Short abstract
New roles for the cytidine deaminase AID and elongator-complex proteins in DNA demethylation.
Abstract
The cytidine deaminase AID and elongator-complex proteins contribute to the extensive removal of DNA methylation in mammalian primordial germ cells and in the paternal pronucleus of the zygote.
In mammalian genomes, DNA methylation is found at cytosine residues that are followed by guanines. This epigenetic modification is essential for the repression of retrotransposons and other elements of foreign origin; it regulates developmental genes, including the pluripotency genes OCT4 and NANOG, and is crucial for genomic imprinting. CpG methylation undergoes dramatic global changes at specific stages of mammalian development. These include acquisition of new methylation patterns early in development, genome-wide removal of DNA methylation in the primordial germ cells (PGCs), and, following fertilization, removal of DNA methylation from the sperm-derived genome ([1] and references therein). Whereas the acquisition of DNA methylation is now well understood, the mechanisms involved in global DNA demethylation in PGCs and the zygote had remained elusive. Two exciting recent studies [2,3] now show that the cytidine deaminase AID contributes to active DNA demethylation in mammals. Another remarkable study has discovered that components of the elongator complex are involved in the process as well [4].
After fertilization, the sperm-derived pronucleus undergoes a rapid, global loss of DNA methylation, which occurs independently of DNA replication. Some genes, however, including imprinted genes, show resistance to this active demethylation process. The maternal pronucleus is also resistant, but undergoes passive, replication-dependent, demethylation during the first few cell cycles of development. Consequently, by the blastocyst stage, both the parental genomes have acquired low levels of methylation. At a later developmental stage, during and following implantation of the embryo, there is extensive acquisition of de novo DNA methylation, so that eventually, 70% or more of all CpGs are methylated [2]. A second round of methylation reprogramming in mammals occurs in the early PGCs of the embryo, between 10.5 and 13.5 days post-coitum (d.p.c.) in the mouse. This wave of DNA demethylation affects the entire genome, although certain sequence elements, including intracisternal A particles (IAPs), are resistant [1]. The removal of DNA methylation in PGCs affects both the parental genomes and, apparently, no genes escape this essential process, which serves to wipe the genome clean of marks so that the germ cells acquire the capacity to support post-fertilization development.
The enzymes that control the acquisition of new DNA methylation are well known and have been studied in detail. Whereas the de novo DNA methyltransferases (DNMTs) DNMT3A and DNMT3B establish new methylation on DNA, DNMT1 maintains patterns of methylation in a replication-dependent manner in all somatic cells ([1] and references therein). In contrast, the enzymatic machineries involved in the active removal of CpG methylation had remained enigmatic in mammals. Important conceptual insights were obtained from flowering plants, though, in which DNA demethylation is mediated by 5-methylcytosine glycosylases. The best studied example of such glycosylases is DEMETER, which mediates the DNA demethylation involved in genomic imprinting in the endosperm, the extra-embryonic part of the developing seed [5]. In mammals, however, this specific class of 5-methylcytosine glycosylases seems not to exist and, therefore, most attention has been focused on cytidine deaminases of the APOBEC family, particularly on Activation-Induced cytidine Deaminase (AID). AID was known to act as a single-strand DNA deaminase in developing B cells, in which it is required for somatic hypermutation and class switch recombination at immunoglobulin genes. In B cells, the deamination of cytosine residues leads to U-G mismatches which can be processed to give rise to double-strand breaks involved in recombination at the immunoglobulin genes.
New roles for AID and components of the elongator complex
Since AID was found to be expressed in PGCs and early embryos, it was suggested that it might be involved in global DNA demethylation [6]. Christian Popp and co-workers [2] tested this intriguing possibility by exploring DNA methylation in PGCs obtained from Aid-/- embryos. In their technically challenging study (mammalian PGCs can be obtained only in small numbers) an unbiased approach was taken that combined bisulphite treatment of genomic DNA with next-generation sequencing. This allowed the authors to assess global levels of DNA methylation. They combined this approach with locus-specific studies in which bisulphite-converted DNA was amplified by PCR followed by methylation analyses by mass spectrometry. In agreement with earlier studies, wild-type PGCs were found to have very low levels of global DNA methylation at 13.5 d.p.c., particularly in female PGCs, which showed less than 10% of methylation globally. The lowest levels of methylation were observed within introns, intergenic regions and repeat elements. PGCs purified from AID-deficient embryos, in contrast, showed higher levels of DNA methylation at these sequences, and globally, demonstrating that AID contributes to the genome-wide demethylation in primordial germ cells.
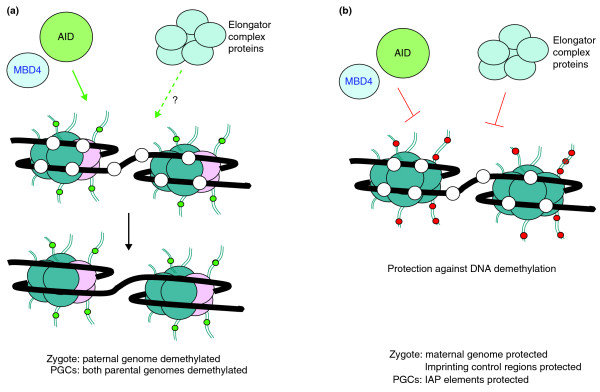
This novel discovery nicely complements a recent study by Bhutani et al. [3] on heterokaryons made by artificially fusing mouse embryonic stem (ES) cells with human fibroblasts. In these heterokaryons, DNA methylation was rapidly removed from the NANOG and OCT4 genes in the fibroblast-derived genome. By using a small interfering RNA (siRNA) approach, the authors showed that this active demethylation requires AID. Concordantly, AID was found to be targeted specifically to the (methylated) NANOG and OCT4 promoters. In combination, the two new studies demonstrate a novel role for AID in mammals: active demethylation of genomic DNA (Figure 1a).
Figure 1.
Active DNA demethylation in mammals. (a) The action of AID on 5-methylcytosine residues (white circles) in DNA (thick black line) gives rise to deaminated 5-methylcytosine, which can be bound by the repair glycosylase MBD4. Through yet-unknown further repair mechanisms, there is conversion into unmethylated cytosines, as shown by the disappearance of the white circles on the lower diagram. The canonical histones found in nucleosomes are colored in blue. In the accessibility model presented here, the presence (green circles) or absence of specific histone tail modifications and/or histone variants (pink spheres) guide the recruitment of the enzymes and other factors involved in the DNA demethylation. It is not yet known whether the requirement for elongator complex proteins is direct or whether they affect DNA demethylation indirectly, by a mechanism unrelated to chromatin. (b) Protection against active DNA demethylation could be linked to the presence of specific histone modifications (red circles). Non-histone proteins could be involved in this process as well.
Intriguingly, however, the Aid-/- PGCs still attained low levels of methylation compared with ES cells and somatic tissues, which indicated that considerable demethylation had occurred even in the absence of AID. Not surprisingly, therefore, Popp et al. [2] did not observe pronounced developmental defects in the offspring of the Aid-/- parent mice. The methylation phenotype in the absence of AID indicates that other factors must also be contributing to the DNA demethylation process.
The question of which other protein factors could be involved was addressed in the third recent study, by Yuki Okada and co-workers [4]. In their elegant study, these authors used the global demethylation in the zygote's paternal pronucleus as a model. Through careful siRNA-mediated knockdown experiments, they tested several candidate proteins. Rather unexpectedly, they discovered that a component of the elongator complex, elongator protein 3 (ELP3), to be required for the removal of DNA methylation in the zygote. The elongator complex was first described as a component of RNA polymerase II holoenzyme in transcriptional elongation, and has histone acetyltransferase activity ([7], and references herein). In particular, a live-cell imaging system allowed these authors to follow global methylation states in zygotes, and showed that knockdown of Elp3 prevented paternal DNA demethylation. Subsequently, the authors showed the same to be true for two other components of the elongator complex, ELP1 and ELP4. These remarkable findings could signify that the whole elongator complex is involved (Figure 1a). Its mode of action in DNA demethylation remains to be discovered.
As is often the case with exciting new discoveries, the recent studies raise many questions. Could transcription be somehow linked to the removal of DNA methylation? Little is known about whether there is actually transcription through genomic regions in PGCs and in the zygote. Embryonic transcription at many genes starts only after the first cell division, but what about transcription across other, non-genic, regions? Could the involvement of elongator proteins be linked to one of their transcription-independent roles, which include modification of tRNAs [7]. Although the RNA polymerase II complex has been shown to interact with AID in B cells, it is not known whether such an interaction could be involved in removing DNA methylation in PGCs and zygotes.
How, in mammals, deamination of 5 meC leads biochemically to DNA demethylation remains unclear. However, a recent study in Zebrafish provides interesting clues [8]. Also in Zebrafish, AID deaminates 5 meC leading to the formation of thymine residues, and hence G:T mismatches. Mismatched Ts are thought to be replaced by cytosines by base excision repair (BER). Methyl binding domain protein 4 (MBD4) is one of the known thymine glycocylases in vertebrates and it recognizes specifically the product of deamination at methylated CpG dinucleotides [9]. Over-expression of MBD4 together with AID in Zebrafish embryos led to partial demethylation of injected methylated DNA fragments. Mbd4 knockdown, in contrast, caused remethylation of DNA [8]. Thus, in Zebrafish, the mismatch-specific thymine glycosylase MBD4 contributes to the demethylation process involving AID. It should be most interesting to explore whether the same is true in mammals.
Is there a correlation between DNA demethylation and histone modifications?
Irrespective of the precise biochemical conversions involved, the new studies raise the question of why certain chromosomal regions lose their DNA methylation and others not. AID and elongator complex proteins are widely expressed, but global DNA demethylation occurs specifically in PGCs and in the zygote. Furthermore, in the zygote the sperm-derived genome undergoes active DNA demethylation, but the maternal genome is resistant. Demethylation of the paternal genome appears to occur after the sperm's protamines have been replaced by histones [10]. At this early time point, however, the newly formed chromatin in the male pronucleus is clearly different from the chromatin of the maternal genome. The histone H3 variant H3.3 is incorporated onto the paternal genome (independent of DNA replication), whereas the maternal genome is already packaged with nucleosomes containing mostly the canonical histone H3.1. At this stage, the histones on the paternal genome show little lysine methylation compared with histones on the maternal genome. The paternal genome is negative for H3 lysine 9 di- and trimethylation, and H3 lysine 27 trimethylation, marks that are present in the maternal pronucleus [10]. One idea, therefore, could be that histone modifications and histone variants determine whether the DNA demethylation machinery (including AID) can access the genomic DNA (Figure 1b).
In early PGCs, there is extensive loss of histone methylation together with the appearance of chaperone proteins that could be involved in incorporating histone variants into chromatin [11]. The nucleosomes and histones are modified around the time that DNA demethylation occurs, so these changes could well be involved in recruiting the DNA demethylation machinery. Certain IAP elements, however, are protected against DNA demethylation in PGCs and it would be interesting to explore the organization of chromatin at these regions.
Research on mammalian DNA demethylation is gaining momentum and, undoubtedly, new players and mechanisms will be revealed during the coming years. Together with the novel discoveries on AID and elongator complex proteins reviewed above, this could provide opportunities to further unravel the biological roles of DNA demethylation in PGCs and in the early embryo.
References
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fisher RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ. Elongator complex: how many roles does it play? Curr Opin Cell Biol. 2007;19:331–336. doi: 10.1016/j.ceb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in Zebrafish involves the coupling of a deaminase, a glycosylase, and Gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]