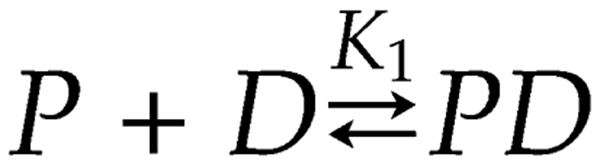Abstract
The enzyme Poly (ADP-Ribose) polymerase-1 (PARP-1) is a global monitor of chromatin structure and DNA damage repair. PARP-1 binds to nucleosomes and poly (ADP-ribosylates) histones and several chromatin-associated factors to expose specific DNA sequences to the cellular machinery involved in gene transcription and/or DNA damage repair. While these processes are critical to genomic stability, the molecular mechanisms of how DNA damage induces PARP-1 activation are poorly understood. We have used biochemical and thermodynamic measurements in conjunction with small angle X-ray scattering to determine the stoichiometry, affinity and overall structure of a human PARP-1 construct containing the entire DNA binding region, the zinc-ribbon domain and automodification domains (residues 1–486). The interaction of this PARP-1 protein construct was evaluated with three different DNA damage models (DNA constructs containing a nick, a blunt end or a 3’-extension). Our data indicates that PARP-1 binds each DNA damage model as a monomer, with similar affinity, and in all cases results in robust activation of the catalytic domain. Using SAXS, we determined that the N-terminal half of PARP-1 behaves as an extended and flexible arrangement of individually folded domains in the absence of DNA. Upon binding DNA, PARP-1 undergoes a conformational change in the area surrounding the zinc-ribbon domain. These data support a model in which upon binding DNA, PARP-1 undergoes a conformational change to become an active nuclear enzyme.
Keywords: Poly(ADP-ribosyl)Polymerase-1, PARP-1 DNA Damage, Small Angle X-ray Scattering
Introduction
The enzyme Poly (ADP-Ribose) polymerase-1 (PARP-1) is a global monitor of chromatin structure and DNA damage. PARP-1 is a member of the large family of enzymes, which are defined by the PARP signature sequence, a 50-amino acid sequence within each member’s enzymatic domain 1. Several of the PARP proteins catalyze the cleavage of NAD+ into nicotinamide and ADP-ribose, using the latter to synthesize branched molecules of poly(ADP-ribose), a substrate that is covalently attached to a variety of nuclear proteins. In eukaryotes, members of the PARP family are best noted for their roles in DNA repair, chromatin remodeling and transcriptional regulation of a variety of genes 1.
PARP-1 is a highly conserved enzyme present in higher eukaryotes and Archaea2 and is responsible for synthesis of the majority of poly(ADP-ribose) (PAR) within eukaryotic cells. Although typically highly expressed at the protein level (200,000-1 million copies per cell 3), PARP-1 enzymatic activity is tightly regulated in the absence of genomic stress 4; 5; 6; 7. Under normal conditions, PARP-1 is found associated with histones, DNA and other chromatin associated factors 8; 9. In response to DNA damage, PARP-1 enzymatic activity is stimulated 10–500 fold. This results in the poly ADP-ribosylation (PAR) of many target proteins including itself, core histones, linker histone H1 and transcription factors. The negative charge associated with PAR renders the target proteins unable to associate with DNA and allows exposure to the cellular DNA repair machinery 9. Interestingly, the PAR moiety itself is bound and hydrolyzed by many other enzymes including macro domain proteins and glycohydrolases3,713,31.
While these processes are critical to genomic stability, the molecular mechanisms of how DNA damage induces PARP-1 activation are poorly understood. Due to the specificity of PARP-1 for non-B-DNA16, we hypothesize that the location and orientation of PARP-1 when it is bound to damaged DNA is critical for subsequent activation of the PARP-1 enzymatic domain.
Human PARP-1 is a modular protein organized into at least 6 domains 10 (Figure 1). Its affinity for damaged DNA is regulated by two unique zinc-finger motifs (CX2CX28–30HX2C) (residues 1–209; referred to as zf-PARP) which are sufficient to target the entire protein to damaged DNA 11; 12. Recently, comparative analysis has defined the zf-PARP fold as an exceptional zinc-finger domain composed of a hydrophobic core with three conserved beta-sheets and two terminal alpha helices16. All zf-PARP family members contain two variable loop regions bridging β2–β3 and α1–α2 that may play a role in determining DNA structure specificity16. Proteins with the zf-PARP fold are evolutionary conserved in the eukaryotic lineage and are associated with many nuclear enzymes outside of the PARP protein family. None of the zf-PARP fingers are known to have sequence specificity, but instead seem to recognize many different DNA structures such as stem-loops, cruciforms, nicked DNA and 3’-extensions. The two PARP-1 zf-PARP domains are known to recognize DNA nicks, overhangs, blunt ends and other forms of damage 12; 13; 14; 15. Thus zf-PARP differ both structurally and in their DNA binding specificity from the abundant and well-studied family of sequence specific C2H2 zinc fingers 16. However, little is known about the interactions of zf-PARP fingers with DNA 16.
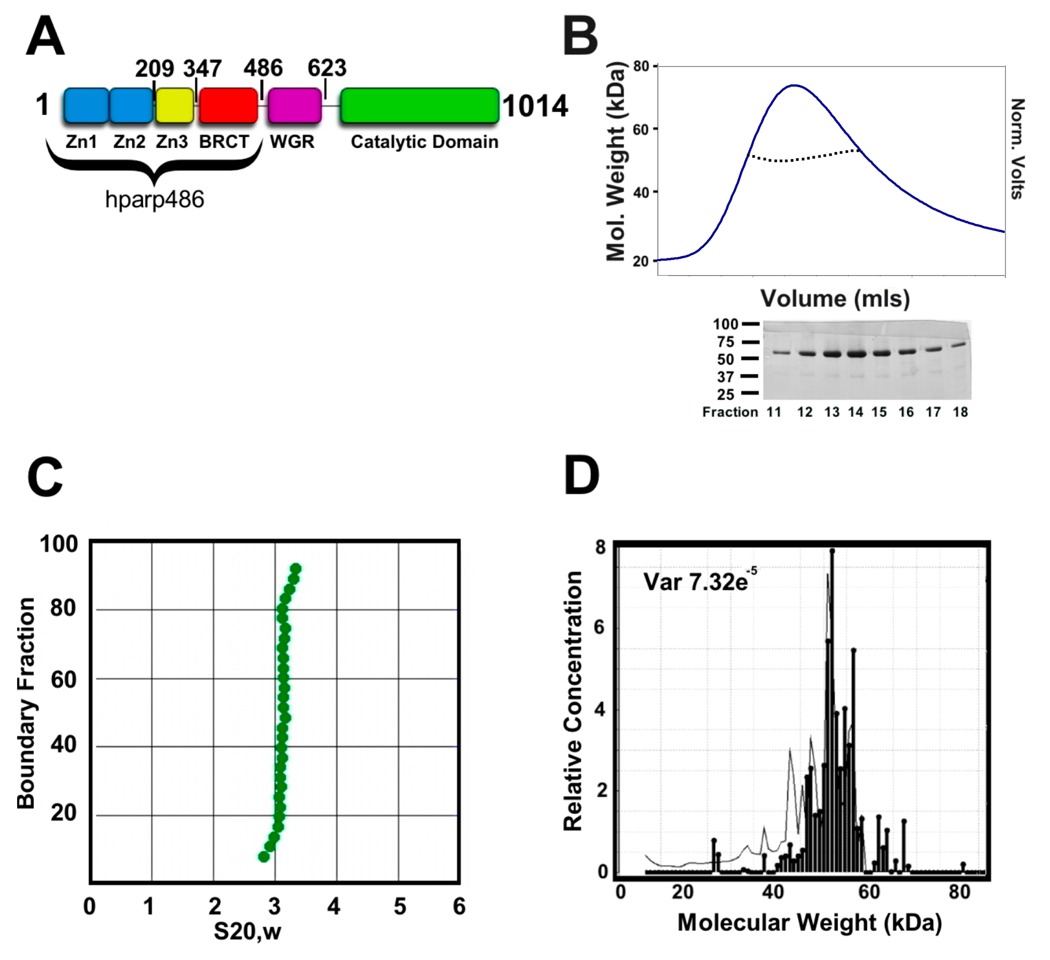
Figure 1. hparp486 is a monomer in the absence of DNA.
A) hparp486 was designed to include the entire DNA binding region of the human PARP-1 protein. The first 486 residues encompass the three N-terminal Zn domains and the BRCT domain. The illustration indicates the C-terminal residue of each domain. B) Top: Size exclusion chromatography and inline static light scattering of hparp486. hparp486 elutes in a single peak from a KW803 size exclusion column. The static light scattering average molecular weight (~57kDa) for the central portion of the peak is illustrated by the black dotted line within the peak. Bottom: SDS-PAGE analysis of fractions from SEC purified hparp486. C) The sedimentation velocity profile of purified are presented as G(s) plots of the integral of S20,w against boundary fraction (%).van Holde - Weischet analysis was used to determine the f/f0 value of 1.8. D) Sedimentation equilibrium ultracentrifugation resolved that over 95% of the purified protein was monomeric at a concentration of 40µM.
Two recent publications have defined a third zinc domain in human PARP-1 10; 17. This zinc ribbon domain resides between residues 233–273 of the protein. Although the zinc ribbon domain does not bind DNA on its own, it is necessary for DNA stimulated activation of the full-length enzyme10. This has lead to speculation that this domain modulates the N-terminal to C-terminal communication (possibly through a conformational change) that leads to DNA dependent activation of PARP-1 10; 17. Nevertheless, in the absence of multidomain PARP-1 structures, no direct evidence of a DNA dependent conformational change has been noted. Moreover, information on length requirements, affinity, stoichiometry and mode of interaction of the DNA sensing region in this abundant enzyme with various DNA damage models is lacking.
Here, we characterize a highly purified fragment of human PARP-1 encompassing the entire DNA binding region, the zinc ribbon domain as well as the auto-modification domain (BRCT), with defined DNA damage models. The BRCT domain of PARP-1 is conserved with the BRCA-1 C-terminal region and is thought to function as a protein-protein interaction module. The structure and function all known human PARP-1 domains are reviewed in1. The PARP-1 construct used in this manuscript was designed based on known domain structures Zn1, Zn2, Zn3 and BRCT (residues 1–486, from now on referred to as hparp486) (pdb IDs: 2dmj, 2cs2, 2jvn, 2cok) (Figure 1A). We find that hparp486 is a monodisperse monomer in solution on its own as well as in complex with blunt ended, nicked, or 3’-overhang DNA. Control experiments indicate that full length PARP-1 is activated to similarly high levels by the different DNA damage model substrates. Small angle X-ray scattering was used to determine the overall shape of hparp486 on its own and in complex with these DNA fragments. Distance distribution functions and ab initio particle reconstructions created from the SAXS data suggest a conformational change occurs in the zinc ribbon domain of the extended polypeptide chain upon DNA binding.
Results
hparp486 is an elongated monomeric particle in solution
An N-terminally 6X His-tagged fusion construct of hparp486 was expressed in E. coli and purified to homogeneity. Purified hparp486 elutes in a single peak from a SHODEX-803 gel filtration column and SDS-page analysis reveals that the protein is ~95% pure (Figure 1B). Static light scattering measurements determined that the purified protein is a monomer with a molar mass of ~57 kDa at concentrations exceeding 2 mg/ml (Figure 1B). Sedimentation velocity experiments determined that hparp486 sediments as a single species of elongated character with an anhydrous frictional ratio (f/f0) = 1.8 (Figure 1C). Further sedimentation equilibrium experiments demonstrate that hparp486 has a molecular mass of 55 – 60 kDa (Figure 1D). These results are in excellent agreement with the calculated mass of a hparp486 monomer, 56.480kDa.
Small angle X-ray scattering (SAXS) is an effective method for determining the low resolution (>10Å) structures of proteins, nucleic acids and their complexes in solution at reasonably low concentrations. After an overall SAXS envelope is determined, hybrid methods can be applied to fit domains of known structure within the SAXS envelope 18. SAXS data were collected on hparp486 at 3, 6 and 9 mg/ml and the scattering curves were overlaid to determine any concentration-dependent effects. To determine the relative amount of folded domains and any aggregation within the hparp486 sample, the data were plotted in a Kratky plot and analyzed by Guinier approximation respectively (Supplemental Figure 1). From this preliminary analysis we determined that the data were deemed of sufficient quality to use the scattering profiles up to qmax=0.205Å−1(where q is the scattering vector). For hparp486, the radius of gyration varies with increasing concentration in the range Rg ~46–48Å (Table 1). Further analysis of the hparp486 scattering data determined that the particles in solution do follow Porod’s law, reviewed in 19. Thus their volume can be calculated from their scattering profile. We established that hparp486 occupies ~103,770 Å3 and has a molecular weight of ~54 kDa (Supplemental Figure 1). This is in good agreement with values that were determined for hparp486 by AUC (~60 kDa) and classical light scattering at lower concentrations (~57kDa; figure 1). Thus, hparp486 exists as a monodisperse monomer over a wide range of concentrations (3–9mg/ml).
Table 1. Overall dimensions hparp486, DNA and hparp486-DNA complexes.
Molar masses were obtained by SEC-MALS, radii of gyration (Rg) and maximum dimension (Dmax) were obtained from SAXS. The calculated molar masses of each molecule and a 1:1 complex of hparp486-DNA are in the second column. The SEC-MALS determined mass listed in the third column. The radius of gyration (Rg) for each molecule at the lowest concentration tested (3 mg/ml) while the highest concentration (9 mg/ml for hparp486 and 6 mg/ml for all other samples) are listed in the fourth and fifth columns respectively. The last column contains the maximum dimension for each molecule as determined by SAXS. Variation of Dmax was negligible for these molecules within this concentration range.
| Molecule | Calculated Mol. Mass |
SEC-MALS Mol. Mass |
SAXS Rg (Å) [3mg/ml] |
SAXS Rg (Å) [6 or 9 mg/ml] |
SAXS Dmax(Å) |
|---|---|---|---|---|---|
| hparp486 | 56.0 | 54.4±0.1% | 46.3 | 48.3 | 150 |
| Hparp209 | 23.0 | 22.5±3% | 29.0 | 29.5 | 115 |
| 30Nick | 18.4 | 22.8±5% | 26.9 | 28.6 | 90 |
| 30Blunt | 18.4 | 22.0±2% | 28.2 | 28.5 | 88 |
| 30Ext | 20.2 | 21.2±0.1% | 30.2 | 29.8 | 100 |
| hparp486-30Nick | 74.4 | 83.5±6% | 63.5 | 64.2 | 195 |
| hparp486-30Blunt | 74.4 | 70.0±6% | 76.5 | 78.7 | 240 |
| hparp486-30Ext | 76.3 | 71.3±1% | 63.5 | 63.0 | 195 |
| Hparp209-30Blunt | 43.1 | --- | 55.2 | 51.6 | 180 |
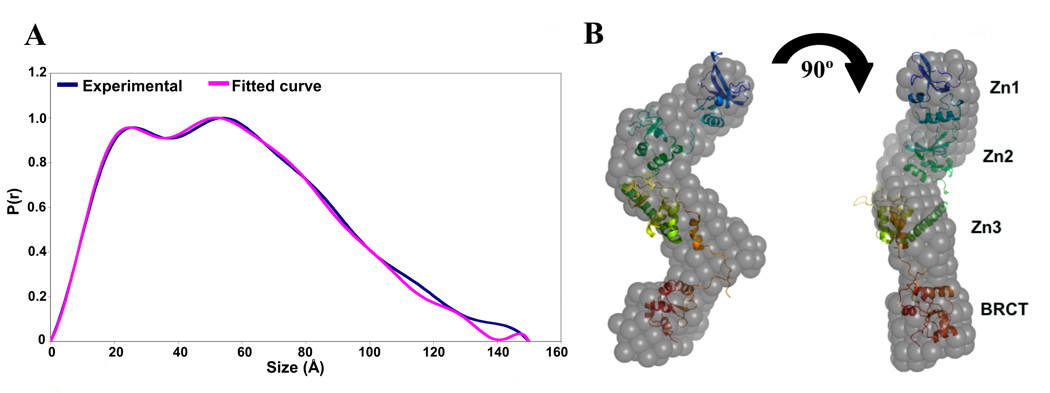
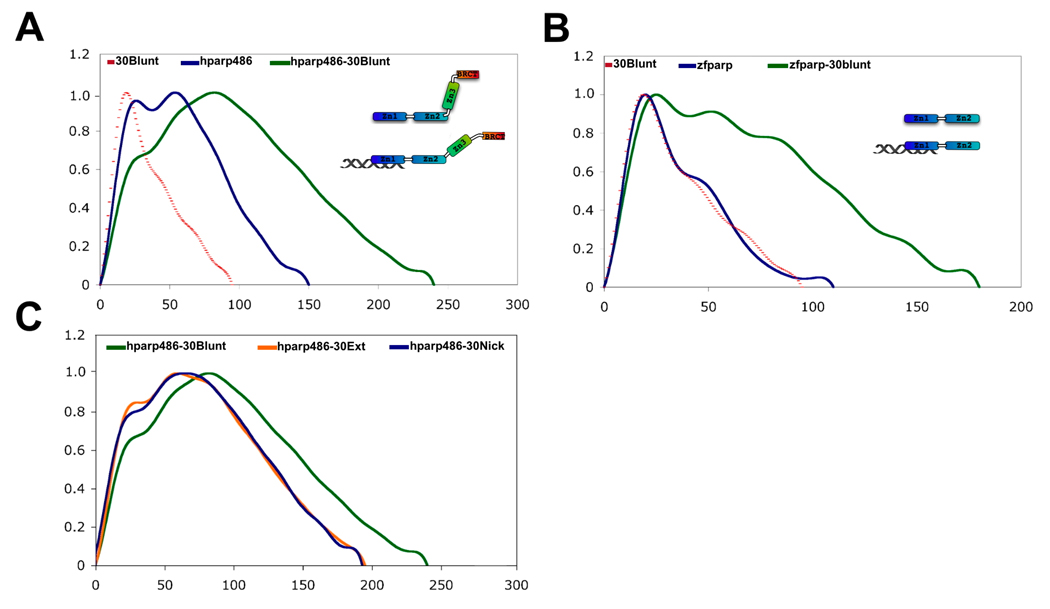
Additional information can be extracted from well-behaved SAXS data. The GNOM program 20 calculates pair distance distribution functions (denoted as P(r) ) from scattering data using an indirect Fourier transformation procedure. For hparp486 we observe two maxima in the P(r) function (Figure 2A). This is characteristic of two distances appearing with a higher probability, and indicates a macromolecule composed of at least two ordered domains connected by a flexible linker. Additionally, the P(r) function shows a shallow descent between 125 and 150Å, which is the signature of an elongated, flexible particle.
Figure 2. The low resolution structure of hparp486 reveals a molecule with conformational flexibility.
A) Superposed experimental and modeled distance distribution functions of hparp486. In blue, the SAXS distance distribution function P(r) for hparp486 is an asymmetric curve with two discrete maxima, representing two ordered regions connected by a flexible linker. The relatively shallow decent towards a maximum size of 150Å is reflective of an elongated, flexible molecule. Illustrated in violet, is the P(r) function resulting from the SASREF tertiary model shown in (B). B) Superposed particle reconstructions and tertiary structure models of hparp486. The DAMMIN particle reconstruction (grey) represents the filtered average of 30 models individually computed from a SAXS scattering curve and fits the SAXS scattering curve with a χ2 of 1.10. The SASREF tertiary structure model was computed by global rigid body modeling of the known domain structures of Zn1 (PDB id 2dmj, blue), Zn2 (PDB id 2cs2, green), Zn3 (PDB id 2jvn orange) and BRCT (PDB id 2cok, red) with steric restraints against the solution scattering data; this model has a χ2 of 1.21. In the orientation shown, the two models have a correlation coefficient of 0.56, while a 180° rotation of the polypeptide resulting in a switch in position of Zn1 and BRCT domains has a correlation coefficient of 0.54. The SASREF model illustrates the extended nature of the polypeptide and minimal inter-domain contacts.
Two models of hparp486 were independently calculated with the programs DAMMIN and SASREF 21; 22. The DAMMIN program was used to calculate 30 ab initio models that fit the SAXS scattering profile of hparp486 with a χ2=1.0–1.1. A superposition of 5 randomly chosen models is shown in supplemental figure 2. The filtered, averaged envelope of the 27 most similar models has a maximum dimension (Dmax) of ~148Å and forms an overall S-shaped structure (Figure 2B). Most residues (432 of 486, or 90%) within the N-terminal half of PARP-1 are represented in one of four domains previously deposited in the Protein Data Bank (PDB IDs: 2dmj, 2cs2, 2jvn, 2cok). The SASREF program performs tertiary structure modeling by reconstructing all or parts of the SAXS model from domains of known structure. The PARP-1 domains were trimmed of extended linkers and subjected to N-C terminus distance restrictions that limited overlapping regions and minimized steric restraints during the modeling process. The domain definitions and optimized restrictions are summarized in the Materials and Methods section. The SASREF model has a Dmax of ~140Å and fits the hparp486 P(r) function with a χ2=1.21 (Figure 2A). The DAMMIN and SASREF models superpose well (Figure 2B), and it is clear that they both represent an overall S-shaped elongated molecule.
The hparp486 interacts with damaged DNA as a monomer
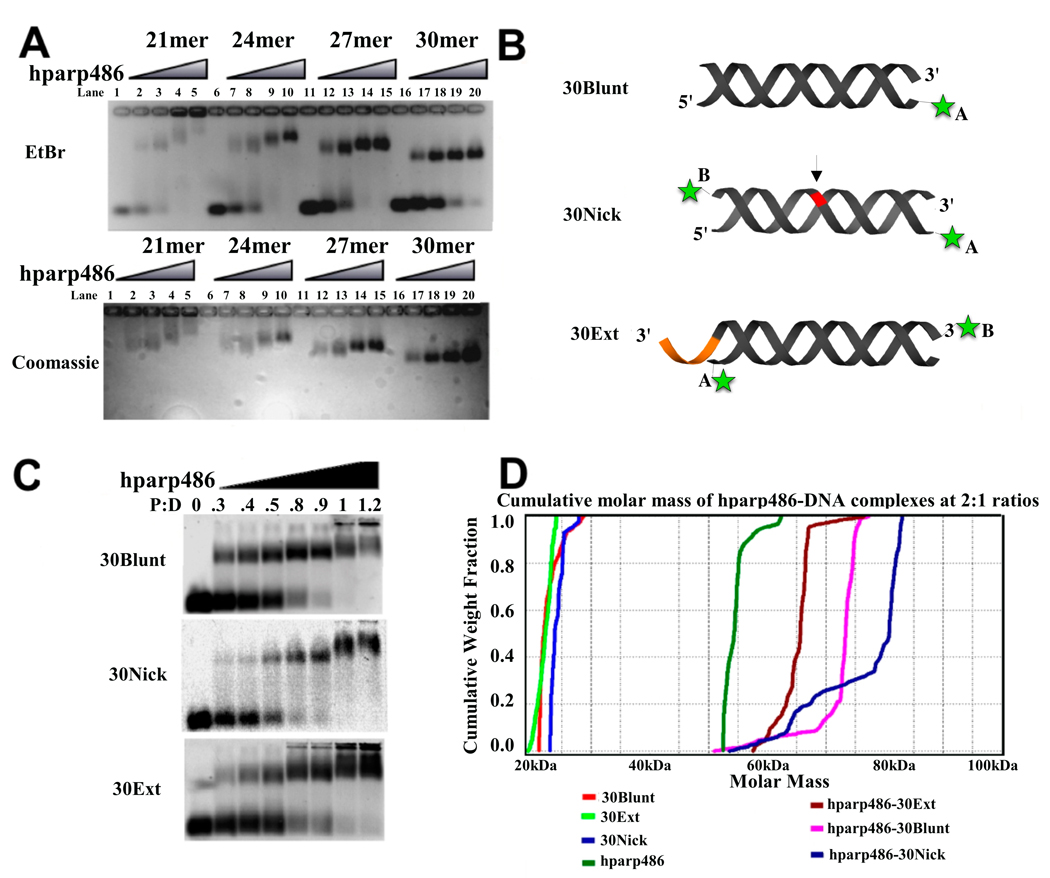
Previous biochemical studies have monitored the interaction of truncated PARP-1 constructs with 55 base pair DNA fragments14; 15. We initially assayed the association of hparp486 to a 53 base pair blunt ended DNA by electrophoretic mobility shift analysis (EMSA) and found that the protein could form either 1:1 or 2:1 complexes with this DNA at hparp486:DNA ratios between 0.5:1 and 2:1 (Supplemental Figure 3). Therefore, we first set out to determine the base pair length requirements needed to promote stable binding in order to investigate whether hparp486 binds DNA as a monomer or dimer. We used an agarose gel-based mobility shift assay in which we visualize the homogeneity of various hparp486-DNA complexes. Shown in Figure 3A is the effect of double-stranded DNA length on the homogeneity of complexes formed between hparp486 and DNA containing 6-nucleotide 3’-overhangs. As the double-stranded DNA length approached 27–30 base pairs, the complex migrates as a sharp single band when visualized by both EtBr and coomassie stains as opposed to a weaker, more diffuse shifting pattern observed with shorter DNA. We concluded from the mobility-shift assay that a 30 base pair double helical region is sufficient to stably bind hparp486 when a 6-base 3’-overhang is present. In order to determine how hparp486 interacted with other forms of DNA damage, we continued the remainder of our experiments to compare DNA of the same base pair length, but with varying ends (blunt ends, and nicked DNA).
Figure 3. hparp486 binds as a monomer to damaged DNA.
A) EtBr and Coomassie stained 1% agarose gel EMSA assay of hparp486-DNA complexes. 1µM DNA was added to each lane. In this assay, all DNA constructs contain a 6-nucleotide 3’ extension, but vary in the length of the double stranded region (21–30 base pairs). hparp486 forms homogenous complexes as the DNA approaches 27–30 base pairs in length. B) A schematic representation of the three DNA damage models tested and the location of the fluorophores (green asterisk) used in this publication. Varying the fluorophore position (either in position A or B) allowed us to test the effect of its position on hparp486 association. The arrow and red region highlight the area of the single stranded break in the 30Nick DNA, while the orange area highlights the 6 nucleotide 3’ extension in 30Ext. C) EMSA of hparp486 association with 2µM fluorescently labeled 30Blunt, 30Nick and 30Ext DNA. hparp486 efficiently shifts the mobility of over 90% of each DNA at a 1:1 ratio, eliminating the possibility of cooperative dimerization under these conditions. D) Cumulative molar mass of DNA, hparp486 and hparp486-DNA complexes as determined by SEC-MALS. 2:1 molar ratios of hparp486:DNA were incubated for 30 minutes prior to subjecting the mixtures to SEC-MALS. The distribution of masses present in the major peaks eluting from the column for DNA, hparp486 or each hparp486-DNA complex is plotted as a fraction of cumulative molecular weight. hparp486 does not stably dimerize on any DNA fragment. Supplemental Figure 4 contains the elution profiles for each solution.
To determine the stoichiometry of the various hparp486-DNA complexes, we used 2 µM fluorescently labeled DNA in an EMSA to quantify the ratio at which all DNA formed a complex with hparp486. DNA molecules with a single label at either location A or B were designed for the purpose of testing the stoichiometry and affinity of hparp486 association (Figure 3B). We determined by EMSA that when using 30 base pair DNA, hparp486 forms homogeneous 1:1 complexes irrespective of nicks, extensions or blunt ends (30Ext, 30Nick, and 30Blunt; Figure 3C). The addition of hparp486 past a 1:1 protein to DNA molar ratio resulted in the retention of both protein and DNA within the wells irrespective of the length of time the gel was run. This may represent either aggregation or a low affinity, possibly anti-cooperative dimerization of hparp486 on these DNA fragments. To overcome the limitation of the mobility shift assay, we confirmed the stoichiometry of hparp486 on DNA in solution by Size Exclusion Chromatography in conjunction with Multi Angle Light Scattering (SEC-MALS). A 2:1 mixture of hparp486 to DNA was loaded on a SEC column that eluted directly into a MALS instrument. An average molecular weight for macromolecules within each peak eluting from the SEC column was calculated from the MALS data. hparp486 has a molar mass of ~56 kDa while each DNA is ~20 kDa, thus 1:1 complexes should be ~76 kDa, while a 2:1 complex would be ~133 kDa. SEC-MALS determined the molar masses for the hparp486 in complex with 30Ext, 30Blunt and 30Nick were 71.3, 74.4 and 84.5 kDa respectively; no evidence for a 2:1 complex was detected (Table 1). In line with this result, the cumulative weight fraction and molar mass vs. volume profiles determined from the SEC-MALS experiments both illustrate the presence of a significant amount of free hparp486 in solution (Figure 3D, Supplemental Figure 4).
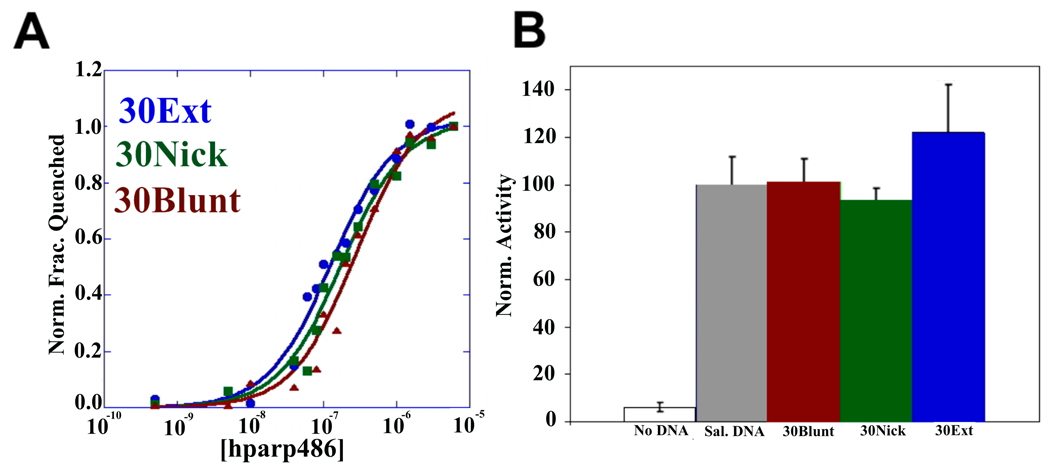
We next determined the affinity of hparp486 for the 30 base pair DNA damage models by monitoring the quenching of the fluorescence of labeled DNA molecules in solution. Using Equation 1 (materials and methods) we fit the single-step scheme (scheme 1, P= hparp486, D=DNA K1=Kd) to the experimental data obtained for each protein-DNA complex. We determined that under uniform conditions, hparp486 associates with 30Ext ≈ 30Nick (Kd of 10 ~100nM) > 30Blunt (Kd of ~300nM), when they all contain the same 30bp ‘DNA core’ (Figure 4A, Table 3). To minimize the possibility of the fluorophore interfering with protein – DNA interactions, we repeated the experiment with 30Ext with the fluorophore at multiple locations without a significant change in affinity (Table 3). However, when the 30Nick DNA was labeled at position A, interacting with hparp486 did not result in sufficient quenching of fluorescence (8%) to determine the Kd (Table 3). Thus hparp486 does not change the environment of the fluorophore at the blunt end when it associates with the region surrounding the nick. We determined that hparp486 associates with each DNA with a Hill coefficient of 0.92–0.99, reflecting the lack of a cooperative association between the protein and any of these DNA damage models (Table 3, Supplemental Figure 5), and further supporting the notion that hparp486 binds as a monomer.
Figure 4. hparp486 has similar affinities for different forms of DNA damage; fulllength human PARP-1 is efficiently activated by DNA containing blunt ends, nicks or a 3’-extension.
A) The affinity of hparp486 for fluorescently labeled 30 base pair DNA: 30Blunt (red), 30Nick (green), 30Ext(red) was determined by measuring the change in fluorescence of 2nM labeled DNA as a function of hparp486 concentration (0.5-6000nM) in solution. The affinity of hparp486 each DNA is shown in Table 3 and is in the range of ~70–300nM. The Hill coefficients calculated for each fluorescent titration were in the range of 0.92–0.99, demonstrating the lack of positive cooperativity in the individual interactions (Table 3). B) The ability of each DNA to activate full-length human PARP-1 was tested with a commercial assay that monitors the incorporation of biotinylated ADP-ribose onto immobilized histones. The activation induced by 1µM of 30Blunt, 30Nick or 30Ext DNA was normalized to the activity stimulated by sheared salmon sperm DNA. Regardless of the DNA damage model tested, PARP-1 was ~40 fold more active than in the absence of DNA.
Table 3. The affinity of hparp486 for blunt, nicked and 3’-extension DNA.
The percent fluorescence quenched, affinity (Kd) and Hill coefficient determined from the interaction of hparp486 with fluorescently labeled blunt ended, nicked or 3’-extension DNA are listed. The DNA constructs were either labeled in Position A or B, as illustrated in Figure 3B. Because of its pseudo symmetry, the 30Blunt construct was only labeled in one location.
| DNA Type | Fluorescence Quenched |
Affinity (Kd) | Hill Coeff. |
|---|---|---|---|
| 30Blunt | 33% | 2.82×10−7±4.0×10−8 | 0.9206 |
| 30Nick (pos. B) | 44% | 1.46×10−7±2.0×10−8 | 0.9670 |
| 30Nick (pos. A) | 8% | ---- | ---- |
| 30Ext (pos. B) | 35% | 1.30×10−7±1.3×10 −8 | 0.9904 |
| 30Ext (pos. A) | 19% | 6.80×10 −8 ±6.0×10−9 | 0.9802 |
hPARP-1 is activated by DNA containing nicks, 3’-extensions and blunt ends
The hPARP-1 enzyme is stimulated 10–500 fold upon binding damaged DNA 23. We used a chemiluminescent hPARP-1 assay kit (Trevigen inc.) to measure hPARP-1 activity in the presence of the various 30 base pair DNA fragments tested above. This assay measures the incorporation of biotinylated Poly (ADP-ribose) onto histone proteins in a 96-well plate format. 30Ext, 30Nick and 30Blunt DNA were each able to stimulate hPARP-1 ~40 fold over the basal activity in the absence of DNA and to the same level as sheared salmon sperm DNA (Figure 4B).
DNA binding induces conformational changes in hparp486
We used SAXS to obtain structural information on the various hparp486-DNA complexes. As a control, we measured the scattering curves for 30Ext, 30Blunt and 30Nick DNA individually at 3 and 6 mg/ml. While small concentration-dependent differences in radius of gyration (Rg) were noticed, superposition of the SAXS scattering profiles and Guinier analysis determined the data are of good quality (Supplemental Figure 5). Charge repulsion energies are known to affect DNA monodispersity in solution, and this may account for the observed concentration dependent effects 24. The analysis of the P(r) function illustrates that all DNA molecules conform to a double helix in either the A or B DNA form in solution, within the resolution of the technique (88–100Å in length). A comparison of the P(r) functions for all datasets obtained for DNA is shown in Supplemental Figure 6. The DAMMIN particle reconstructions of each DNA molecule are illustrated in Supplemental Figure 7.
Despite similarities in binding affinities, it is possible that the various DNA damage models bind differently to hparp486. We collected SAXS data on hparp486 in complex with 30Nick, 30Ext or 30Blunt DNA. After initial protein and DNA concentrations were determined, each complex was further titrated to homogeneity as determined by mobility shift assays. SAXS data for hparp486 in complex with various DNA fragments were initially analyzed the same way as was done for data from hparp486 or DNA fragments alone: The dependence of the nature of scattering on concentration was studied by comparing scattering profiles (Supplemental Figure 8A), the scattering behavior at low angle was studied by Guinier analysis (Supplemental Figure 8C), the protein folding was checked by representing the data in a Kratky plot (Supplemental Figure 8B) and the pair distance distribution functions were determined by indirect Fourier Transform (Figure 5). The results show that there is minimal concentration dependence for the SAXS data; the Kratky plot is typical for partially flexible molecules.
Figure 5. The distance distribution functions of hparp486-DNA complexes portray a conformational change within hparp486 upon binding DNA.
A) The P(r) functions of 30Blunt (dotted line), hparp486 (blue) and a 1:1 complex of hparp486-30Blunt complex (green). Upon binding 30Blunt, the profile of hparp486 in complex changes significantly when compared to the normalized profile of hparp486 in the absence of DNA. In particular, interacting with DNA changes the P(r) function from two discrete maximum of similar height to a single dominant maximum of 240Å, reflecting an increase in Dmax of 90Å over hparp486. We interpret this to represent a migration from a conformation containing two ordered regions connected by an internal flexible linker to an elongated, dynamic complex that does not have an internal highly flexible region. B) The P(r) function of zf-parp (blue), 30Blunt (dotted line) and a 1:1 complex of zf-parp-30Blunt. In contrast to hparp486, the P(r) function of zf-parp is characterized by a distinct single maxima, which reflects the lack of a high degree of internal flexibility. Upon binding 30Blunt DNA, no evidence of a conformational change is noted and with a maximum dimension of 180Å, the complex reflects an increase in size of only 65Å. C) The SAXS scattering profiles of solutions containing 1:1 ratios of hparp486 in complex with 30Blunt, 30Nick or 30Ext DNA were analyzed to determine the general shape and size of the complexes (green, blue, orange respectively). The P(r) functions of the complexes illustrate that hparp486 forms complexes of similar maximum size with 30Nick and 30Ext DNA (~200Å), but forms a much longer complex with 30Blunt DNA (~240Å maximum length).
The SAXS scattering curves for each hparp486-DNA complex are depicted in Supplemental Figure 8. The P(r) functions hparp486, 30Blunt and the hparp486-30Blunt complex allowed us to qualitatively compare the overall shape of particles in solution and quantitatively compare Rg and Dmax values with the relatively rigid DNA fragment (Figure 5A, Table 2). A comparison of the P(r) functions indicates a significant change in the particle nature upon binding of hparp486 to 30Blunt DNA. hparp486 alone consists of at least two folded domains (of ~40 and 60 Å in size) connected by a flexible linker. On the other hand, the P(r) function of hparp486-30Blunt DNA is distinct in almost all of its characteristics when compared to hparp486 on its own (Figure 5A). This profile has a sharp maximum at ~100Å and a Dmax of 240Å (an increase of 90Å over hparp486 on its own). In contrast to the double maxima of the hparp486, the P(r) function of the complex contains a single maximum, which could be reflective of a structural change within the protein that occurs upon binding DNA.
Table 2. SASREF domain definitions and distance restrictions.
The solution structures for the human PARP-1 domains Zn1, Zn2, Zn3 and BRCT (PDB entries 2dmj, 2cs2, 2jvn, 2cok, respectively) were truncated of N- and C-terminal flexible regions prior to tertiary model reconstruction by the program SASREF. The residues included and C-terminal distance restrictions used in the tertiary model are listed for each domain.
| Domain | Modeled Residues | C-term distance restriction |
|---|---|---|
| Zn1 | 1–193 | 20Å |
| Zn2 | 103–207 | 7Å |
| Zn3 | 225–359 | 30Å |
| BRCT | 380–486 | ---- |
To identify if the changes in the P(r) function were reflective of a conformational change taking place in the Zn3-BRCT region of the protein, we repeated the experiment using the zinc finger region of PARP-1 (residues 1–209, referred to as hparp209). We found hparp209 behaved as a homogenous monomer by SEC-MALS (Supplementary Figure 9A), and bound 30blunt DNA as a monomer by mobility shift assay (data not shown). The raw SAXS scattering profile and Kratky plot of hparp209 are shown in Supplementary Figures 9B–C. The P(r) function of hparp209 contains a single discreet maximum and a smaller shoulder peak at ~50Å in size (Fig. 5B). Particle reconstructions of hparp209 clearly depict a flexible terminal region of the protein, but otherwise superpose well with each other (supplementary figure 9D). The hparp209-30Blunt complex maintains the same overall profile as hparp209 on its own but only increases in size by 65Å to a Dmaxof 180Å (Fig. 5B). Thus, in the absence of Zn3-BRCT, hparp209 retains the ability to interact with DNA, but does not retain the characteristic change in shape that we identified in hparp486.
Inspection of the P(r) function allows us to compare the complex formed between hparp486, 30Ext, 30Nick DNA and 30Blunt (Fig. 5C). We determined a similar Dmax for the 30Nick and 30Blunt complexes, but note some differences in the profile of the P(r) function for each complex. We found that both particles have a Dmax of ~190–210Å and exhibit an overall elongated shape. Similarly, both complexes have a broad peak in the P(r) plot with a maximum at ~80–100Å in size. At approximately 190–210Å in Dmax, these complexes are ~20% shorter than the sum of Dmax obtained for hparp486 and DNA individually and the hparp486-30Blunt complex.
Discussion
The specific molecular mechanisms that lead to PARP-1 enzymatic activation in the presence of DNA damage have yet to be identified. Our understanding of the enzymatic complexity of PARP-1 relies not only on the structure and function of individual domains, but requires knowledge of the inter-domain communications that regulate the enzyme. Here we propose that PARP-1 zf-parp domains communicate the presence of DNA-damage by inducing a conformational change that leads to activation of the PARP-1. Additionally, we have established many basic features of DNA recognition by PARP-1 that were previously unknown, for example length requirements, affinity, stoichiometry, and the mode of interaction.
Notably, our SAXS data have provided the first low-resolution structure of the N-terminal half of PARP-1 (Figure 2). The most striking feature of this model is interior structural flexibility, (likely in the form of hinges connecting two structured domains or sets of domains), illustrated by the double maxima in the hparp486 P(r) function. The region of interior flexibility is ~55Å from either the C or N-terminus of hparp486. After comparing the particle reconstruction, tertiary model and the dimensions of the individual PARP-1 domains, we propose two models for the sequences which might constitute the hinge region. The first would be a region immediately following the second zinc finger domain (residues ~208–220). The distance across the zinc-binding face of the first two zinc finger domains is ~25Å, and when including a small linker region between the domains the total distance between the N-terminus of zinc finger 1 to the C-terminus of zinc finger 2 is ~50–60Å. An alternative model places the hinge region between residues 370–385. This region is ~55Å away from the BRCT domain C-terminus in the hparp486 SASREF model. Interestingly, either model supports the notion that the zinc ribbon domain functions to relay the DNA damage signal from the zf-PARP region to the enzymatic domain. Moreover, both models are supported by the P(r) function of hparp209, which does not contain this interior flexible region.
We found that PARP-1 has similar affinities for the three DNA damage forms tested. This may be a result of PARP-1 having evolved as a general sensor of DNA ends and not as a specialized ‘nick’ sensor. The ~3 fold reduced affinity of PARP-1 for blunt ended DNA may be due to the increased rigidity of this DNA fragment, as two zf-PARP containing enzymes have been shown favor large DNA distortions 25; 26.
The SAXS data of the PARP-1-DNA complexes depict an overall elongated, asymmetric structure. In line with previous results27, the Dmax of the PARP-1-DNA complexes suggest significant contacts are made between the protein and double stranded DNA in the three models that were tested. The asymmetric form of binding with which PARP-1 associates to the region surrounding the DNA damage is reminiscent of the DNA damage recognition of HU-like proteins within bacteria 28; 29. Recently, HU-like proteins have been shown to associate with cruciforms, nicks and other high affinity DNA targets of PARP-128; 29. This asymmetric association to regions of DNA damage may represent a conserved mechanism that exposes the lesion while protecting the surrounding undamaged region.
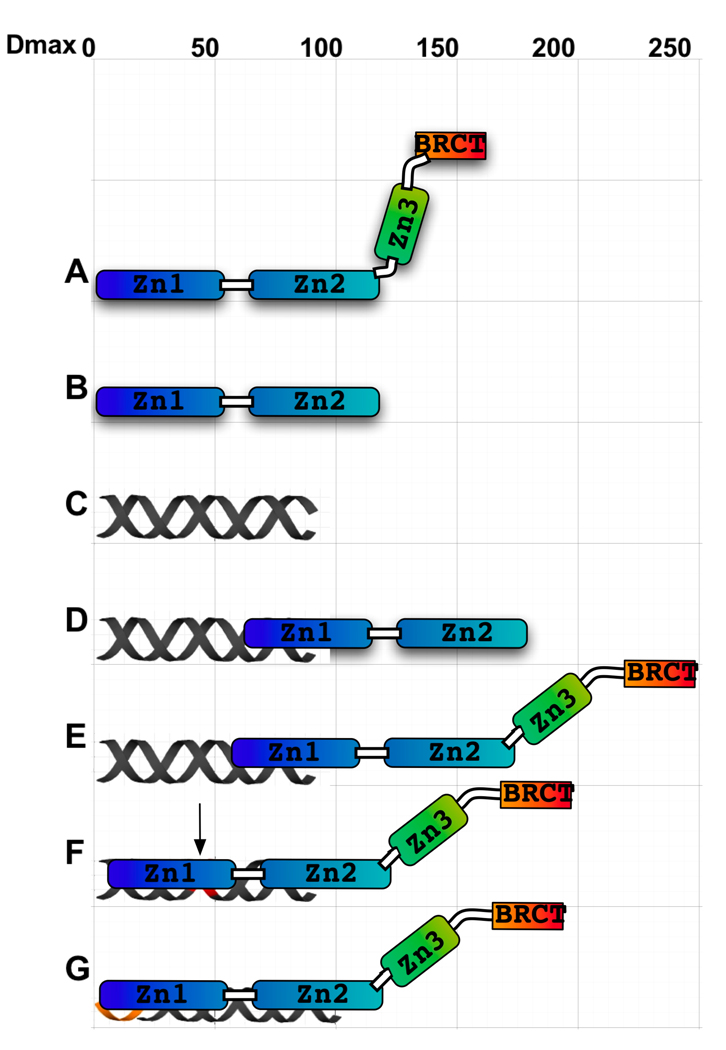
Two recent structural studies of the PARP-1 zinc-ribbon domain have revealed that this domain is required for DNA-mediated activation of PARP-1 by either catalyzing dimerization of the full-length protein or inducing a conformational change10; 17. In light of our SAXS data and in the absence of any data suggestive of DNA-mediated dimerization, we support the notion that a DNA-mediated conformational change in the region surrounding zinc-ribbon domain leads to PARP-1 activation. We propose that the conformational change that occurs upon DNA association is communicated across the hinge region, and ultimately leads to activation of the PARP-1 enzymatic domain. We have summarized an interpretation of our findings in Figure 6. The model is drawn to scale to reflect total maximum dimension of individual hparp486 domains, DNA, and the hparp486-DNA complexes. We propose that hparp486 interacts with DNA containing overhangs or nicks through an increased area of contact compared to its interaction with blunt end DNA in interactions that lead to the decreased Dmax for these complexes. Taking into account the single maximum revealed from the SAXS data, we have modeled this complex as an extended form of hparp486 bound to the area surrounding the DNA damage forming an elongated and asymmetric complex. In conclusion, we believe it is unlikely that PARP-1 is activated by DNA-dependent dimerization, but instead propose a conformational change upon interaction with damaged DNA results in its activation.
Figure 6. A model for the interactions of hparp486 with damaged DNA.
The overall shape and Dmax of hparp486 (A), zf-parp (B), 30Blunt DNA (C) and their complexes (D and E) have been experimentally determined by SAXS. The Dmax of 180Å (a 65Å increase) and asymmetric shape of the zf-parp-30Blunt complex is consistent with a single parp zinc finger interacting the blunt end (D). The hparp486-30Blunt complex is also elongated (E), but increases in size by 90Å (to 240Å). To account for both the change in profile of the P(r) function (Fig. 5 A, C) and greater Dmax,we model this with a conformational change that occurs in the Zn3-BRCT region of the protein. In contrast, when interacting with nicked DNA (F) or a 3’ extension region (G), hparp486 forms a more compact complex but a conformational change still needs to be taken into account to reach a Dmax of ~200Å. We propose that ultimately the conformational change that occurs during binding of DNA leads to activation of the PARP-1 enzymatic domain.
Materials and Methods
Cloning, expression and purification of hparp486
DNA encoding the first 486 residues of human PARP-1 (codon optimized for E. coli) was cloned into the pET28a vector system (Novagen). hparp486 was expressed (via IPTG induction) in E. coli BL21 (DE3) (Novagen) for 6 hours at 25°C. Following cell lysis, the protein was bound to a Nickel-NTA fast flow column (Qiagen) and washed with 3 volumes of resuspension buffer prior to being eluted with 300 mM imidazole. hparp486 was further purified by S200 size exclusion and Hitrap SP cation exchange columns (GE Healthcare).
Cloning, expression and purification of Hparp209
The DNA sequence encoding the first 209 residues of human PARP-1 (codon optimized for insect cells) were cloned into pAB-GST (AB-Vector) baculovirus transfer vectors and co-transfected with linearized Pro-fold C1 (AB-Vector) baculovirus DNA. The transfected virus was expanded and plaque assays were used to determine the isolates that best expressed hparp209. The resulting infectious viruses express hparp209 under the control of the very-late Polyhedron promoter. One liter of SF9 insect cells were grown to a density of 1×106 cells/ml in Ex-cell 420 serum free media (Sigma-Aldritch) and infected with an MOI of 3:1 for 72 hours at 28° C. At this point cell pellets were spun down and frozen at −20°C until further use. Upon thawing, the cell pellets were resuspended in lysis buffer (25 mM Tris HCl, 200 mM NaCl) and sonicated for 2 minutes. The suspension was spun at 10,000 g for 20 minutes and the supernatant was incubated with 10 mls of a Glutathione resin slurry (Qiagen) for 1 hour before washing with 50 mls of lysis buffer. The GST moiety was removed by incubating the protein with 100 units of PreScission Protease (GE Healthcare) overnight while rocking at 4°C. The protein was purified to homogeneity by HPLC chromatography over a Heparin HP ion exchange column. The protein bound at pH 7.0, 100mM NaCl and was eluted at pH 7.0 300mM NaCl (GE Healthcare). Size exclusion chromatography (S200; GE Healthcare) was used as a final purification step. The resulting protein preparation yields approximately 2.5 mg of hparp209 per liter of culture and the protein was stable for several weeks at a concentration of 1mg/ml when kept at 4°C.
Light Scattering Measurements
2mg/ml samples of either hparp486, DNA or the hparp486-DNA complex in 75 mM Tris/HCl pH 8.0, 300 mM NaCl and 1 mM TCEP were loaded onto an ÄKTA purifier HPLC system. Samples were characterized over a SHODEX-803 size exclusion column at a flow rate of 0.5 mls/minute prior to flowing into an online Dawn Heleos II (Wyatt Technologies) multiangle light scattering instrument followed by a REx refractive index detector (Wyatt Technologies). A differential index of diffraction (dn/dc) value of 0.185, 0.170, 0.180 were used to determine the concentration of hparp486, DNA and the hparp486-DNA complex respectively.
Analytical ultracentrifugation
Samples were dialyzed extensively against 25 mMTris pH 8.0, 200 mM NaCl and 1 mM TCEP prior to sedimentation. All experiments were performed in a Beckman XL-I using the absorbance optical system and a 4-hole, AN60-Ti rotor. Sedimentation velocity (SV) was performed in a 1.2 cm, 2-sector EPON centerpiece, while sedimentation equilibrium (EQ) was performed in a 1.2 cm, 6-sector centerpiece. For SV, 400 µl samples of hparp486 were sedimented at 50,000 rpm for 4 hours at 20°C, with a radial step size of 0.002cm in the continuous scanning mode. For EQ, 100 µl, samples were spun to equilibrium at 17,000, 24,000, 30,000 and 35,000 RPM.
DNA oligomer preparations
The oligomer sequences for the 30Ext primer were 5’-ATCAGATAGCATCTGTGCGGCCGCTTAGGGTTAGGG-3’ for the overhang strand and 5’-CCCTAAGCGGCCGCACAGATGCTATCTGAT-3’ for the shorter strand. Other DNA constructs were generated by adding or deleting nucleotides from the 3’ region for the sequences shown above. Oregon Green labeled 3’- and 5’- labeled and unlabeled sequences were ordered from IDT DNA. Various 3’ overhangs, nicked and blunt ended double stranded DNAs were created by annealing equimolar amounts of oligomer. To anneal DNAs, the oligomer were mixed in an Eppendorf tube that was inserted in a 1 liter boiling water bath for 5 minutes, then allowed to cool to room temperature over night.
Mobility shift assays
Protein-DNA complexes were assayed for homogeneity and stoichiometry by analyzing their mobility on 0.8–1.2% native agarose gels stained with Coomassie blue and ethidium bromide to visualize protein and DNA respectively. Low EEO agarose was obtained from Fisher. Gels were run in 25mM Tris pH. 8.0, 20 mM Glycine, at 60 volts for 60 minutes at 4°C.
Thermodynamic Measurements
Affinity measurements of human parp486-DNA complexes were carried out in the Perkin Elmer Victor 3V plate reader. Corning 384 well non-binding polystyrene plates were used to minimize nonspecific binding of the protein complex to the chamber surfaces. In these experiments, human parp486 and DNA were resuspended into 75 mM Tris pH 8.0, 300 mM NaCl and 1 mM TCEP. 2 nM Oregon Green labeled DNA, 0.005-6µM PARP was added to each sample. The fluorophores were excited at 488 nm and the quenching of fluorescence at 530 nanometers was monitored. Triplicate samples were averaged and plotted with Kaleidagraph.
| Equation 1 |
Data were fit to Equation 1, derived from reaction scheme 1 (see text) to the normalized fraction quenched (F.C.) which was observed as a function of P, where Pt is the total concentration of protein titrated, NH is the Hill coefficient, and Kd is the apparent dissociation constant. The NH was assumed to be one unless the data dictated otherwise.
Small Angle X-ray Scattering
SAXS data for human parp486, DNA constructs and their complexes were measured at the SIBYLS beamline (12.3.1) at the ALS using a Mar CCD area detector (165 mm diameter) at room temperature. The intensity curves were measured at concentrations of human parp486 at 9mg/ml, 6mg/ml, and 3mg/ml; DNA samples at 6mg/ml and 3mg/ml; and human parp486-DNA complex samples equal to 6mg/ml and 3mg/ml. Data images were subjected to circular integration, normalization, and subtraction of sample and buffer image files. The Rg for each particle was approximated using PRIMUS (Kovnarev et al., 2003) to evaluate the Guinier equation and GNOM (Svergun, 1992) to evaluate the P(r) function. The value of the maximum diameter of the particle, Dmax, was determined empirically by examining the quality of the fit to the experimental data for a range of Dmax values.
Ab initio particle reconstructions were built by the program DAMMIN 21. For each final model, thirty independent DAMMIN reconstructions were aligned and averaged with the programs DAMAVER and SUPCOMB to reduce the likelihood that the models represent local minima 19; 30. The most typical averaged model was filtered to an appropriate volume with the program DAMFILT.
In an independent approach, a model of human parp486 in solution was generated by rigid-body refinement from the high-resolution structures of the individual domains, obtained from the Protein Data Bank. The program SASREF 22 uses simulated annealing to find an optimal configuration of the domains by simultaneous fitting of the SAXS data to a model consisting of the coordinate files from Zn1, Zn2, Zn3 and BRCT domains (PDB entries 2dmj, 2cs2, 2jvn, 2cok, respectively). The coordinate files were initially truncated to minimize extended loops at either N or C terminal regions.
Supplementary Material
Acknowledgments
We thank Drs. Andy Andrews, Mary Robinson and Michal Hammel for their support. These studies were supported by funding from the American Cancer Society to WL, by NIH GM061909 and by the Howard Hughes Medical Institute. Dr. Lilyestrom is an American Cancer Society Postdoctoral Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 3.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobson EL, Antol KM, Juarez-Salinas H, Jacobson MK. Poly(ADP-ribose) metabolism in ultraviolet irradiated human fibroblasts. J Biol Chem. 1983;258:103–107. [PubMed] [Google Scholar]
- 5.Oleinick NL, Evans HH. Poly(ADP-ribose) and the response of cells to ionizing radiation. Radiat Res. 1985;101:29–46. [PubMed] [Google Scholar]
- 6.Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 7.McCurry LS, Jacobson MK. Poly(ADP-ribose) synthesis following DNA damage in cells heterozygous or homozygous for the xeroderma pigmentosum genotype. J Biol Chem. 1981;256:551–553. [PubMed] [Google Scholar]
- 8.Nolan NL, Butt TR, Wong M, Lambrianidou A, Smulson ME. Characterization of poly(ADP-ribose)--histone H1 complex formation in purified polynucleosomes and chromatin. Eur J Biochem. 1980;113:15–25. doi: 10.1111/j.1432-1033.1980.tb06133.x. [DOI] [PubMed] [Google Scholar]
- 9.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 10.Tao Z, Gao P, Hoffman DW, Liu HW. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry. 2008;47:5804–5813. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber V, Molinete M, Boeuf H, de Murcia G, Menissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. Embo J. 1992;11:3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradwohl G, Ménissier de Murcia JM, Molinete M, Simonin F, Koken M, Hoeijmakers JH, de Murcia G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Murcia G, Schreiber V, Molinete M, Saulier B, Poch O, Masson M, Niedergang C, Menissier de Murcia J. Structure and function of poly(ADP-ribose) polymerase. Mol Cell Biochem. 1994;138:15–24. doi: 10.1007/BF00928438. [DOI] [PubMed] [Google Scholar]
- 14.Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 15.Pion E, Ullmann GM, Ame JC, Gerard D, de Murcia G, Bombarda E. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation. Biochemistry. 2005;44:14670–14681. doi: 10.1021/bi050755o. [DOI] [PubMed] [Google Scholar]
- 16.Petrucco S, Percudani R. Structural recognition of DNA by poly(ADP-ribose) polymerase-like zinc finger families. Febs J. 2008;275:883–893. doi: 10.1111/j.1742-4658.2008.06259.x. [DOI] [PubMed] [Google Scholar]
- 17.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 18.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 19.Koch MH, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 20.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. Journal of Applied Crystallography. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dantzer F, Ame JC, Schreiber V, Nakamura J, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 24.Bai Y, Das R, Millett IS, Herschlag D, Doniach S. Probing counterion modulated repulsion and attraction between nucleic acid duplexes in solution. Proc Natl Acad Sci U S A. 2005;102:1035–1040. doi: 10.1073/pnas.0404448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrucco S. Sensing DNA damage by PARP-like fingers. Nucl. Acids Res. 2003;31:6689–6699. doi: 10.1093/nar/gkg890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Cam E, Fack F, Menissier-de Murcia J, Cognet JA, Barbin A, Sarantoglou V, Revet B, Delain E, de Murcia G. Conformational analysis of a 139 base-pair DNA fragment containing a single-stranded break and its interaction with human poly(ADP-ribose) polymerase. J Mol Biol. 1994;235:1062–1071. doi: 10.1006/jmbi.1994.1057. [DOI] [PubMed] [Google Scholar]
- 27.Pion E, Bombarda E, Stiegler P, Ullmann GM, Mely Y, de Murcia G, Gerard D. Poly(ADP-ribose) polymerase-1 dimerizes at a 5' recessed DNA end in vitro: a fluorescence study. Biochemistry. 2003;42:12409–12417. doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- 28.Kamashev D, Balandina A, Mazur AK, Arimondo PB, Rouviere-Yaniv J. HU binds and folds single-stranded DNA. Nucl. Acids Res. 2007 doi: 10.1093/nar/gkm667. gkm667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. Embo J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. Journal of Applied Crystallography. 2001;34:33–41. [Google Scholar]
- 31.Nusinow D, Hernandez-Munoz I, Fazzio T, Shah G, Kraus L, Panning B. Poly(ADP-ribose) Polymerase 1 Is Inhibited by a Histone H2A Variant, MacroH2A, and Contributes to Silencing of the Inactive X Chromosome. Journal of Biological Chemistry. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.