD4+ T cells. Constitutive expression of Blimp-1 blocks Bcl-6 expression and TFH differentiation but does not block the proliferation and differentiation of non-TFH CD4+ cells2. Like Bcl6−/− CD4+ T cells, Blimp-1-expressing CD4+ T cells cannot induce germinal center formation, which results in considerably reduced antibody titers2. Conversely, Blimp-1-deficient CD4+ T cells preferentially differentiate into TFH cells in vivo2. Therefore, in CD4+ T cells, Blimp-1 is a physiological regulator of Bcl-6 expression, and vice versa2.
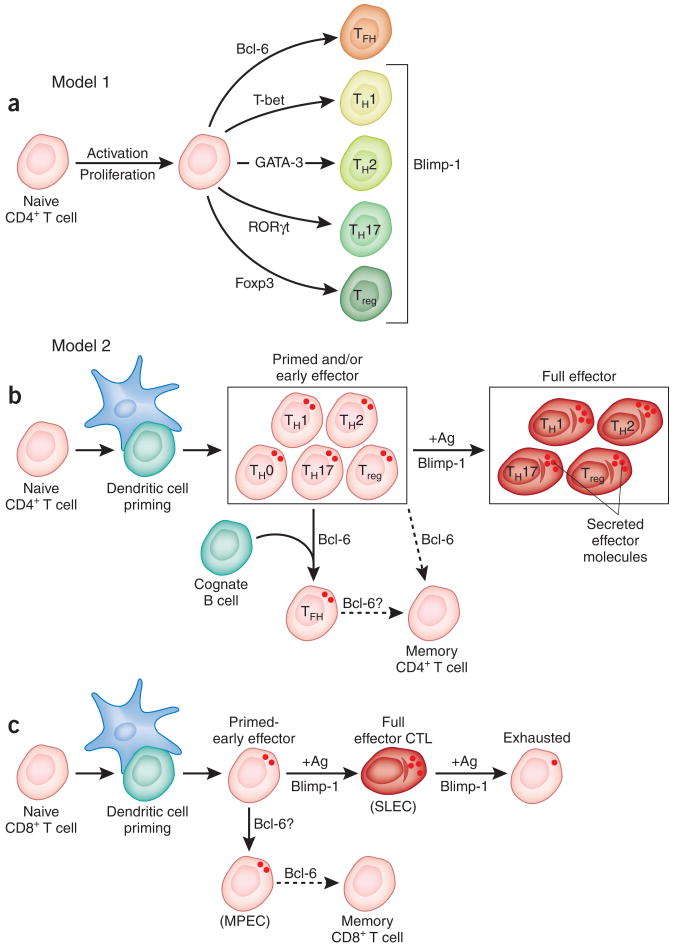
There are two models for interpreting the Bcl-6 and Blimp-1 results outlined above in the context of CD4+ T cell lineage differentiation (Fig. 3a,b). The simplest model proposes that TFH differentiation occurs via a fully independent differentiation pathway controlled by Bcl-6, analogous to the differentiation pathways for TH1, TH2, TH17 and inducible Treg (iTreg) cells37 (model 1; Fig. 3a). A second model posits that TFH cells are a distinct type of effector CD4+ T cell but that the TFH differentiation pathway is not fully independent of TH1, TH2, TH17 or iTreg differentiation2,45 (model 2; Fig. 3b). In this second model, a CD4+ T cell is primed by a dendritic cell and acquires early TH1, TH2, TH17, iTreg or unbiased TH0 cell characteristics (Fig. 3b). The primed CD4+ T cell (early effector) can then further polarize into a Blimp-1hi full effector TH1 cell2, TH2 cell2,43, TH17 cell or iTreg cell46 (Fig. 3b). Alternatively, the primed CD4+ T cell may encounter a B cell—which induces high Bcl-6 expression—and differentiate into a TFH cell (Fig. 3b). In support of model 1, there is evidence that Bcl-6 can inhibit TH1, TH2 and/or TH17 gene expression3,4,17,47, and this model fits well with conventional helper T cell schematics. However, several lines of evidence indicate that the second model is probably a more accurate representation of the in vivo biology. First, TFH cells produce TH1 cytokines2,48–50, TH2 cytokines,43,48,50–52 or TH17 cytokines53. The production of TH1, TH2 or TH17 cytokines probably enables TFH cells to specify the antibody isotypes produced during the B cell response50. Nevertheless, TFH cells produce lower amounts of TH1, TH2 or TH17 cytokines than do non-TFH cells2,4,43, which express Blimp-1 (refs. 2,43). Therefore, Bcl-6-expressing TFH cells seem to have some TH1, TH2 or TH17 characteristics acquired at the initial dendritic cell priming (Fig. 3b), but those characteristics are partially subdued in the presence of Bcl-6. Second, dendritic cell priming is insufficient for TFH differentiation in vivo, as TFH cells are not observed in the absence of cognate B cells2,54, which indicates that the interaction of a primed CD4+ T cell with a cognate B cell is a key second signal for the induction of TFH differentiation2,45,54. TFH differentiation can be restored in B cell–deficient mice by the expression of Bcl-6 in antigen-specific CD4+ T cells, which suggests that cognate B cells specifically induce Bcl-6 expression in CD4+ T cells2 (Fig. 3b).
Figure 3.
Roles of Bcl-6 and Blimp-1 in T cell differentiation. (a,b) Two proposed models of effector CD4+ T cell differentiation: model 1 (a) and model 2 (b). in b, the partial TH1, TH2, Treg and TH17 characteristics of TFH cells are not shown, for simplicity. (c) A model of CD8+ T cell effector and memory differentiation highlighting the similarities to CD4+ T cells. Dotted lines and question marks indicate uncertainty. +Ag, antigen stimulation; SLeC, short-lived effector cell; MPeC, memory precursor effector cell.
Blimp-1 is expressed late in CD4+ T cell differentiation in vitro25,55. Blimp-1 expression is therefore associated with highly polarized effector CD4+ T cells, late in differentiation, with a high cytokine-secretion capacity (Fig. 3b). This is intriguingly similar to the role of Blimp-1 in effector CD8+ T cell differentiation (Fig. 3c; discussed below) and similar to (although less extreme than) its role in plasma cells, which have high expression of Blimp-1 and an enormous secretion capacity (Fig. 4). Furthermore, the highly polarized Blimp-1-expressing effector CD8+ T cells or plasma cells have limited or no proliferative capacity, respectively, which suggests that Blimp-1hi effector CD4+ T cells may have similar properties (Fig. 4). Blimp-1-deficient T cells exhibit hyperproliferation in vivo, which results in autoimmunity46,56. Early CD4+ T cell effector differentiation depends on neither Bcl-6 nor Blimp-1 (refs. 3,46,56). Those findings are consistent with our model, in which Blimp-1 is not required for the priming and development of CD4+ T cell effector functions (cytokine secretion) but is critical for full effector cell differentiation and inhibition of the proliferation of those effector cells (Fig. 3b).
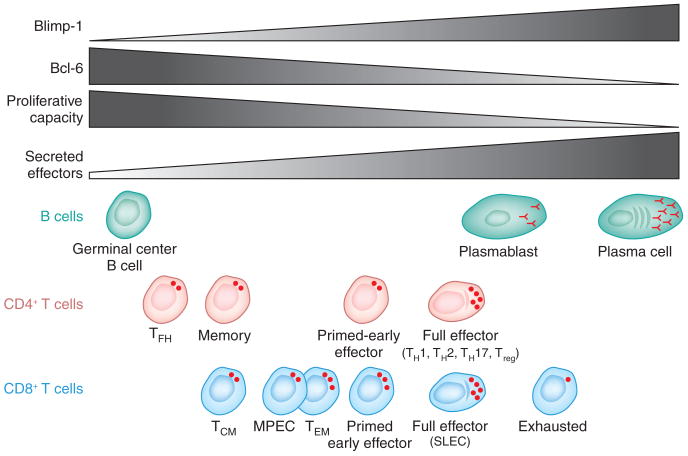
Figure 4.
Functional effects of different amounts of Bcl-6 and Blimp-1 expression. The amount of Bcl-6 and Blimp-1 expression is balanced differently in different lymphocytes at distinct differentiation stages. Lymphocytes with higher expression of Bcl-6 exhibit greater proliferative capacity but less secretory capacity. Lymphocytes with higher expression of Blimp-1 exhibit lower proliferative capacity and greater secretory capacity. Although exhausted CD8+ T cells have large amounts of Blimp-1 and lower proliferative capacity, they do not have more secreted effectors. TCM, central memory T cell; TEM, effector memory T cell.
Although Bcl-6 and Blimp-1 are clearly central to effector CD4+ T cell differentiation (and possibly memory, as discussed below), many aspects of their involvement remain unclear. In particular, how Bcl-6 controls TFH differentiation is an area of intensive study. Initial reports have indicated that Bcl-6 can regulate key cytokine receptors (such as the interleukin 6 receptor (IL-6R) or IL-21R)3 and suppress an important cluster of microRNAs4, as well as suppress TH1, TH2 and TH17 gene expression3,4,44. The specific roles of Blimp-1 in CD4+ T cell effector differentiation remain to be extensively explored.
Bcl-6 and Blimp-1 in CD8+ T cell differentiation
Naive CD8+ T cells differentiate into cytotoxic effectors (cytotoxic T lymphocytes) that are critical for the control and clearance of intracellular pathogens. Recent studies have shown that Blimp-1 is expressed in effector and memory CD8+ T cells46,56,57 and that the absence of Blimp-1 can result in excessive proliferation and increased numbers of memory CD8+ T cells56. Recent data have now convincingly demonstrated that Blimp-1 is an important regulator of effector CD8+ T cell function, proliferation and conversion into memory cells5–7. Virus-specific Blimp-1-deficient CD8+ T cells fail to differentiate into KLRG1hiIL-7Rlo cells6,7, also known as short-lived effector cells because of their effector functions and limited ability to survive and convert into memory cells58. Instead, Blimp-1-deficient CD8+ T cells preferentially differentiate into KLRG1loIL-7Rhi memory precursor effector cells6,7, which exhibit better survival than short-lived effector cells and have the potential to convert into memory CD8+ T cells58 (Fig. 3c). Blimp-1-deficient CD8+ T cells have lower expression of effector molecules important for cytotoxicity, such as granzyme B5–7. Nonetheless, Blimp-1-deficient CD8+ T cells have sufficient effector functions to control and clear acute infection with lymphocytic choriomeningitis virus or influenza virus6,7. Together these studies elegantly demonstrate that Blimp-1 expression is required for the terminal differentiation of effector CD8+ T cells6,7.
Blimp-1-deficient CD8+ T cells express more Bcl-6 than do wild-type CD8+ T cells6,7. Constitutive Bcl-6 expression suppresses granzyme B expression59. Conversely, Bcl6−/− CD8+ T cells express more granzyme B59. Bcl-6 and Blimp-1 are therefore probably reciprocal regulators of CD8+ T cell effector differentiation.
Certain commonalities can be gleaned from the CD8+ and CD4+ T cell studies about how Blimp-1 and Bcl-6 regulate these cell types. In both CD8+ and CD4+ T cells, Blimp-1 is critical for most terminal effector cell differentiation (Fig. 3b,c). Terminal differentiation is characterized by low proliferative potential and high effector molecule secretion (Fig. 4). Blimp-1-deficient CD8+ T cells are diverted to a memory-precursor differentiation pathway with enhanced proliferative potential and have lower effector molecule production (Figs. 3 and 4). Blimp-1-deficient CD4+ T cells are diverted away from terminal TH1 or TH2 differentiation and toward TFH differentiation2 (Fig. 3) and exhibit greater proliferative potential46,56. These proliferation and secretion commonalities of Blimp-1-expressing T cells are again shared with the B cell lineage, in which B cells with high Blimp-1 expression are terminally differentiated effectors (plasma cells) with low proliferative potential and high effector molecule production (Fig. 4).
Regulation of T and B cell memory by Bcl-6 and Blimp-1
Blimp-1-deficient CD8+ T cells preferentially differentiate into memory CD8+ T cells6,7,56, and Bcl-6 is upregulated in memory CD8+ T cells60. Blimp-1 is also expressed differently by the two main memory CD8+ T cell subsets6,7. Effector memory T cells, which are functionally potent memory CD8+ T cells that patrol the periphery but proliferate poorly58,61, have relatively high Blimp-1 expression. In contrast, central memory T cells have relatively low Blimp-1 expression6,7 (Fig. 4) and are characterized by lower immediate effector functions and high proliferative potential58,61.
Bcl-6-deficient CD8+ T cells proliferate poorly and are less able to acquire a memory-like phenotype62. Conversely, CD8+ T cells overexpressing Bcl-6 generate a larger memory cell population63. There is also some limited evidence that Bcl-6 has a role in memory CD4+ T cells. Bcl-6-deficient CD4+ T cells have an impaired ability to persist as memory cells after immunization64. Consistent with that idea, Blimp-1 expression is lower in memory CD4+ T cells than in effector CD4+ T cells46. Given that Bcl-6 promotes proliferation, Bcl-6 expression may be a key attribute of CD8+ and CD4+ T cell memory differentiation that Blimp-1 inhibits to direct cells toward terminal effector differentiation.
A simple model is that expression of Bcl-6 and Blimp-1 is a bimodal, self-reinforcing switch between effector and memory T cell differentiation, which is conserved between CD8+ and CD4+ T cells. However, this is an oversimplification, because among effector CD4+ T cells, only TFH cells depend on Bcl-6 expression2–4 and the absence of Blimp-1 (ref. 2). Furthermore, effector CD8+ T cells have a substantial amount of functionality in the absence of Blimp-1 (refs. 5–7,56). A somewhat more sophisticated model (Fig. 3b,c) incorporates the multiple stages of these CD4+ and CD8+ T cell differentiation pathways and highlights possible parallels in the activities of Bcl-6 and Blimp-1 in each lineage. Of course, many other proteins are also important participants in CD4+ and CD8+ T cell differentiation and functionality, but here we are focusing only on the contributions of Bcl-6 and Blimp-1.
The parallels between the functions of Blimp-1 and Bcl-6 in CD4+ and CD8+ T cells also raise a challenging question about the relationship between memory CD4+ T cells and the TFH and non-TFH effector CD4+ T cell lineages (Fig. 3b,c). Do Bcl-6hiBlimp-1lo TFH cells convert more readily into memory cells than do Bcl-6loBlimp-1hi effector CD4+ T cells? Additional studies of the mechanistic role of Bcl-6 in CD8+ and CD4+ T cell memory differentiation are much needed.
The role of Bcl-6 in memory B cell differentiation is controversial13,65,66. Bcl-6 is generally considered crucial for the generation of B cell memory, as long-lived plasma cells and memory B cells are mainly products of germinal centers (Fig. 2). It has been proposed that Bcl-6 expression is necessary to prevent germinal center B cells from differentiating into plasma cells, allowing them to differentiate into memory B cells instead65,67. Human memory B cells do not express Blimp-1 (refs. 66,68). It has also been shown that Bcl-6 expression can enhance B cell survival and thus may regulate memory B cell self-renewal19,69. However, Bcl-6 mRNA expression is not correlated with memory B cell differentiation in mice70 or humans66, and some memory B cells can develop in Bcl-6-deficient mice in the absence of germinal centers19. High Bcl-6 expression sustains a germinal center B cell phenotype and inhibits human memory B cell differentiation in vitro66. It remains to be determined what transcription factor induces memory B cell differentiation from Bcl-6-expressing germinal center B cells.
Blimp-1 in exhausted CD8+ T cells
Effector CD8+ T cells can become exhausted by persistent antigen presentation during chronic viral infection71,72. Chronic viral infections associated with T cell exhaustion, such as infection with human immunodeficiency virus or hepatitis C virus, are the cause of enormous human suffering and loss of life73. Exhausted CD8+ T cells are functionally impotent and proliferate poorly72, but when exhaustion is reversed in mice by therapeutic treatment, proliferation and effector functions are impressively restored73–77. Exhausted CD8+ T cells in mice chronically infected with lymphocytic choriomeningitis virus have abnormally high Blimp-1 expression5. In contrast, Blimp-1-deficient CD8+ T cells resist exhaustion and exhibit improved cell numbers5. Despite this less exhausted state, mice with conditional knockout of Blimp-1 are unable to efficiently clear the virus5. This is probably because Blimp-1 is necessary for effector functions and terminal differentiation, as has been shown in mice with CD8+ T cells heterozygous for Prdm1 deletion, which clear virus more rapidly than do wild-type mice5.
Mechanisms of action
Although Bcl-6 and Blimp-1 are clearly powerful regulators of effector and memory cell fate decisions by CD4+ T cells, CD8+ T cells and B cells, it is less clear how Bcl-6 and Blimp-1 accomplish these functions at the level of transcriptional repression. It is also unclear to what extent the gene targets of Bcl-6 and Blimp-1 remain constant across different types of lymphocytes.
Bcl-6 targets are best understood in B cells. Bcl-6 suppresses a collection of DNA-damage–response genes and inducers of apoptosis and cell cycle arrest (p53, cyclin D2 and ATR) while facilitating AID expression in germinal center B cells11 (Fig. 1). AID induces class-switch recombination and somatic hypermutation16, which would result in cell cycle arrest and apoptosis if these normal DNA-damage responses were not suppressed by Bcl-6 (ref. 11). These Bcl-6 targets are B cell specific and are not shared by CD4+ or CD8+ T cells (although, curiously, some AID expression is observed in TFH CD4+ T cells2 and Blimp-deficient CD8+ T cells6).
A key aspect of Bcl-6 biology is that Bcl-6 dimers repress transcription only in combination with corepressors, and there are many corepressors that Bcl-6 can partner with. Most corepressors bind to or near the Bcl-6 bric-a-bric, tramtrack, broad complex–poxvirus zinc-finger domain, including BCoR78,79, N-CoR80,81, SMRT80,81, CtBP31, BAZF82, PLZF83, MIZ1 (encoded by Zbtb17)84 and others. At least some of these partners can also form mixed complexes31,83. The corepressor MTA3 binds to a second domain of the Bcl-6 protein called RDII (refs. 32,85). The corepressor ETO (encoded by Runx1t1) binds to the zinc-finger domain of Bcl-686. The use of these many different corepressors allows combinatorial targeting of Bcl-6 to different collections of genes in different cell types at different times. For example, Bcl-6 inhibits Blimp-1 via interactions with MTA3 (refs. 32,87; Fig. 1) and AP-1 (ref. 88). Bcl-6 protein inhibition of the Bcl6 gene instead depends on CtBP31 (Fig. 1). Whole-genome chromatin immunoprecipitation plus micrarray analysis has revealed that Bcl-6 regulates a very different set of genes in primary germinal center B cells than in a diffuse large B cell lymphoma33. Of 3,345 genes bound by Bcl-6, less than half of those targets are bound in both cell types33, which indicates that differences in corepressor availability probably have considerable effects on Bcl-6 gene targeting in different lymphocyte types at different differentiation stages.
There are only a smattering of obvious gene-expression changes conserved between Bcl-6-expressing TFH CD4+ T cells and Bcl-6-expressing germinal center B cells, beyond Blimp-1 inhibition2,89. This indicates that different Bcl-6 corepressors are present in TFH cells and germinal center B cells, which act together to suppress different constellations of genes in the two cell types. There are also only limited similarities in the gene-expression changes in Bcl-6-expressing TFH CD4+ T cells and Blimp-1-deficient CD8+ T cells (which have higher expression of Bcl-6)2,6, which again suggests that the specific gene-expression changes controlled by the Bcl-6–Blimp1 regulatory axis are more different than similar in B cells, CD4+ T cells and CD8+ T cells and are therefore probably highly influenced by the available corepressors. The present understanding of the genes and gene networks controlled by Bcl-6 remains limited, particularly in T cells, and this is an area that now needs extensive examination, given the importance of Bcl-6 in T cell functions.
It is also important to note that Bcl-6 mRNA quantities are a poor indicator of Bcl-6 protein expression90 and a poor indicator of Bcl-6 function, as Bcl-6 is controlled by a wide array of post-transcriptional regulatory mechanisms, including absence of translation at the mRNA level90, as well as protein phosphorylation91, acetylation92 and cofactor-mediated degradation93. This allows intense signal integration by Bcl-6, as the functional amount of Bcl-6 protein in a cell can be heavily influenced by many transcriptional and post-transcriptional activities, in combination with changes in the availability of corepressors.
The inhibition of Blimp-1 by Bcl-6 is a common feature in germinal center B cells and CD4+ TFH cells2,20,31 and probably in differentiating CD8+ T cells6,7. Because Bcl-6 and Blimp-1 are reciprocal antagonists, it is challenging to disentangle their direct effects versus their effects achieved via inhibition of each other. How much of the effect of Bcl-6 expression in a given cell type is via inhibition of Blimp-1 and vice versa? These and related questions need to be addressed.
Like Bcl-6 target genes, Blimp-1 target genes are best characterized in B cells. Blimp-1 targets in B cells can be placed into three functional categories: inhibition of proliferation (c-Myc30,94 and E2F1 (ref. 27)), induction of secretory machinery (XBP-1 (ref. 27)), and inhibition of the germinal center B cell program (Bcl-6 (ref. 27), Pax5 (ref. 95) and CIITA96,97). Inhibition of proliferation is a common feature of B cells, CD4+ T cells and CD8+ T cells with high expression of Blimp-1 (it is important to note that the antiproliferative effects of Blimp-1 are dose dependent2,26,55). The gene encoding Id3, a pro-proliferation transcription factor98 expressed in TFH cells2, germinal center B cells27 and Blimp-1-deficient CD8+ T cells6, seems to be one common Blimp-1 target in both B cells and T cells and may be a key component of the antiproliferative effects of Blimp-1.
Induction of the secretory apparatus by Blimp-1 (via XBP-1 and possibly other mechanisms) is most dramatic for plasma cells23 but also occurs in T cells99, as terminally differentiated effector T cells are specialized producers of cytokines and other secreted products. In both plasma cells and terminally differentiated effector T cells, the cellular metabolism is optimized for protein production and secretion instead of DNA synthesis and proliferation (Fig. 4).
One of the most interesting targets of Blimp-1 that has been characterized in T cells is the gene encoding IL-2 (refs. 25,55,100), a cytokine critical for T cell and B cell proliferation and differentiation. TFH cells have been observed to produce more IL-2 than do non-TFH cells2,4, consistent with the smaller amount of Blimp-1 in TFH cells2. Blimp-1-deficient CD8+ T cells also exhibit greater IL-2 production6,7. Memory CD8+ T cells are also frequently characterized by their ability to produce large amounts of IL-2. Given the importance of IL-2 for T cell function and memory differentiation, Il2 is a key Blimp-1 target gene.
Concluding remarks
Transcription factors drive B lymphocytes and T lymphocytes to differentiate into a variety of different effector and memory cells. Bcl-6 and Blimp-1 are unusual in that they are not unique to one lineage but are common to B cells, CD4+ T cells and CD8+ T cells. Although the roles of Bcl-6 and Blimp-1 in these cell lineage programs share general features—Bcl-6 frequently sustains proliferative potential but not terminal effector function, Blimp-1 enables secretion and effector functions but inhibits proliferation, and the two genes serve as a bimodal self-reinforcing genetic switch—the genes regulated by Bcl-6 and Blimp-1 to produce those phenotypes seem to be predominantly distinct in B cells, CD4+ T cells and CD8+ T cells, which highlights the complexity of the gene programs involved and the importance of additional layers of gene-expression regulation.
Acknowledgments
Supported by a Pew Scholar Award and the National Institutes of Health (072543 and 063107 to S.C.).
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Schoenberger SP, Crotty S. In: Fundamental Immunology. 6th. Paul WE, editor. Lippincott Williams & Wilkins; Philadelphia: 2008. pp. 862–898. [Google Scholar]
- 2.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurieva R, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8+ T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Ye BH, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 9.Baron BW, et al. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci USA. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerckaert JP, et al. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- 11.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 12.Jardin F, Ruminy P, Bastard C, Tilly H. The BCL6 proto-oncogene: a leading role during germinal center development and lymphomagenesis. Pathol Biol. 2007;55:73–83. doi: 10.1016/j.patbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Tangye S, Tarlinton D. Memory B cells: Effectors of long-lived immune responses. Eur J Immunol. 2009;39:9–11. doi: 10.1002/eji.200939531. [DOI] [PubMed] [Google Scholar]
- 14.Fairfax KA, Kallies A, Nutt SL, Tarlinton DM. Plasma cell development: from B-cell subsets to long-term survival niches. Semin Immunol. 2008;20:49–58. doi: 10.1016/j.smim.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Allen CDC, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 17.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 18.Ye BH, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 19.Toyama H, et al. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 2002;17:329–339. doi: 10.1016/s1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- 20.Tunyaplin C, et al. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 21.Cattoretti G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 24.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 25.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 26.Kallies A, et al. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med. 2004;200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaffer AL, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 30.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 31.Mendez LM, et al. CtBP is an essential corepressor for BCL6 autoregulation. Mol Cell Biol. 2008;28:2175–2186. doi: 10.1128/MCB.01400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita N, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Cimmino L, et al. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J Immunol. 2008;181:2338–2347. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 34.Kallies A, et al. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555–566. doi: 10.1016/j.immuni.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopf M, Le Gros G, Coyle AJ, Kosco-Vilbois M, Brombacher F. Immune responses of IL-4, IL-5, IL-6 deficient mice. Immunol Rev. 1995;148:45–69. doi: 10.1111/j.1600-065x.1995.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 37.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsiagbe VK, Thorbecke GJ. In: The Biology of Germinal Centers. Thorbecke GJ, Tsiagbe VK, editors. Springer-Verlag; Berlin: 1998. pp. 1–103. [Google Scholar]
- 39.Chtanova T, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 40.Kim CH, et al. Unique gene expression program of human germinal center T helper cells. Blood. 2004;104:1952–1960. doi: 10.1182/blood-2004-03-1206. [DOI] [PubMed] [Google Scholar]
- 41.Rasheed AU, Rahn HP, Sallusto F, Lipp M, Müller G. Follicular B helper T cell activity is confined to CXCR5hiICOShi CD4 T cells and is independent of CD57 expression. Eur J Immunol. 2006;36:1892–1903. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- 42.King C, Tangye SG, Mackay CR. T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu Rev Immunol. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 43.Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma CS, et al. Early commitment of naive human CD4+ T cells to the T follicular helper (TFH) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 45.McHeyzer-Williams LJ, Pelletier N, Mark L, Fazilleau N, McHeyzer-Williams MG. Follicular helper T cells as cognate regulators of B cell immunity. Curr Opin Immunol. 2009;21:266–273. doi: 10.1016/j.coi.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martins GA, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 47.Dent AL, Hu-Li J, Paul WE, Staudt LM. T helper type 2 inflammatory disease in the absence of interleukin 4 and transcription factor STAT6. Proc Natl Acad Sci USA. 1998;95:13823–13828. doi: 10.1073/pnas.95.23.13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith KM, et al. Th1 and Th2 CD4+ T cells provide help for B cell clonal expansion and antibody synthesis in a similar manner in vivo. J Immunol. 2000;165:3136–3144. doi: 10.4049/jimmunol.165.6.3136. [DOI] [PubMed] [Google Scholar]
- 49.Smith KM, Brewer JM, Rush CM, Riley J, Garside P. In vivo generated Th1 cells can migrate to B cell follicles to support B cell responses. J Immunol. 2004;173:1640–1646. doi: 10.4049/jimmunol.173.3.1640. [DOI] [PubMed] [Google Scholar]
- 50.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaretsky AG, et al. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J Exp Med. 2009;206:991–999. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King IL, Mohrs M. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J Exp Med. 2009;206:1001–1007. doi: 10.1084/jem.20090313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu HC, et al. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol. 2008;9:166–175. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- 54.Haynes NM, et al. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 55.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 56.Kallies A, et al. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 57.Intlekofer AM, et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med. 2007;204:2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida K, et al. Bcl6 controls granzyme B expression in effector CD8+ T cells. Eur J Immunol. 2006;36:3146–3156. doi: 10.1002/eji.200636165. [DOI] [PubMed] [Google Scholar]
- 60.Fukuda T, et al. The murine BCL6 gene is induced in activated lymphocytes as an immediate early gene. Oncogene. 1995;11:1657–1663. [PubMed] [Google Scholar]
- 61.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 62.Ichii H, et al. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 63.Ichii H, Sakamoto A, Kuroda Y, Tokuhisa T. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol. 2004;173:883–891. doi: 10.4049/jimmunol.173.2.883. [DOI] [PubMed] [Google Scholar]
- 64.Ichii H, et al. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int Immunol. 2007;19:427–433. doi: 10.1093/intimm/dxm007. [DOI] [PubMed] [Google Scholar]
- 65.Scheeren FA, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 66.Kuo TC, et al. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J Exp Med. 2007;204:819–830. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fearon DT, Manders P, Wagner SD. Arrested differentiation, the self-renewing memory lymphocyte, and vaccination. Science. 2001;293:248–250. doi: 10.1126/science.1062589. [DOI] [PubMed] [Google Scholar]
- 68.Blink EJ, et al. Early appearance of germinal center-derived memory B cells and plasma cells in blood after primary immunization. J Exp Med. 2005;201:545–554. doi: 10.1084/jem.20042060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shvarts A, et al. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19ARF-p53 signaling. Genes Dev. 2002;16:681–686. doi: 10.1101/gad.929302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tomayko MM, et al. Systematic comparison of gene expression between murine memory and naive B cells demonstrates that memory B cells have unique signaling capabilities. J Immunol. 2008;181:27–38. doi: 10.4049/jimmunol.181.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller SN, Ahmed R. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc Natl Acad Sci USA. 2009;106:8623–8628. doi: 10.1073/pnas.0809818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin H, Wherry EJ. CD8 T cell dysfunction during chronic viral infection. Curr Opin Immunol. 2007;19:408–415. doi: 10.1016/j.coi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Ha SJ, West EE, Araki K, Smith KA, Ahmed R. Manipulating both the inhibitory and stimulatory immune system towards the success of therapeutic vaccination against chronic viral infections. Immunol Rev. 2008;223:317–333. doi: 10.1111/j.1600-065X.2008.00638.x. [DOI] [PubMed] [Google Scholar]
- 74.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 75.Ejrnaes M, et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203:2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brooks DG, et al. IL-10 and PD-L1 operate through distinct pathways to suppress T-cell activity during persistent viral infection. Proc Natl Acad Sci USA. 2008;105:20428–20433. doi: 10.1073/pnas.0811139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 79.Ghetu AF, et al. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dhordain P, et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dhordain P, et al. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okabe S, et al. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol Cell Biol. 1998;18:4235–4244. doi: 10.1128/mcb.18.7.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dhordain P, et al. Colocalization and heteromerization between the two human oncogene POZ/zinc finger proteins, LAZ3 (BCL6) and PLZF. Oncogene. 2000;19:6240–6250. doi: 10.1038/sj.onc.1203976. [DOI] [PubMed] [Google Scholar]
- 84.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 85.Jaye DL, et al. The BCL6-associated transcriptional co-repressor, MTA3, is selectively expressed by germinal centre B cells and lymphomas of putative germinal centre derivation. J Pathol. 2007;213:106–115. doi: 10.1002/path.2199. [DOI] [PubMed] [Google Scholar]
- 86.Chevallier N, et al. ETO protein of t(8;21) AML is a corepressor for Bcl-6 B-cell lymphoma oncoprotein. Blood. 2004;103:1454–1463. doi: 10.1182/blood-2003-06-2081. [DOI] [PubMed] [Google Scholar]
- 87.Parekh S, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 89.Ci W, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Allman D, et al. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 91.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 93.Hirata Y, et al. BCL6 degradation caused by the interaction with the C-terminus of pro-HB-EGF induces cyclin D2 expression in gastric cancers. Br J Cancer. 2009;100:1320–1329. doi: 10.1038/sj.bjc.6605010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin KI, Lin Y, Calame K. Repression of c-myc is necessary but not sufficient for terminal differentiation of B lymphocytes in vitro. Mol Cell Biol. 2000;20:8684–8695. doi: 10.1128/mcb.20.23.8684-8695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Piskurich JF, et al. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 97.Ghosh N, Gyory I, Wright G, Wood J, Wright KL. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J Biol Chem. 2001;276:15264–15268. doi: 10.1074/jbc.M100862200. [DOI] [PubMed] [Google Scholar]
- 98.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 99.Kamimura D, Bevan MJ. Endoplasmic reticulum stress regulator XBP-1 contributes to effector CD8+ T cell differentiation during acute infection. J Immunol. 2008;181:5433–5441. doi: 10.4049/jimmunol.181.8.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med. 2008;205:1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]