Abstract
Actin filaments and microtubules polymerize and depolymerize by adding and removing subunits at polymer ends, and these dynamics drive cytoplasmic organization, cell division and cell motility. Since Wegner proposed the treadmilling theory for actin in 1976, it has largely been assumed that the chemical state of the bound nucleotide determines the rates of subunit addition and removal. This chemical kinetics view is difficult to reconcile with observations revealing multiple structural states of the polymer that influence polymerization dynamics, but are not strictly coupled to the bound nucleotide state. We refer to these phenomena as “structural plasticity”, and discuss emerging evidence that they play a central role in polymer dynamics and function.
Introduction
Pioneering observations of cell division using polarization microscopy showed that protein polymers in the cell undergo rapid exchange with soluble subunits, and can generate force by subunit addition (polymerization) and loss (depolymerization) (1). Subsequent work revealed that polymerization dynamics of actin filaments, microtubules, and their prokaryotic cousins, indeed play central roles in diverse physiological processes, including cell shape control, cell motility and chromosome segregation (2–4). Understanding the mechanisms by which these cytoskeletal polymers polymerize and depolymerize is critical for understanding how they spatially organize and promote motility. The field of cytoskeletal polymer research has traditionally adopted a chemical kinetics view of polymerization dynamics, which posits that the chemical state of the subunit-bound nucleotide uniquely controls association and dissociation rates of polymer subunits. Accumulating evidence has questioned the purely chemical kinetics view, and points to an important role for structural plasticity, defined here as change in the structural state of a polymer without change in the chemical state of its bound nucleotide, in modulating polymer dynamics. Structural plasticity is likely to play a major role in modulating polymer behavior in cells, and a full understanding of polymerization dynamics will require its integration with chemical kinetics. Here we review basic structural and biochemical properties of actin and tubulin, and models for their polymerization dynamics that are rooted in chemical kinetics theory. We then review evidence for the existence of structural plasticity in these cytoskeletal polymers and discuss implications for their dynamics inside cells.
End-dependent dynamics and nucleotide hydrolysis
In eukaryotes, both actin and tubulin assemble into multi-stranded, polar polymers. Actin filaments consist of two strands that intertwine to form a double helical structure. Microtubules, the polymers of tubulin, usually consist of 13 parallel strands (or protofilaments) that associate laterally to form a sheet-like lattice. Along the microtubule length, this sheet curves around and closes on itself, giving rise to a hollow tubular structure. The structures of prokaryotic actin and tubulin relatives are currently a topic of intense investigation (5). Multi-stranded polymer architecture has two important consequences: it provides mechanical strength, and it largely restricts subunit association and dissociation to polymer ends, because subunits at ends make fewer contacts with neighbors. This end-independence enables cells to control the assembly of long (micron-scale) polymers using localized (nanometer-scale) biochemical reactions at polymer ends, allowing the precise spatial control of polymerization necessary for cell polarity and motility.
Actin and tubulin subunits, as well as their prokaryotic relatives, bind nucleotide triphosphate (NTP), ATP for actin, GTP for tubulin, and polymerize preferentially in their NTP-bound form. Shortly after polymerization, subunits hydrolyze NTP to nucleotide diphosphate (NDP), releasing phosphate (Pi) and retaining NDP in the polymer. The resultant NDP-bound polymer is weaker than an NTP-bound polymer and consequently depolymerizes, releasing individual subunits for another round of polymerization and depolymerization. In this scheme, the free energy of NTP hydrolysis does not catalyze polymerization per se, but instead drives depolymerization, enabling polymers to undergo continuous non-equilibrium turnover in cells. This turnover in turn allows polymers to assemble in some places in the cell while they disassemble in others, and to perform mechanical work by pushing or pulling, or by bending in the case of FtsZ, a bacterial tubulin homolog that helps divide the bacterial cell in two at the end of the cell cycle (6). Understanding how polymers use the energy of nucleotide hydrolysis to promote turnover and perform mechanical work is a central theme in cytoskeleton research.
The chemical kinetics view of polymerization dynamics
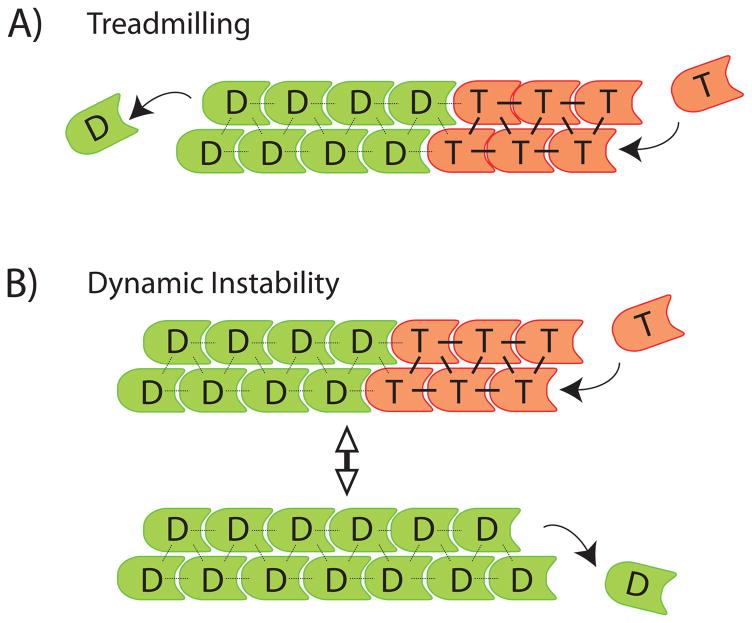
While work in the 1960s and 70s demonstrated a role for NTP hydrolysis in actin and tubulin polymerization, how exactly NTP hydrolysis could drive polymer turnover remained unclear. A solution was proposed in Wegner’s influential treadmilling theory for actin turnover (7). At the heart of this theory was the concept that the chemical state of the nucleotide bound to a subunit influences its association and dissociation rates at the polymer end. Broadly speaking, NTP-bound subunits preferentially associate at ends, whereas NDP-bound subunits preferentially dissociate from ends. In conjunction with polarity-dependent rate constants, this concept gives rise to a mode of polymer turnover where actin filaments grow from one end whilst shrinking from the other (Figure 1A). In the Wegner model, treadmilling is driven by hydrolysis of polymer-bound ATP, which allows ADP-bound subunits to dissociate preferentially from one polymer end whilst ATP-bound subunits are associating preferentially onto the other. This model of polymer turnover was first tested in solutions of actin (7), and was later confirmed and extended by direct observations of single-filaments using timelapse imaging (8).
Figure 1.
Classical chemical kinetics models of polymerization dynamics. T – NTP-bound subunit (red); D – NDP-bound subunit (green). A) Treadmilling. Arrows indicate NTP-subunit association (T, right), and NDP-subunit dissociation from the opposite end (D, left). B) Dynamic instability. Arrows at polymer ends indicate NTP-subunit association (top) and NDP-subunit dissociation (bottom). Bidirectional arrow indicates reversible transitions of the polymer between growing and shrinking states.
After Wegner’s work, microtubules were also proposed to treadmill (9), but later it was found that their dominant kinetic behavior is dynamic instability, where individual ends alternate between bouts of growth and shrinkage (10). Dynamic instability was initially rationalized using a chemical kinetics model, where a lag between tubulin subunit association and subsequent GTP hydrolysis was proposed to generate a cap of GTP-bound tubulin that stabilizes growing ends (Figure 1B, top). Loss of the cap by GTP hydrolysis initiates depolymerization (Figure 1B, bottom). A similar model was proposed recently to account for dynamic instability of ParM, a prokaryotic actin relative whose polymerization generates pushing force to segregate plasmids (11).
Models based purely on chemical kinetics, like those in figure 1, continue to dominate the polymerization dynamics field, in part because they employ simple kinetic equations that can successfully rationalize a lot of experimental data and account for observed polymer behaviors. However, these models are becoming increasingly difficult to reconcile with studies revealing multiple structural states of microtubules and actin filaments that strongly influence dynamics, but are not strictly coupled to the chemical state of the bound nucleotide – i.e, structural plasticity. These studies do not invalidate the chemical kinetics viewpoint, but they demand a more sophisticated analysis that takes structural plasticity into account.
Structural plasticity in microtubule dynamics
For microtubules, multiple structural states at polymer ends have long been observed. Early negative stain electron microscopy (EM) images of microtubules coated with associated proteins showed that depolymerizing ends are frayed, inconsistent with depolymerization by simple reversal of polymerization (12, 13). Later, rapid freezing and cryo-EM of pure tubulin microtubules revealed relatively straight open sheets extending from ends of growing microtubules (Figure 2A), and frayed, outwards-curling protofilaments at ends of shrinking microtubules (Figure 2B) (14, 15). These images began to transform our view of microtubule polymerization dynamics from one dominated by chemical kinetics (the GTP cap view) to one dominated by structural transitions at polymer ends. Outwards curling of protofilaments in shrinking microtubules may reflect a more bent conformation of the GDP-tubulin subunit compared to a straighter GTP subunit, as seen in structural studies (16, 17). An alternative view suggests that GTP and GDP subunits may be similarly bent in solution, and that subunits straighten only after their incorporation into growing microtubules (18, 19). In the current, structural plasticity-based view of dynamic instability (20–22), GTP tubulin subunits add to open sheets at the end of growing microtubules, perhaps straightening during addition. A few seconds later, the sheet closes into a tube. Sheet closure may be facilitated in cells by microtubule tip-binding proteins such as EB1 (23), which can modulate the rates of transition between growing and shrinking states. However, it is still unclear how tip-binding proteins modulate chemical or structural dynamics at microtubule ends. At some point after polymerization – we do not know exactly when – GTP is hydrolyzed and phosphate released. The resulting GDP protofilaments prefer to be curved outwards, either because their GDP-tubulin subunits are more stable in the bent conformation (19), and/or because they make weaker interactions with neighbors (18). The preference of GDP protofilaments for an outwards-curved configuration causes mechanical stress to accumulate in the microtubule lattice. While the microtubule keeps growing, this stress cannot be released, because protofilaments are trapped in the straight configuration by interactions with their neighbors. However, stress makes the lattice fragile. If a crack develops between protofilaments at the end, it allows protofilaments to peel apart and curl outwards, finally releasing the stored free energy from GTP hydrolysis. The force of outwards curling causes the crack to propagate down the microtubule, releasing stress and triggering depolymerization as it travels. This stress storage-crack propagation model provides a structural explanation for how ends can persist in growing or shrinking states. It differs from a GTP-cap model in that depolymerization is initiated not by hydrolysis of GTP bound to terminal tubulin subunits (Figure 1B), but by crack formation between neighboring protofilaments near the polymer end. It has interesting implications for how depolymerizing microtubules in the mitotic spindle might pull on attached chromosomes during chromosome segregation. A number of recent models suggest that the outwards curling of protofilaments directly generates pulling forces that, through different coupling mechanisms, are transduced from the tip of the depolymerizing microtubule to the attached chromosome (20, 21, 22, 24).
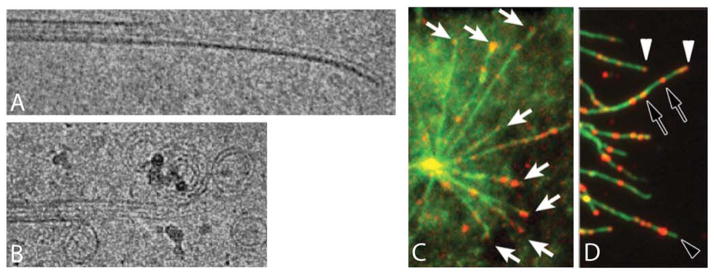
Figure 2.
Microtubule structural plasticity. A) Cryo-EM images of a growing microtubule end showing a curved, open sheet. B) Cryo-EM image of a shrinking microtubule end showing outwards curled protofilaments. C,D) Localization of microtubule segments with a stable lattice structure, recognized by a recombinant antibody. C. Pure tubulin microtubules (green) growing from a centrosome stained with an antibody that recognizes a stable structural state of the microtubule lattice (red). Note staining of growing tips (white arrows). D) Microtubules in a cell (green) stained with the antibody (red). Note tip staining, presumably on growing microtubles (white arrowhead), lack of tip staining, presumably on shrinking microtubules (empty arrowhead) and internal segments recognized by the antibody (empty arrows). A,B courtesy of T. Hyman, MPI Dresden. C, D adapted from (26).
Although microtubule dynamics are dominated by behavior at ends, structural plasticity in the middle of microtubules may also regulate dynamics in important ways. The polymer lattice places strong constraints on subunit conformation, but comparative EM of microtubules with bound GDP vs. GMPCPP (a GTP-analog) showed that their lattices differ subtly, with the inter-subunit spacing ~0.4nm shorter along the microtubule axis in GDP microtubules (25). Recently, an antibody was generated that recognizes a stable state of the microtubule lattice, that is present in microtubule stabilized by GMPCPP or the drug taxol, but not in unstable GDP-bound microtubules (26). The antibody stained segments a few hundred nm long at the plus ends of growing microtubules, both with pure tubulin (Figure 2C) and in cells (Figure 2D), consistent with the classical GTP cap model (Figure 1), but not ruling out alternative models positing structural plasticity at ends. Intriguingly, the antibody also stained short segments in the middle of cellular microtubules (Figure 2D), which were shown to act as reflecting boundaries to bouts of depolymerization. Whether these stable internal segments contain GTP-tubulin (a chemical kinetics view) or GDP-tubulin in an alternative, stable structural state of the lattice (a structural plasticity view) remains to be determined. The stable-state antibody is an exciting new probe that will stimulate interest in studying the mechanisms of structural plasticity in regulating microtubule dynamics.
Structural plasticity in actin dynamics
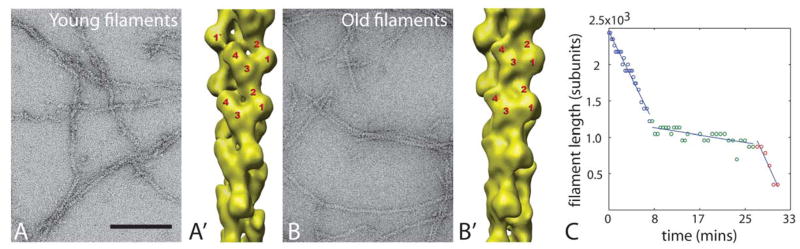
The structure of the actin filament was first determined by fitting a high resolution crystal structure of an actin monomer into a filament model to fit x-ray fiber diffraction data from an oriented actin gel (27, 28), and recently refined using improved versions of these methods (29). This canonical double helical structure is supported by numerous mutagenesis and EM studies (30). However, EM studies are increasingly showing that the actin helix can exhibit multiple structural states, and can transition between states in a highly cooperative manner (31). When pure actin filaments were analyzed by EM within ~2 minutes of polymerization, they were found to exhibit several different structural states, including some where individual subunits were substantially tilted away from the canonical helical orientation (32, 33). After aging in solution for 2 hours, filaments mostly adopted the canonical helical structure (Figure 3A, B) (33). The chemical kinetics view predicts that newly-polymerized filaments are more stable than aged filaments, since they contain a larger proportion of bound ATP (or ADP.Pi, the chemical state after hydrolysis but before phosphate release) (34, 35). However, the EM structures seem to contradict this view, since the tilted state present in young filaments appears to have less stable inter-subunit contacts compared to the canonical helical state in aged filaments (36). We directly compared the stability of young and aged pure actin filaments using single filament imaging and kinetic assays in bulk solution (37), and found that newly-polymerized filaments depolymerized faster than aged filaments, inconsistent with the chemical kinetics view, but consistent with EM structural observations. Time-dependent switching from fast to slow depolymerization was seen by live imaging of single filaments (Figure 3C, blue-green transition). Less frequently, aged, stable filaments reverted back to the unstable, fast-shrinking state (Figure 3C, green-red transition). The majority of aged filaments were in a slow-shrinking state, and this state dominated bulk depolymerization kinetics measured in solution (37). In light of EM data, we hypothesize that time-dependent stabilization arose from cooperative and reversible transitions of actin filaments from a disordered or tilted state in young filaments, which depolymerizes rapidly, to the canonical helical structure in older filaments, which depolymerizes slowly.
Figure 3.
Actin filament structural plasticity. A) Negative-stain EM images of pure actin filaments two minutes after initiation of polymerization. Note ragged appearance. A′) 3D reconstruction of Cryo-EM images of actin filaments, revealing a structural state in which subunits (numbered) are tilted ~30o relative to the canonical helix. This state is enriched in newly-polymerized filaments. B) EM image of actin filaments aged for 2 hrs. Note regular appearance. B′) 3D reconstruction of Cryo-EM images of actin filaments, revealing a canonical helical state. This canonical state is enriched in aged filaments. Scale bar = 100 nm in both EM images. (Images courtesy of A. Orlova, and E. Egelman, University of Virginia. Reconstructions courtesy of V. Galkin, A. Orlova and E. Egelman, University of Virginia).- again permissions required C) Length vs. time trace for a single fluorescently-labeled actin filament depolymerizing in buffer. Adapted from (36), © 2008, National Academy of Sciences USA. Filament switches from fast to slow depolymerization, and then back to fast. We propose that these switches are caused by spontaneous structural transitions possibly between those shown in A, B.
Structural plasticity appears to be an important determinant of polymerization dynamics for pure actin, but is it important in cells, where dynamics are regulated by many other proteins? Among the most important dynamics regulators are proteins in the Actin Depolymerizing Factor (ADF)/cofilin family, which promote actin turnover in diverse eukaryotic cells (38). ADF/cofilins selectively bind ADP-actin filaments and catalyze their depolymerization (39, 40), consistent with a chemical kinetics view of their function. A number of different mechanisms have been reported by which ADF/cofilins promote filament turnover: acceleration of subunit loss from ends (37, 40), severing of filaments away from ends (41), and disassembly of long filament segments in abrupt bursts, a pathway observed in the presence of co-factors Aip1 and Coronin (42). These alternative depolymerization pathways are difficult to explain from a pure chemical kinetics viewpoint, but structural plasticity begins to reconcile them. Binding of ADF/cofilin is known to induce, or stabilize, alternative structures of the actin filament that can be both hyper-twisted (43) and tilted (33, 36). The tilted state induced by ADF/cofilin was similar to that seen in the young filaments of pure actin, so ADF/cofilin binding can be thought of as ‘rejuvenating’ aged, stable filaments (33). Alternative depolymerization pathways can then be viewed as consequences of ADF/cofilin induced structural transitions that weaken the filament. Because ADF/cofilin binding is promoted by ATP hydrolysis and phosphate release, these proteins may restore time-dependent destabilization to a polymer where structural plasticity promotes time-dependent stabilization.
The concept that binding proteins influence actin dynamics by exploiting intrinsic structural plasticity of the filament may extend to other proteins; recent EM work showed that binding of fimbrin, a cross-linking protein, induces changes in filament internal structure (44). We hypothesize that tropomyosin, a coiled-coil protein that binds cooperatively along the filament and antagonizes cofilin binding (45), might function by inducing (or stabilizing) the more ordered, canonical helical state. Even end-binding proteins might function in part by inducing structural changes in the filament. Two barbed-end binding proteins, formins and gelsolin, both nucleate filaments with altered internal structures (46, 47). If diverse actin binding proteins take advantage of intrinsic structural plasticity of the filament to promote their own cooperative binding, and to regulate interaction of the filament with other proteins, then we expect a rich variety of higher-order behaviors to emerge, which could promote diversity in structure and function of cellular actin assemblies.
Future directions
For both microtubules and actin filaments, the case for structural plasticity regulating dynamics is strong. We now need to determine exactly how these multiple structural states of the polymer influence dynamics, how they interconvert, and how they influence, or are influenced by, the chemical kinetics of nucleotide hydrolysis. We can then integrate structural data with chemical kinetics into full models of polymerization dynamics. For microtubules, relatively simple chemical kinetics models have so far sufficed to account for dynamic instability behavior in cells (48, 49). As we seek to model complex assemblies, understand how microtubules pull on chromosomes, and elucidate mechanisms of proteins that regulate polymerization dynamics, we will need models that integrate structural transitions with GTP hydrolysis kinetics. To achieve this, we will need to learn exactly when nucleotide hydrolysis and phosphate release occur at growing microtubule plus ends, how these chemical events relate to sheet growth and tube closure, and how tip binding proteins such as EB1 influence, and are influenced by, structural and chemical transitions. For actin, the field continues to rely on chemical kinetics models that neglect structural plasticity (50, 51), despite the EM data. As we seek to understand local regulation of actin turnover in cells, and how actin and its binding proteins function at the systems level, we will need to understand better how filament structure influences dynamics, and the extent to which filament binding proteins control dynamics by modulating filament structure. For both microtubules and actin filaments, we will need probes that recognize different structural states of the polymer in cells. Antibodies that recognize specific structural states of the polymer are a promising start (26, 52, 53), but a more general solution may come from binding factors that have evolved to recognize different chemical or structural states.
For prokaryotic relatives of tubulin and actin there are many outstanding questions, including in most cases basic descriptions of polymer architecture, dynamics and biological function. Once these have been answered, we can go on to investigate possible roles of structural plasticity. A purely chemical kinetics model for dynamic instability of ParM filaments was recently proposed (11), but alternate models, which take into account the observed structural plasticity of ParM filaments (54), may be worth considering. Prokaryotes apparently lack motor proteins and possess a much smaller complement of polymer binding proteins, making their cytoskeletal systems apparently simpler than those in eukaryotes. Will prokaryotic polymers consequently be more, or less, complicated in terms of their structure and dynamics? Given the diversity of tubulin and actin relatives among prokaryotes, it seems likely that we will see entirely new polymer structures and dynamics, where both nucleotide chemistry and structural plasticity are put to new uses.
Summary.
Actin and tubulin polymers exhibit multiple structural states that strongly influence their polymerization dynamics, but are not directly coupled to the chemical state of the polymer-bound nucleotide
Acknowledgments
We thank E. Egelman and S. Dumont for insightful comments on this manuscript, as well as B. Brieher, D. Needleman, and other members of the Mitchison lab for discussions. This work was supported by NIH grant GM 23928. HYK is a Howard Hughes Medical Institute predoctoral fellow.
References and Notes
- 1.Inoue S, Sato H. J Gen Physiol. 1967 Jul;50(Suppl):259. [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cell. 2003 Feb 21;112:453. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Mitchison TJ, Salmon ED. Nat Cell Biol. 2001 Jan;3:E17. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- 4.Dye NA, Shapiro L. Trends Cell Biol. 2007 May;17:239. doi: 10.1016/j.tcb.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Egelman EH. Curr Opin Struct Biol. 2003 Apr;13:244. doi: 10.1016/s0959-440x(03)00027-7. [DOI] [PubMed] [Google Scholar]
- 6.Osawa M, Anderson DE, Erickson HP. Science. 2008 May 9;320:792. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wegner A. J Mol Biol. 1976 Nov;108:139. doi: 10.1016/s0022-2836(76)80100-3. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara I, Takahashi S, Tadakuma H, Funatsu T, Ishiwata S. Nat Cell Biol. 2002 Sep;4:666. doi: 10.1038/ncb841. [DOI] [PubMed] [Google Scholar]
- 9.Margolis RL. Proc Natl Acad Sci U S A. 1981 Mar;78:1586. doi: 10.1073/pnas.78.3.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchison T, Kirschner M. Nature. 1984 Nov 15–21;312:237. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 11.Garner EC, Campbell CS, Mullins RD. Science. 2004 Nov 5;306:1021. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 12.Kirschner MW, Williams RC, Weingarten M, Gerhart JC. Proc Natl Acad Sci U S A. 1974 Apr;71:1159. doi: 10.1073/pnas.71.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandelkow EM, Mandelkow E. J Mol Biol. 1985 Jan 5;181:123. doi: 10.1016/0022-2836(85)90330-4. [DOI] [PubMed] [Google Scholar]
- 14.Mandelkow EM, Mandelkow E, Milligan RA. J Cell Biol. 1991 Sep;114:977. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chretien D, Fuller SD, Karsenti E. J Cell Biol. 1995 Jun;129:1311. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller-Reichert T, Chretien D, Severin F, Hyman AA. Proc Natl Acad Sci U S A. 1998 Mar 31;95:3661. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HW, Nogales E. Nature. 2005 Jun 16;435:911. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice LM, Montabana EA, Agard DA. Proc Natl Acad Sci U S A. 2008 Apr 8;105:5378. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buey RM, Diaz JF, Andreu JM. Biochemistry. 2006 May 16;45:5933. doi: 10.1021/bi060334m. [DOI] [PubMed] [Google Scholar]
- 20.Molodtsov MI, Grishchuk EL, Efremov AK, McIntosh JR, Ataullakhanov FI. Proc Natl Acad Sci U S A. 2005 Mar 22;102:4353. doi: 10.1073/pnas.0501142102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh JR, et al. Cell. 2008 Oct 17;135:322. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grishchuk EL, et al. Proc Natl Acad Sci U S A. 2008 May 13;105:6918. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitre B, et al. Nat Cell Biol. 2008 Mar 23;10:415. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- 24.Westermann S, et al. Nature. 2006 Mar 23;440:565. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 25.Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Mol Biol Cell. 1992 Oct;3:1155. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimitrov A, et al. Science. 2008 Oct 16; [Google Scholar]
- 27.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Nature. 1990 Sep 6;347:37. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 28.Holmes KC, Popp D, Gebhard W, Kabsch W. Nature. 1990 Sep 6;347:44. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 29.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. Nature. 2009 Jan;457:441. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 30.Reisler E, Egelman EH. J Biol Chem. 2007 Oct 26; doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- 31.Orlova A, Prochniewicz E, Egelman EH. J Mol Biol. 1995 Feb 3;245:598. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- 32.Steinmetz MO, Goldie KN, Aebi U. J Cell Biol. 1997 Aug 11;138:559. doi: 10.1083/jcb.138.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orlova A, et al. Proc Natl Acad Sci U S A. 2004 Dec 21;101:17664. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Combeau C, Carlier MF. J Biol Chem. 1988 Nov 25;263:17429. [PubMed] [Google Scholar]
- 35.Pollard TD. J Cell Biol. 1986 Dec;103:2747. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. J Cell Biol. 2001 Apr 2;153:75. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kueh HY, Brieher WM, Mitchison TJ. Proc Natl Acad Sci U S A. 2008 Oct 17; doi: 10.1073/pnas.0807394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maciver SK, Hussey PJ. Genome Biol. 2002;3:3007. doi: 10.1186/gb-2002-3-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanchoin L, Pollard TD. J Biol Chem. 1999 May 28;274:15538. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 40.Carlier MF, et al. J Cell Biol. 1997;136:1307–22. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrianantoandro E, Pollard TD. Mol Cell. 2006 Oct 6;24:13. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Kueh HY, Charras GT, Mitchison TJ, Brieher WM. J Cell Biol. 2008 Jul 28;182:341. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGough A, Pope B, Chiu W, Weeds A. J Cell Biol. 1997 Aug 25;138:771. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galkin VE, Orlova A, Cherepanova O, Lebart MC, Egelman EH. Proc Natl Acad Sci U S A. 2008 Feb 5;105:1494. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono S, Ono K. J Cell Biol. 2002 Mar 18;156:1065. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papp G, et al. Biophys J. 2006 Oct;91:2564. doi: 10.1529/biophysj.106.087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prochniewicz E, Zhang Q, Janmey PA, Thomas DD. J Mol Biol. 1996 Aug 2;260:756. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 48.Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. J Cell Biol. 1992 Sep;118:1097. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vorobjev IA, Rodionov VI, Maly IV, Borisy GG. J Cell Sci. 1999 Jul;112(Pt 14):2277. doi: 10.1242/jcs.112.14.2277. [DOI] [PubMed] [Google Scholar]
- 50.Bindschadler M, Osborn EA, Dewey CF, Jr, McGrath JL. Biophys J. 2004;86:2720–39. doi: 10.1016/S0006-3495(04)74326-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vavylonis D, Yang Q, O’Shaughnessy B. Proc Natl Acad Sci U S A. 2005 Jun 14;102:8543. doi: 10.1073/pnas.0501435102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Egelman EH. Nat Rev Mol Cell Biol. 2003 Aug;4:621. doi: 10.1038/nrm1176. [DOI] [PubMed] [Google Scholar]
- 53.Schoenenberger CA, et al. J Struct Biol. 2005 Dec;152:157. doi: 10.1016/j.jsb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Orlova A, et al. Nat Struct Mol Biol. 2007 Sep;14:921. doi: 10.1038/nsmb1300. [DOI] [PMC free article] [PubMed] [Google Scholar]