Abstract
Previously, we prepared monoclonal antibodies (mAbs) by immunizing rats with the recombinant fusion proteins of mouse Langerin/CD207, which contained a flexible linker sequence from E. coli OmpF and a FLAG epitope. We found many of new rat mAbs were not reactive to mouse Langerin, and here we identify the epitopes of two of these IgG mAbs, L2 and L5, and assess their efficacy in various immunodetection methods. MAb L5 is a rat IgG mAb against the FLAG epitope, which detected both N-terminal and C-terminal FLAG tagged protein 2 to 8 times better than the conventional anti-FLAG mAb M2 by Western blot. For mAb L2, we found its epitope to be a 14 amino acid sequence SGFANELGPRLMGK which consisted of both sequences from the OmpF derived linker and mouse Langerin. This epitope sequence was named OLLAS (E. coli OmpF Linker and mouse Langerin fusion Sequence), and mAb L2 as mAb OLLA-2. When the OLLAS sequence was inserted into recombinant proteins at N-terminal, C-terminal, or internal sites, the OLLAS tag was detected by mAb OLLA-2 with very high sensitivity compared to other conventional epitope tags and anti-tag mAbs. MAb OLLA-2 recognized OLLAS tagged proteins with at least 100-fold more sensitivity than anti-FLAG M2 and anti-V5 mAbs in Western blot analyses. We also find the OLLAS epitope to be superior in immunoprecipitation and other immunodetection methods, such as fluorescent immunohistochemistry and flow cytometry. In the process, we successfully utilized the OLLAS epitope sequence as an internal linker for fusion between the engineered mAb and the antigen, and thus achieved improved immunodetection.
Keywords: Monoclonal Antibody, Protein Tagging, FLAG Tag, OLLAS Tag
1. Introduction
Epitope tagging has become an important tool for detecting, localizing and purifying expressed recombinant proteins (Nygren et al., 1994). This methodology involves the fusion of the tag amino acid sequence to the amino or carboxy terminus of a protein of interest, and then identifying the tag with a monoclonal antibody (mAb). For most biochemical applications, the use of epitope tags eliminates the need to generate an antibody to the specific protein that is to be detected and/or purified.
Currently, there are several validated mAb epitopes in wide use for protein tagging, including the FLAG peptide epitope (8 amino acid residues reactive to mAbs M1 and M2) (Hopp et al., 1988), the V5 epitope (14 amino acids found in the P and V proteins of paramyxovirus, Simian Virus 5) (Southern et al., 1991), the myc epitope (10 amino acids derived from the c-myc proto-oncogene product) (Evan et al., 1985), the HA epitope (9 amino acids from the hemagglutinin of influenza virus) (Wilson et al., 1984), and the 6×His epitope (Lindner et al., 1997). However, the choice of an epitope tag depends on the application, because not all tags and in particular the corresponding mAbs are equally suitable for all immunodetection methods, e.g. Western blotting, immunofluorescence staining, immunoprecipitation, and flow cytometry.
Previously, we generated a large panel of mAbs to the extracellular domain (ECD) of mouse Langerin/CD207 (Cheong et al., 2007). In the process, we immunized rats with two different forms of fusion proteins for mLangerin ECD expressed and purified from culture supernatants of stably transfected Chinese hamster ovary (CHO) cells (Cheong et al., 2007). The two fusion partners of mLangerin ECD were chosen for the convenience of affinity purification, i.e. human IgG1 Fc fusion for Protein A column purification and a FLAG epitope fusion for mAb M1 column purification. In both fusion forms of mLangerin ECD proteins, a flexible linker sequence was inserted between the mLangerin ECD and the fusion partner in order to facilitate the folding of fusion proteins and thus increase secretion from the cells. The linker sequence consisted of 17 amino acid residues from the highly flexible domain in E. coli OmpF protein, named OFL (OmpF linker). Following immunization of rats with these two fusion proteins of mLangerin ECD, we generated and screened 78 rat mAb hybridomas positive on ELISA with FLAG/OFL tagged mLangerin ECD protein. Only 10 of the hybridomas turned out to be reactive to mLangerin ECD (Cheong et al., 2007), while all the remaining mAbs were by-products without a known binding specificity.
Amongst the by-product rat IgG mAbs, mAbs L2 and L5 were unique in terms of strong affinity and high specificity to the FLAG/OFL tagged mLangerin ECD protein. In this report, we identify the epitopes of mAbs L2 and L5 and characterize the usefulness of these new rat mAbs in comparison with other current mAbs to epitope tag. We find that newly generated mAb L5 is specific to the FLAG epitope and binds with higher affinity than mAb M2, a widely used ANTI-FLAG® reagent. For mAb L2, we identify the epitope of a 14 amino acid sequence residing in the junction between OFL and mLangerin ECD, named OLLAS (E. coli OmpF Linker and mouse Langerin fusion Sequence) epitope. When compared to other epitope mAbs, mAb L2 (renamed as mAb OLLA-2) specifically recognizes the OLLAS tagged proteins with very high sensitivity, and we demonstrate that mAb OLLA-2 and the OLLAS epitope tag are effective tools for diverse immunodetection methods. The new anti-OLLAS mAb OLLA-2 and anti-FLAG mAb L2 are both rat IgG's.
2. Materials and methods
2.1. Animals and cell lines
Wistar Furth rats were purchased from Charles River Laboratories (Wilmington, MA). C57BL/6 and BALB/c mice were purchased from Taconic Farms (Hudson, NY) and Charles River Labs, and used at 6–8 wks of age. All animals were maintained under specific pathogen-free conditions. Animal care and experiments were conducted according to institutional guidelines of the Rockefeller University and Memorial Sloan-Kettering Cancer Center.
Chinese hamster ovary (CHO) cells and 293T cells were cultured in DMEM with 7 % FBS or 5 % Ultra-Low IgG FBS supplemented with 2mM glutamine (Gibco BRL, Invitrogen, Carlsbad, CA), antibiotics (Invitrogen) and non-essential amino acids (Invitrogen).
2.2. MAb hybridoma production
Generation and screening of mAb hybridomas against FLAG/OFL tagged mLangerin ECD were described previously (Cheong et al., 2007). In brief, Wistar Furth rats were immunized with the purified proteins of mLangerin ECD fused with OFL and human IgG1 Fc (GenBank accession no. DQ917567) and FLAG/OFL tagged mLangerin ECD (GenBank accession no. DQ917568), followed by the injection of mouse dendritic cells enriched from ear skin organ cultures (Cheong et al., 2007). Then, the cells from spleen were used for hybridoma fusion at the Monoclonal Antibody Core Facility of the Rockefeller University and Memorial Sloan Kettering Cancer Center. Culture supernatants from the hybridomas were screened on ELISA with FLAG/OFL tagged mLangerin ECD protein as previously described (Cheong et al., 2007).
2.3 Reagents
To purify mAbs L2, L5, L31 and NLDC145, the supernatants of individual hybridomas were cultured and the mAbs were purified with protein G (Pierce, Rockford, IL; GE Healthcare, Piscataway, NJ) column, according to the manufacture's instructions. The following reagents were purchased; ANTI-FLAG® M2, DAPI (Sigma-Aldrich, St. Louis, MO), anti-beta actin (Abcam Inc., Cambridge, MA), anti-human CD8 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), pEGFP-N1, anti-GFP (Clontech, Mountain View, CA), anti-B220 (BD Biosciences, San Jose, CA), anti-V5 (Invitrogen), HRP-conjugated anti-mouse IgG and HRP- or PE-conjugated anti-rat IgG (Southern Biotech, Birmingham, AL), PE-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), Alexa350-, Alexa488- or Alexa647-conjugated anti-rat IgG and Alexa647-conjugated anti-mouse IgG (Molecular Probes, Invitrogen).
2.4. Vector constructions and expression of recombinant proteins
The construction, expression and purification of murinized heavy and light chains for anti-DEC205 mAb NLDC145 (mNLDC145) and isotype control mAb have been described (Hawiger et al., 2001). The sequences of OLLAS peptide (SGFANELGPRLMGK; GenBank accession no. EF635496) or OLLAS-tagged ovalbumin (GenBank accession no. EF635488) were inserted at the carboxy-terminus in heavy chains of mNLDC145 or isotype control mAbs. 293T cells were transfected with the expression vectors for mNLDC145 or isotype control mAbs carrying OLLAS-tagged inserts. Then, the culture supernatants or mAbs purified by Protein G affinity column were used for further analyses. Soluble FLAG-mLangerin.ECD protein was purified from culture supernatants of the stable CHO/FLAG-mLangerin.ECD cells by ANTI-FLAG® M1 Affinity Gel (Sigma-Aldrich) following the manufacturer's instruction.
The generation of expression vectors for the open reading frames (ORFs) of full-length mLangerin and soluble fusion mLangerin ECD with FLAG/OFL was described previously (Cheong et al., 2007). The cDNAs for epitope tags or subdomains of individual genes were generated by PCR, sequenced, and ligated to make the ORFs for respective recombinant fusion proteins. Then, the ORFs were cloned into pCMV mammalian expression vector (Clontech) and stably transfected into CHO cells or transiently transfected to 293T cells. The expression vectors for the ORFs of following recombinant proteins were generated; FLAG-mLangerin.ECD (GenBank accession no. EF635489), FLAG-OFL/2-mLangerin.ECD (GenBank accession no. EF635490), FLAG-mSIGN-R1.ECD (GenBank accession no. EF635491), rat P11-FLAG.HA (GenBank accession no. EF635492), human CD8.ECD-OFL-mLangerin.ECD (GenBank accession no. EF635493), hCD8.ECD-OFL-mLangerin.ECD.deletion.#1 (GenBank accession no. EF635494), hCD8.ECD-OFL-mLangerin.ECD.deletion.#2 (GenBank accession no. EF635495), OT1-EGFP-OLLAS (GenBank accession no. EF635497), FLAG.OLLAS-EGFP (GenBank accession no. EF635498). V5-hIgG1Fc-OLLAS was generated from V5-hIgG1Fc-OFL-mLangerin.ECD (GenBank accession no. DQ917567; Cheong et al., 2007) by replacing OFL-mLangerin.ECD with OLLAS. OLLAS-EGFP and mSIGN-R1.Cytosol-OLLAS-EGFP were generated by inserting OLLAS or mSIGN-R1.Cytosol-OLLAS (GenBank accession no. EF635499) sequences into the multi-cloning site of pEGFP-N1 vector (Clontech).
2.5. Characterization of mAbs
(i) SDS-PAGE and Western blot analysis
293T cells were transfected with the expression vectors for recombinant fusion proteins, harvested at day 2, lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1% Nonident P-40, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with protease inhibitor cocktails (Sigma-Aldrich), and stored at −20 °C. Each lysed sample was mixed with an equal volume of 2× SDS PAGE sample buffer and boiled at 95 °C for 5 min. Then the samples were separated in 12 or 15% SDS-PAGE and transferred onto PVDF membranes, followed by incubation with antibodies. Antibody-reactive bands on the blots were visualized by incubation with peroxidase-labeled secondary Ab followed by treatment with ECL Plus™ reagents (GE Healthcare). The isotypes of mAb heavy and light chains were determined by the Western blot analysis using mAb supernatant as primary Ab and rat isotype specific HRP-conjugated Abs (Southern Biotech) as secondary Ab.
(ii) Immunofluorescence
Stably transfected CHO cells and lymph nodes sections were examined for immunofluorescence as described previously (Kang et al, 2003; Kang et al, 2004). Cells cultured on slides were washed with PBS, fixed in acetone for 5 min, and subsequently washed and blocked with PBS with 3% BSA for 1 hr. Then, after incubation with primary Abs for 1 hr at room temperature, cells were washed and incubated with fluorochrome-labeled secondary Abs. For tissue staining, peripheral lymph nodes from C57BL/6 mice were collected and embedded in Tissue-Tek OCT® (optimal cutting temperature) compounds (Sakura Finetek USA, Torrance, CA) before freezing at −80 °C. Frozen tissues were sectioned 10 μm in thickness on a microtome and fixed in cold acetone for 15 min. Sections were incubated with primary Abs at room temperature in a humidified chamber for 1 hr, washed and then incubated with fluorochrome-labeled secondary Abs. Sections were mounted in Aqua-Poly Mount (Polysciences, Warrington, PA) and were stored at 4°C until microscopic examination. The images were acquired with a deconvolution microscopy (Olympus, Melville, NY) or with a Zeiss LSM 510 system (Carl Zeiss MicroImaging, Thornwood, NY) at the Rockefeller University Bio-Imaging Resource Center.
(iii) FACS analysis
After detaching with 1 mM EDTA in PBS for 10 min, CHO/mDEC205 cells were incubated with primary Abs for 15 min at 4 °C. Cells were washed, detected by Alexa 647 conjugated secondary Abs for 15 min at 4 °C, and analyzed with FACSCalibur flow cytometer (BD Biosciences) at the Rockefeller University Flow Cytometry Resource Center.
2.6. Sequence analysis
The sequences of OFL and OLLAS were analyzed by PSIPRED (Bryson et al., 2005; McGuffin et al., 2000) using the PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/), and the NCBI BLAST database (http://www.ncbi.nlm.nih.gov/BLAST/) were searched for the sequence similarities.
3. Results
3.1. MAb L5 recognizes the FLAG epitope and L2 recognizes an OFL dependent tag
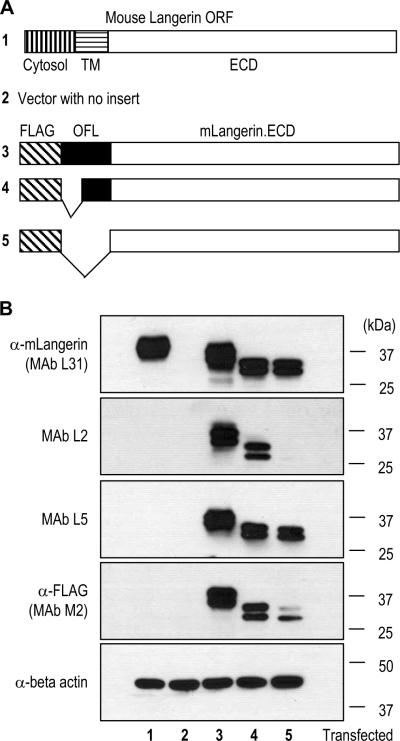
To identify the epitopes of newly generated rat IgG mAbs L2 and L5, we first made different forms of recombinant mLangerin proteins with/without E. coli OmpF derived flexible linker (OFL) sequences (Figure 1A), which consisted of 17 amino acid residues, NATPITNKFTNTSGFAN. FLAG epitope tags with full or half-deleted OFL or without OFL were fused to the N-terminus of mLangerin extracellular domain (ECD) for which a specific L31 mAb was recently obtained and described (Cheong et al., 2007). These constructs were cloned into CMV mammalian expression vectors and transfected to 293T cells. The cell lysates were subjected to Western blot analyses (Figure 1B), using mAbs L2 and L5 in comparison with L31 (anti-mLangerin; Cheong et al., 2007) and the commercial mAb M2 (ANTI-FLAG® from Sigma Aldrich). The results indicated that, while anti-mLangerin mAb L31 recognized all the recombinant proteins containing mLangerin ECD, mAb L2 only detected the two recombinant proteins containing OFL sequences (Figure 1B, lanes 3 & 4). Since constructs containing mLangerin ORF (Figure 1B, lane 1) or FLAG only (Figure 1B, lane 5) were not detected by mAb L2, the epitope of mAb L2 is different from the epitopes identified by anti-mLangerin L31 and anti-FLAG M2. Interestingly, mAb L2 could detect the construct containing half-deleted OFL sequence (Figure 1B, lane 4) where two N-glycosylation sites in OFL were removed (Figure 3C).
Figure 1.
Newly generated rat IgG monoclonal antibodies (mAbs) recognize tags expressed as fusions with mouse Langerin (mLangerin). (A) Schematic view of different forms of recombinant mLangerin proteins. Cytosol, transmembrane (TM) and extracellular domain (ECD) of the mLangerin open reading frame (ORF) are indicated. A FLAG epitope tag and/or an E. coli OmpF derived flexible linker (OFL) sequences were fused to the N-terminus of the mLangerin ECD. (B) Different mLangerin constructs cloned into a CMV mammalian expression vector were transfected into 293T cells, followed by the Western blot analyses with newly generated rat IgG mAbs, i.e. L31 specific for the mLangerin ECD, and the new L2 and L5 MAbs specific for the sequence tags, and anti-FLAG (M2; mouse IgG mAb from Sigma-Aldrich) and anti-beta-actin.
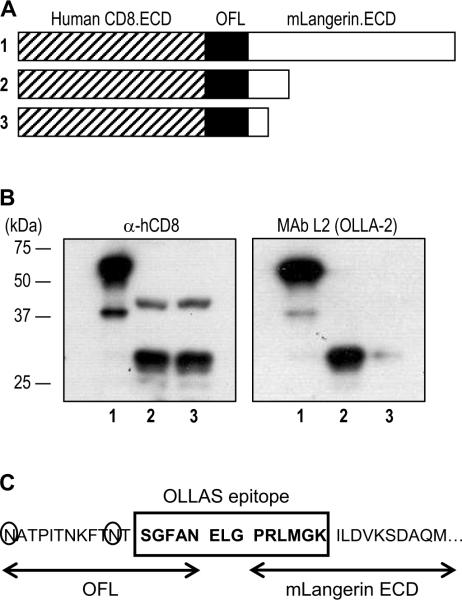
Figure 3.
The epitope of mAb L2 (renamed OLLA-2) is a fusion sequence between OFL and mLangerin ECD. (A) Schematic view of serial deletions in hCD8.ECD-OFL-mLangerin.ECD fusion proteins. (B) The series of C-terminal deletion constructs in (A) were transfected into 293T cells, followed by Western blot analyses with anti-hCD8 (left panel) and mAb OLLA-2 (right panel). (C) The 14 amino acid sequence, named OLLAS (E. coli OmpF Linker mouse Langerin fusion Sequence), residues from both OFL and mLangerin ECD was identified as the epitope recognized by mAb OLLA-2. The 3 amino acid residues (ELG) in the middle of OLLAS epitope correspond to the junction sequence from the ligation between DNAs for OFL and mLangerin ECD. The 2 Asn (N) residues circled are potential sites for N-glycosylation in OFL.
Another newly generated mAb L5 was specifically reactive to all the recombinant proteins containing a FLAG sequence, but not mouse Langerin itself, similarly to anti-FLAG mAb M2 (Figure 1B). Thus, mAb L5 is a new rat IgG mAb against the FLAG epitope. We also have used mAb L5 efficiently in the immunoprecipitation and immunofluorescent detection of FLAG tagged recombinant proteins (data not shown).
3.2. Comparison of anti-FLAG binding sensitivity between rat IgG mAb L5 and a commercially available mouse IgG mAb M2
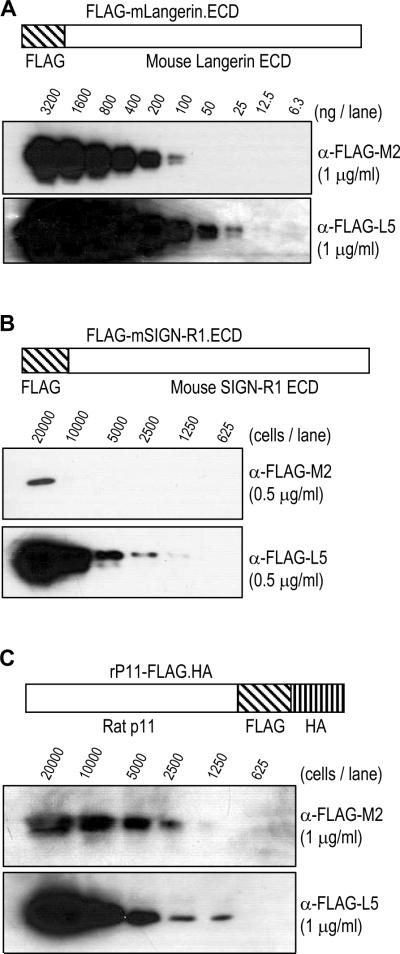
To compare the binding sensitivity to the FLAG epitope between mouse IgG mAb M2 and the new rat IgG mAb L5, we performed Western blot analyses with different FLAG tagged proteins. First, we loaded the purified protein of N-terminal FLAG tagged mLangerin ECD in serial, two-fold dilutions (Figure 2A). The serially diluted samples were blotted in 1 μg/ml of anti-FLAG mouse IgG mAb M2 or rat IgG mAb L5, followed by detection with secondary anti-mouse IgG or anti-rat IgG antibodies respectively. The results with the two anti-FLAG mAbs indicated that mAb L5 could detect the FLAG tagged protein at 4 fold lower amounts than M2 mAb (Figure 2A). We also performed Western blot analyses with FLAG-mSIGN-R1.ECD, another N-terminal FLAG tagged recombinant protein. The cell lysate of 293T cells transfected with FLAG-mSIGN-R1.ECD construct was analyzed in serial, two-fold dilution with mAbs M2 and L5 as above. Similar to the results from purified, N-terminal FALG tagged protein, mAb L5 could detect anti-FLAG signals in the cell lysates with 8 fold fewer cells (Figure 2B). To test whether the location of FLAG epitope tag could affect the binding sensitivity, we made rP11-FLAG.HA construct, in which the FLAG epitope was inserted at the C-terminus of rat p11 ORF (Figure 2C), and transfected into 293T cells. Again in this C-terminal FLAG tagged recombinant protein, mAb L5 could detect FLAG in cell lysates from as little as 1250 cells, whereas the detection of mAb M2 required cell lysates from 2500 cells (Figure 2C). Therefore, mAb L5 detects both N-terminal and C-terminal FLAG tagged proteins 2–8 times better than the conventional anti-FLAG mAb M2.
Figure 2.
Comparison of binding sensitivity to FLAG epitope tagged proteins between mouse mAb M2 and the new rat mAb L5 under similar conditions. (A) Purified protein comprised of mLangerin ECD with an N-terminal FLAG tag (structure shown in upper panel) was diluted as indicated and separated in SDS-PAGE gels followed by blotting with 1 μg/ml of mouse anti-FLAG mAb M2 (middle panel) and rat anti-FLAG mAb L5 (lower panel). (B) 293T cells were transfected with pCMV-FLAG-mSIGN-R1.ECD (structure shown in upper panel), lysed, diluted as indicated, and separated in SDS-PAGE gels followed by blotting with 0.5 μg/ml of mouse anti-FLAG mAb M2 (middle panel) and rat anti-FLAG mAb L5 (lower panel). (C) 293T cells were transfected with pIRES.Neo3-rP11-FLAG.HA (structure shown in upper panel), lysed, diluted as indicated, and separated in SDS-PAGE gels followed by blotting with 1 μg/ml of mouse anti-FLAG mAb M2 (middle panel) and rat anti-FLAG mAb L5 (lower panel).
3.3. Mapping of the epitope detected by mAb L2, renamed OLLA-2
As shown in Figure 1, mAb L2 recognized an undefined epitope in OFL-mLangerin ECD, indicating the C-terminal half of OFL was required to be detected by mAb L2. To map the epitope recognized by OLLA-2, we designed serial deleted constructs. First, mLangerin ECD was fused with human CD8 ECD with OFL, and serial deletions were made from the C-terminus (Figure 3A). 293T cells were transfected with each construct, lysed, followed by the Western blotting with anti-hCD8 (Figure 3B, left panel) and mAb L2 (Figure 3B, right panel). The results showed that the epitope of mAb L2 included the N-terminal region of the mLangerin ECD (Figure 3B, right panel, lanes 2 & 3). Subsequently, we defined the 14 amino acid peptide epitope recognized by mAb L2 as SGFANELGPRLMGK (Figure 3C), and named the tag, OLLAS (E. coli OmpF Linker and mouse Langerin fusion Sequence). We also renamed mAb L2 as mAb OLLA-2.
3.4. The OLLAS epitope as a superior tag for immunodetection and immunoprecipitation of recombinant EGFP proteins
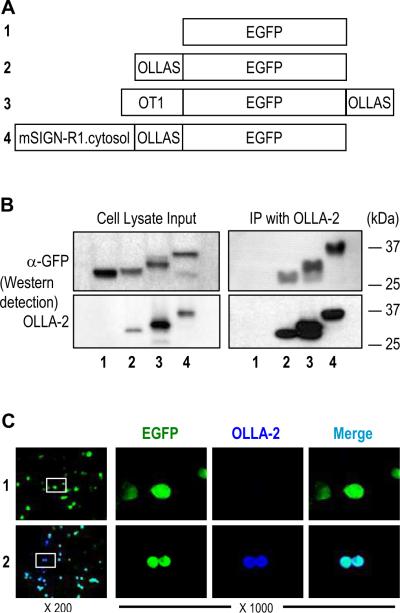
To confirm the use of the new OLLAS epitope as a tag to detect fusion proteins, we fused the OLLAS sequence (SGFANELGPRLMGK) to the N-terminus, the C-terminus, and internal sites of recombinant EGFP proteins (Figure 4A). The expression vectors for these recombinant EGFP proteins were transfected into 293T cells. The cell lysates were subjected to SDS-PAGE followed by Western blot detection with anti-GFP and mAb OLLA-2 (Figure 4B, left panels). Then, these cell lysates were subjected to immunoprecipitation with mAb OLLA-2 followed by the Western blot detection of the immunoprecipitates with anti-GFP and mAb OLLA-2 (Figure 4B, right panels). Detection by anti-GFP or mAb OLLA-2 was clearly visible by Western blot analyses of the cell lysates regardless of the location of the OLLAS epitope (Figure 4B, left panels). Detection of the immunoprecipitates by anti-GFP indicated that successful immunoprecipitation was also achieved with OLLAS epitope tagged proteins by mAb OLLA-2 (Figure 4B, right panels). In addition, the expression vectors for EGFP alone and OLLAS tagged EGFP were transfected into CHO cells to test immunofluorescent staining of cells with mAb OLLA-2. Only the OLLAS epitope tagged EGFP was co-stained with mAb OLLA-2 (Figure 4C, lower panels), indicating that the OLLAS epitope is suitable for the fluorescent immunocytochemistry.
Figure 4.
Immunodetection of OLLAS tagged EGFP proteins. (A) Schematic view of 4 recombinant EGFP proteins without a tag (#1) or with OLLAS epitope tag attached to the N-terminus (#2), C-terminus (#3), or internal site (#4). The OT1 peptide, a ligand of mouse MHC I from ovalbumin, were also present in the N-terminus of EGFP protein #3. The 53 amino acid full-length cytosolic domain from mouse SIGN-R1 was present at the N-terminus of EGFP protein #4. (B) Expression vectors for the 4 different recombinant EGFP proteins were transfected into 293T cells. Cell lysates (left) were immunoprecipitated (IP) with mAb OLLA-2 (right). Then, all samples were separated in SDS-PAGE gels followed by blotting with anti-GFP (upper panels) and mAb OLLA-2 (lower panels). (C) Expression vectors for EGFP (#1; upper panels) and EGFP with OLLAS tag at the N-terminus (#2; lower panels) were transfected into CHO cells. CHO cells were visualized with the signals for EGFP (green) and immunofluorescence staining for OLLA-2 (blue). Insets in left panels are shown at 1000 fold magnification in the right.
3.5. Comparison of binding sensitivity of mAb OLLA-2 and other anti-epitope tag mAbs by Western blot analyses
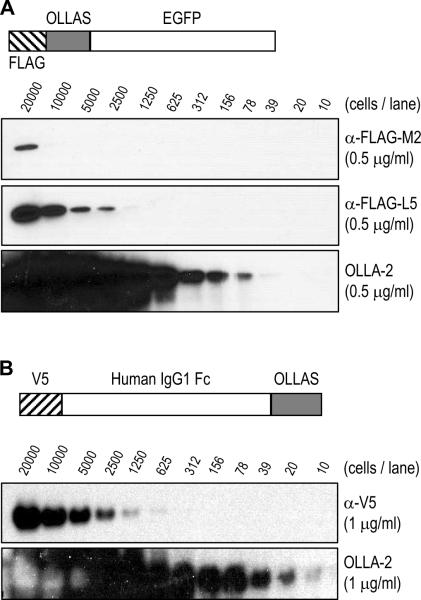
To compare the binding sensitivity of anti-OLLAS tag mAb OLLA-2 with anti-FLAG tag mAbs M2 and L5, EGFP protein was fused with FLAG and OLLAS tag in tandem at the N-terminus (Figure 5A). 293T cells were transfected with this FLAG.OLLAS-EGFP expression vector, lysed, and analyzed in serial, two-fold dilutions by Western blotting with 0.5 μg/ml of anti-FLAG mAbs M2 and L5 as well as mAb OLLA-2 (Figure 5A). The result shows that mAb OLLA-2 binds to its OLLAS epitope with at least 100-fold more sensitivity than anti-FLAG mAb M2 and with at least 30-fold more sensitivity than anti-FLAG mAb L5.
Figure 5.
Comparison of binding sensitivity of mAb OLLA-2 and other anti-epitope tag mAbs. (A) 293T cells were transfected with pCMV-FLAG.OLLAS-EGFP (structure shown in upper panel), lysed, diluted as indicated, and separated in SDS-PAGE gels followed by blotting with 0.5 μg/ml of anti-FLAG mAb M2 (second panel), the new anti-FLAG mAb L5 (third panel), and mAb OLLA-2 (lower panel). (B) 293T cells were transfected with pCMV-V5-hIgG1Fc-OLLAS (structure shown in upper panel), lysed, diluted as indicated, and separated in SDS-PAGE gels followed by blotting with 1 μg/ml of anti-V5 mAb (Invitrogen; middle panel) and mAb OLLA-2 (lower panel).
Next, to compare the binding sensitivity of mAb OLLA-2 with anti-V5 mAb (Invitrogen catalog number R960), human IgG1 Fc protein was fused with V5 epitope at the N-terminus and with OLLAS epitope at the C-terminus (Figure 5B). 293T cells were transfected with this V5-hIgG1Fc-OLLAS expression vector, lysed, and analyzed in serial, two-fold dilutions for the Western blotting with 1 μg/ml of anti-V5 mAb and mAb OLLA-2 (Figure 5B). The results showed that mAb OLLA-2 bound to its OLLAS epitope with at least 100-fold more sensitivity than anti-V5 mAb. These findings indicate that the OLLA-2 mAb possesses very strong affinity for the OLLAS epitope, i.e. SGFANELGPRLMGK, and reacts specifically with recombinant proteins containing this epitope.
3.6. OLLAS epitope as a tag on engineered mAb for immunodetection
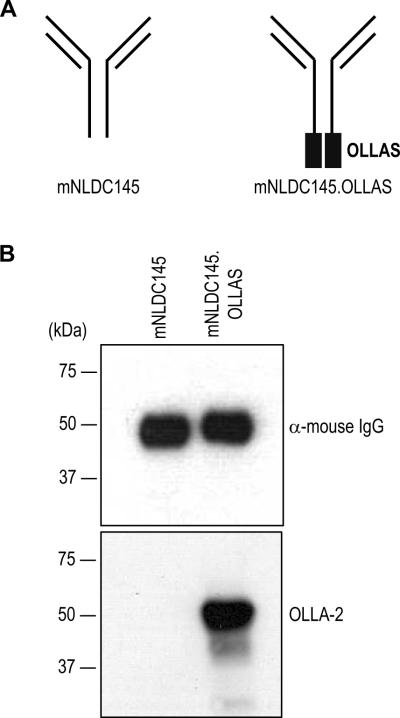
To test whether the OLLAS epitope was suitable as a tag for engineered mAbs, the sequence was fused to the C-terminus of the heavy chain of murinized NLDC145 (mNLDC145; Figure 6A), a cloned and engineered anti-mouse DEC205 mAb (Hawiger et al., 2001). The culture supernatants from 293T cells, which had been transiently transfected with mNLDC145 or mNLDC145.OLLAS expression vectors, were collected and separated in SDS-PAGE gels followed by Western blotting with anti-mouse IgG (Figure 6B, upper panel) and mAb OLLA-2 (Figure 6B, lower panel). The OLLA-2 mAb was specific for the engineered mNLDC145.OLLAS mAb.
Figure 6.
Production of murinized anti-mouse DEC205 mAb mNLDC145 expressing the OLLAS tag. (A) Schematic view of the anti-mDEC205 mAb mNLDC145 engineered so that all rat constant domains are replaced with mouse IgG1 sequences (left) and OLLAS tagged mAb mNLDC145.OLLAS (right). (B) Expression vectors for mNLDC145 and mNLDC145.OLLAS were transfected into 293 cells. In 2 days, the culture supernatants from transiently transfected cells were collected and separated in SDS-PAGE gels followed by blotting with anti-mouse IgG (upper panel) and mAb OLLA-2 (lower panel).
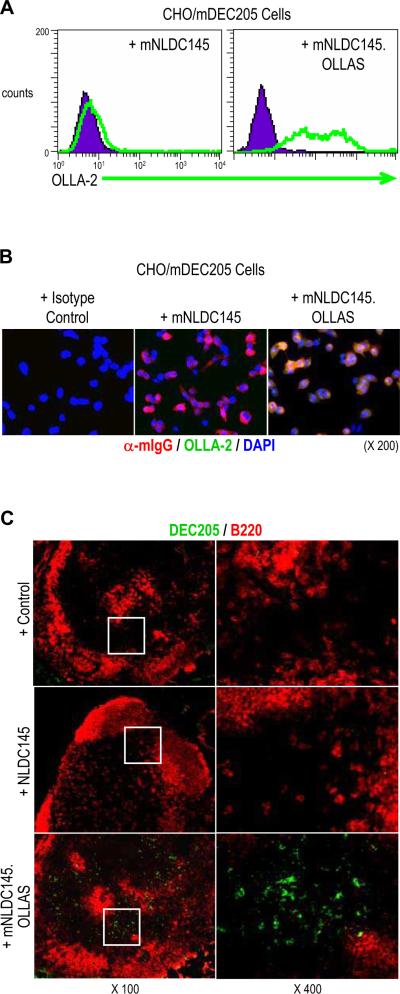
To confirm the usage of OLLAS epitope as a tag for engineered mAbs, the culture supernatants from 293T cells transfected with mNLDC145 or mNLDC145.OLLAS expression vectors were used for FACS analysis. Stable CHO cells expressing mDEC205 (CHO/mDEC205 cells) were incubated with each supernatant and stained with mAb OLLA-2 followed PE-conjugated anti-rat IgG prior to flow cytometry (Figure 7A). The result showed that only mNLDC145.OLLAS with mAb OLLA-2 was able to stain the CHO/mDEC205 cells. For fluorescent immunocytochemistry, CHO/mDEC205 cells were incubated with the culture supernatants containing mNLDC145 or mNLDC145.OLLAS (Figure 7B). Then, cells were stained with anti-mouse IgG (red) and mAb OLLA-2 with anti-rat IgG (green). The result showed that the OLLA-2 signal (green) co-localized with anti-mouse IgG, i.e. mNLDC145, signal (red) only when mNLDC145.OLLAS supernatant was used (Figure 7B, right panel).
Figure 7.
Immunodetection of mDEC205 with OLLAS epitope tag. (A) Expression vectors for mNLDC145 and mNLDC145.OLLAS as in figure 6 were transfected into 293 cells. Stable CHO/mDEC205 cells expressing cell surface mDEC205 were incubated with the culture supernatants containing mNLDC145 (left panel) or mNLDC145.OLLAS (right panel). Then, the cells were further incubated with mAb OLLA-2 followed by PE-conjugated anti-rat IgG prior to flow cytometry. (B) Stable CHO/mDEC205 cells were immunostained with either mouse control IgG (left panel), mNLDC145 (middle panel), or mNLDC145.OLLAS (right). Then, the cells were probed with anti-mouse IgG (red), OLLA-2/anti-rat IgG (green), and DAPI (blue). (C) Mouse lymph nodes were immunostained with rat control IgG (upper panels), rat IgG mAb NLDC145 (middle panels), and mNLDC145.OLLAS (lower panels). Then, the tissues were stained with mAb OLLA-2 followed by anti-rat IgG labeled with Alexa488 (green) and anti-B220 (red). Insets in left panels are shown at 400 fold magnification in the right.
For fluorescent immunohistochemistry, mouse lymph nodes were incubated with rat IgG control, rat IgG mAb NLDC145, or engineered mNLDC145.OLLAS respectively. Subsequently, all lymph nodes were further stained with mAb OLLA-2 and anti-rat IgG labeled with Alexa488 (green) and anti-B220 (red). These results showed that mAb OLLA-2 was specific and sensitive to the OLLAS epitope and that there was no non-specific, background signals in the tissue staining of lymph nodes. These findings confirm that the OLLAS epitope and mAb OLLA-2 is broadly useful in various immunodetection methods, including FACS analysis and fluorescent immunostaining.
3.7. Improved immunodetection with OLLAS epitope tag
As shown in Figure 7C, murinized mAb mNLDC145.OLLAS was able to detect mDEC205 in sections of lymph node tissues (Figure 7C, lower panels), whereas rat IgG mAb NLDC145 was not sensitive enough (Figure 7C, middle panels). This might be explained because the detection of mNLDC145.OLLAS required the secondary mAb OLLA-2 followed by the fluorescent labeled tertiary anti-rat IgG Ab, while the detection of NLDC145 required only the fluorescent labeled anti-rat IgG Ab.
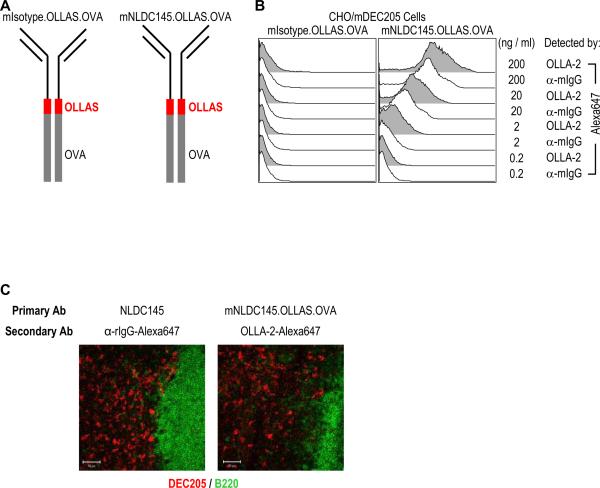
In order to compare directly the use of mNLDC145.OLLAS and mAb OLLA-2, we generate two engineered mAbs with the OLLAS epitope tag (Figure 8A). Expression vectors for murinized isotype control mAb fused with OLLAS epitope and ovalbumin (mIsotype.OLLAS.OVA) and mAb mNLDC145 fused with OLLAS epitope and ovalbumin (mNLDC145.OLLAS.OVA) were transfected into 293T cells. Then, mIsotype.OLLAS.OVA and mNLDC145.OLLAS.OVA mAbs were purified from culture supernatants. MAb OLLA-2 were also purified and then labeled with Alexa647 fluorochrome. As shown in Figure 8B, under the same conditions, CHO/mDEC205 cells incubated with mNLDC145.OLLAS.OVA were better detected by Alexa647 labeled OLLA-2 than Alexa647 labeled anti-mouse IgG.
Figure. 8.
Immunodetection of engineered mAbs with protein fusion by the use of OLLAS epitope tag/linker and mAb OLLA-2. (A) Schematic view of engineered mAbs fused with ovalbumin where OLLAS epitope tag was used as a linker. Murinized IgG isotype control mAb, mIsotype.OLLAS.OVA (left), and anti-mDEC205 mAb mNLDC145.OLLAS.OVA (right) are shown. (B) CHO/mDEC205 cells were incubated for 15 min with 2 μg/ml of mAb mNLDC145.OLLAS.OVA (right panel) or the corresponding isotype control, mIsotype.OLLAS.OVA (left panel). After washing, the cells were incubated with different doses of Alexa647 labeled OLLA-2 or anti-mouse IgG respectively. The analysis was performed by FACS. The data are representative of two experiments. (C) Mouse lymph nodes were immunostained with rat IgG mAb NLDC145 (left panel) and murinized mAb mNLDC145.OLLAS.OVA (right panel). Then, the tissues were stained with Alexa647 labeled anti-rat IgG (red, left panel) or Alexa647 labeled OLLA-2 (red, right panel) and anti-B220 (green). Bar scales equal 50 μm.
Then, we examined the lymph node tissues stained with either engineered mAb mNLDC145.OLLAS.OVA or rat mAb NLDC145 followed by visualization with Alexa647 labeled OLLA-2 and Alexa647 labeled anti-rat IgG, respectively (Figure 8C). Unlike the previous result of lymph modes staining by mAb NLDC145 with Alexa488 labeled anti-rat IgG (Figure 7C, middle panels), mAb NLDC145 with Alexa647 labeled anti-rat IgG was able to detect mDEC205 in sections of lymph node tissues (Figure 8C, left panel) as efficiently as mAb mNLDC145.OLLAS.OVA with Alexa647 labeled OLLA-2 (Figure 8C, right panel). From the results, we conclude that OLLAS epitope is a useful tag for engineered mAbs, which then can be used better than or as efficiently as original mAbs in various applications of immunodetection. We also found that the use of OLLAS peptide as an inter-molecular linker between two recombinant proteins, such as in mNLDC145.OLLAS.OVA and mIsotype.OLLAS.OVA, improved the expression of recombinant fusion proteins (data not shown) without losing its role as a highly effective epitope tag in various immunodetections.
3.8. Secondary structure analysis
We utilized a secondary structure prediction program called PSIPRED (Bryson et al., 2005; McGuffin et al., 2000) to characterize possible conformations of the OFL and OLLAS sequences used in-between the fused proteins that we had generated successfully in previous (Galustian et al., 2004) and current studies. As shown in Figure 9, we included 20 amino acids before and after the OFL and OLLAS linkers into the query sequences to increase the accuracy of the predicted secondary structures. The OFL linker mostly adopts a random coil conformation, while the OLLAS linker shows a strong propensity to form an α-helical conformation.
Figure. 9.
Secondary structure (SS) analysis of the amino acid (AA) sequences of OFL (solid box) and OLLAS (dashed box) linkers located in-between the fused proteins. Straight lines in SS represent random coil conformations, cylinders helical conformations, and an arrow extended (sheet) conformation. Note that both OFL and OLLAS sequences coexist in mouse IgG1-OFL-mLangerin.ECD.
4. Discussion
Here we described two novel rat mAbs (mAbs L5 and OLLA-2) against synthetic tags. Monoclonal antibodies against epitope tags are an efficient, convenient and rapid method for detecting recombinant protein expression (Jarvik and Telmer, 1998). If there is no antibody against the protein of interest, adding an epitope tag to this protein allows for protein detection with an antibody against the epitope sequence. For example, affinity tags such as a FLAG-tag appended to recombinant proteins have traditionally been used as a way of purifying proteins using standard conditions rather than developing individual biochemical purifications based on each protein's physical characteristics (Hopp et al., 1988).
Previously, we immunized rats to produce anti-mLangerin/CD207 antibody using two different forms of fusion proteins for mLangerin ECD expressed and purified from culture supernatants of stably transfected CHO cells (Cheong et al., 2007). As fusion partners of mLangerin ECD, FLAG epitope and flexible linker sequences from E.coli OmpF protein (OFL) were used. FLAG epitope was chosen for column purification and OFL was chosen to facilitate the folding of fusion proteins and to increase secretion from the cells. We initially employed the OFL sequence as a linker because the OFL sequence was viewed as highly flexible based on the molecular dynamics simulation of OmpF from E. coli (Im and Roux, 2002). Although the OFL sequence forms a beta-hairpin loop in OmpF, the secondary prediction by PSIPRED (Bryson et al., 2005; McGuffin et al., 2000) indicates that the OFL exists as a flexible coil in-between the fused proteins. This prediction is corroborated by the successful expressions of fusion proteins for C-type lectins we generated, which were functional in sugar binding activities (Galustian et al., 2004) as well as immunogenic in mAb productions (Kang et al., 2004; Cheong et al., 2007).
In the process, we found that new mAbs L5 and OLLA-2 were specific to the FLAG/OFL tagged mLangerin ECD protein, not to mLangerin ECD. In this report, we demonstrate that L5 is a new anti-FLAG mAb (Figure 1) and OLLA-2 is a highly reactive mAb against a new epitope tag (Figure 1 and 3). The new anti-FLAG L5 mAb shows high sensitivity for the FLAG epitope and specificity for both N-terminally and C-terminally tagged FLAG epitope. In Western bolt analyses, L5 can detect as little as 25 ng of N-terminally FLAG tagged purified protein (Figure 2A) and lysates from 2,500 cells transfected with N-terminally FLAG tagged recombinant protein construct (Figure 2B). Conventional anti-FLAG M2 can detect 100 ng of N-terminally FLAG tagged purified protein (Figure 2A) and lysates from 20,000 cells transfected with N-terminally FLAG tagged recombinant protein construct (Figure 2B). Besides highly enhanced reactivity against the FLAG epitope, the new anti-FLAG L5 is a rat IgG mAb and provides an additional option in immunostaining since all other conventional anti-FLAG antibodies are mouse monoclonals or rabbit polyclonals.
MAb L2 or OLLA-2 is a novel rat IgG mAb against the newly identified epitope (SGFANELGPRLMGK) named OLLAS, with many advantages in recombinant protein engineering and immunodetection. First, this novel pair of tag and anti-tag mAb has a remarkable sensitivity and thus enhances the performance of a variety of immunodetection methods, including Western blot, immunocytochemistry, immunohistochemistry, flow cytometry, and immunoprecipitation. Especially, mAb OLLA-2 can detect the OLLAS epitope tagged proteins by Western blotting with more than 100 fold higher sensitivity than anti-FLAG M2 and anti-V5 mAbs. Second, the new OLLAS epitope can be fused to the C-terminal, the N-terminal, and the internal (or linker) sites of recombinant and fusion proteins including the engineered mAbs. The OLLAS epitope does not interfere with the biological activities of the recombinant proteins where it was inserted, such as fluorescence of EGFP and binding of antibodies, and can improve the expression of recombinant proteins as an inter-molecular linker between two recombinant proteins, as shown in the engineering of anti-mDEC205 mAb fused with ovalbumin. Third, mAb OLLA-2, as a rat IgG, can be used for the immunostaining of mouse and human samples without direct labelling of fluorochrome or enzyme. Because anti-tag mAbs and Abs are mostly made from mouse and rabbit, the anti-OLLAS tag mAb OLLA-2 from rat IgG will provide an extra advantage in immunodetection.
As indicated in Figure 3C, the OFL sequence contains two N-glycosylation sites. It is possible that the N-glycosylation in OFL region could interfere with the structural integrity of the fusion proteins and/or the antibody responses against the fusion proteins where the OFL linker was used. However, the newly identified OLLAS epitope sequence does not have any N-glycosylation site. Unlike OFL, when the OLLAS are used as a linker in the fused proteins, it is predicted to adapt an alpha-helical conformation by PSIPRED (Figure 9). It has been suggested that, in linker engineering, the helical linkers like OLLAS are better than flexible linkers like OFL to maximize the desired function of engineered fusion proteins (Arai et al., 2001). As demonstrated in Figure 8, the OLLAS sequence appears to function properly as a linker in the engineered mAbs fused with antigens. In addition, the OLLAS sequence is a superior protein-tagging epitope to other currently existing tag epitopes. It should be stressed that the OLLAS, generated by fusion between the sequences from E. coli and mouse, is a synthetic sequence different from any of the known protein sequences in nature, indicated by the NCBI BLAST search. The uniqueness of its sequence may make the OLLAS tag more useful for many different applications in immunodetection, especially in vivo applications in diverse organisms.
In summary, the data presented here demonstrate that mAb L5 is a new anti-FLAG with higher sensitivity, and that the novel OLLAS epitope and mAb OLLA-2 are superior tag and anti-tag mAb in terms of highly sensitive detection, broader range of applications, sequence uniqueness, and potential use as a linker. These new epitope tag and anti-tag mAbs will improve the current immunodetection methods for recombinant proteins.
Acknowledgments
We thank Judy Adams for preparing the figures. We were supported by NIH Grants to RMS (AI 13013, AI 40045, AI 057158) and CGP (AI 057158) and a grant from the Histiocytosis Association of Canada.
Abbreviations
- CHO cells
Chinese hamster ovary cells
- DCs
dendritic cells
- ECD
extracellular domain
- FBS
fetal bovine serum
- mAb
monoclonal antibody
- OFL
E. coli OmpF derived linker
- OLLAS
E. coli OmpF linker and mouse Langerin fusion sequence
- ORF
open reading frame
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14:529. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33(Web Server issue):W36. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C, Idoyaga J, Do Y, Pack M, Park SH, Lee H, Kang YS, Choi JH, Kim JY, Bonito A, Inaba K, Yamazaki S, Steinman RM, Park CG. Production of monoclonal antibodies that recognize the extracellular domain of mouse Langerin/CD207. J. Immunol. Methods. 2007;324:48. doi: 10.1016/j.jim.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 1985;5:3610. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galustian C, Park CG, Chai W, Kiso M, Bruening SA, Kang YS, Steinman RM, Feizi T. High and low affinity carbohydrate ligands revealed for murine SIGN-R1 by carbohydrate array and cell binding approaches, and differing specificities for SIGN-R3 and langerin. Int. Immunol. 2004;16:853. doi: 10.1093/intimm/dxh089. [DOI] [PubMed] [Google Scholar]
- Hawiger D, Inaba K, Dorsett Y, Guo M, Mahnke K, Rivera M, Ravetch JV, Steinman RM, Nussenzweig MC. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J. Exp. Med. 2001;194:769. doi: 10.1084/jem.194.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp TP, Prickett KS, Price V, Libby RT, March CJ, Cerretti P, Urdal DL, Conlon PJ. Synthesis of protein with an identification peptide (proteins) Bio/Technology. 1988;6:1205. [Google Scholar]
- Im W, Roux B. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J. Mol. Biol. 2002;319:1177. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- Jarvik JW, Telmer CA. Epitope tagging. Annu. Rev. Genet. 1998;32:601. doi: 10.1146/annurev.genet.32.1.601. [DOI] [PubMed] [Google Scholar]
- Kang YS, Kim JY, Bruening SA, Pack M, Charalambous A, Pritsker A, Moran TM, Loeffler JM, Steinman RM, Park CG. The C-type lectin SIGN-R1 mediates uptake of the capsular polysaccharide of Streptococcus pneumoniae in the marginal zone of mouse spleen. Proc. Natl. Acad. Sci. U.S.A. 2004;101:215. doi: 10.1073/pnas.0307124101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner P, Bauer K, Krebber A, Nieba L, Kremmer E, Krebber C, Honegger A, Klinger B, Mocikat R, Pluckthun A. Specific detection of his-tagged proteins with recombinant anti-His tag scFv-phosphatase or scFv-phage fusions. Biotechniques. 1997;22:140. doi: 10.2144/97221rr01. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Nygren PA, Stahl S, Uhlen M. Engineering proteins to facilitate bioprocessing. Trends Biotechnol. 1994;12:184. doi: 10.1016/0167-7799(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Southern JA, Young DF, Heaney F, Baumgartner WK, Randall RE. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 1991;72:1551. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]