Abstract
Methionine residues are susceptible to oxidation, but this damage may be reversed by methionine sulfoxide reductases MsrA and MsrB. Mammals contain one MsrA and three MsrBs, including a selenoprotein MsrB1. Here, we show that MsrB1 is the major methionine sulfoxide reductase in liver of mice and it is among the proteins that are most easily regulated by dietary selenium. MsrB1, but not MsrA activities, were reduced with age, and the selenium regulation of MsrB1 was preserved in the aging liver, suggesting that MsrB1 could account for the impaired methionine sulfoxide reduction in aging animals. We also examined regulation of Msr and selenoprotein expression by a combination of dietary selenium and calorie restriction and found that, under calorie restriction conditions, selenium regulation was preserved. In addition, mice overexpressing a mutant form of selenocysteine tRNA reduced MsrB1 activity to the level observed in selenium deficiency, whereas MsrA activity was elevated in these animals. Finally, we show that selenium regulation in inbred mouse strains is preserved in an outbred aging model. Taken together, these findings better define dietary regulation of methionine sulfoxide reduction and selenoprotein expression in mice with regard to age, calorie restriction, dietary Se, and a combination of these factors. Antioxid. Redox Signal. 12, 829–838.
Introduction
For many organisms, selenium (Se) is an important micronutrient. In mammals, this trace element is biologically active, essential during development, and has been reported to possess cancer prevention activity (7, 13). It is thought that the majority of biological effects of Se are exerted by selenoproteins, which contain a selenocysteine (Sec) residue. This rare amino acid is inserted into proteins co-translationally in response to the codon UGA (41). Mammalian Sec insertion machinery includes an RNA stem-loop structure known as the SECIS element (3, 4, 21), a Sec-specific tRNA(Ser)Sec (19), an elongation factor EFsec (15, 44), a SECIS-binding protein 2 (12, 31), and several additional factors (10).
In selenoproteins, Sec is often located in catalytic sites and serves an oxidoreductase function. Thus, selenium and redox processes are tightly associated. Two major cellular redox systems in mammals, thioredoxin and glutathione systems (20), utilize Se through thioredoxin reductases (TRs) and glutathione peroxidases (GPxs), respectively. These proteins have been the major focus of selenium research because of their abundance, apparent antioxidant function, and regulation by dietary Se (1, 16, 26). In addition, GPxs, together with selenoprotein P, have been used in determining selenium requirement in the diets of animals and humans (8).
As biological effects of selenium and its impact on human health are largely due to selenoproteins, it is of interest to elucidate how additional selenoproteins are regulated by this dietary factor. It was found that selenoprotein methionine-R-sulfoxide reductase (MsrB1) is responsive to selenium status in the diet (35, 36). This protein is one of the enzymes that can reduce oxidized methionine residues back to methionines (5, 28, 46). Many amino acids in proteins are susceptible to oxidative modifications, but oxidation of sulfur-containing amino acids, cysteine and methionine, is of particular importance (5, 46).
Oxidation of methionine residues leads to a mixture of S- and R-epimers of methionine sulfoxide. Methionine sulfoxide reductase A (MsrA) can stereospecifically reduce methionine-S-sulfoxide, whereas MsrB can only reduce methionine-R-sulfoxide (5). Methionine sulfoxide reduction has been implicated in regulating numerous biological processes, including lifespan in animals and progression of neurological disorders (42). Recent studies revealed a possible role of methionine sulfoxide reduction in regulating lifespan in animals (25, 32, 38, 39). Although the effect of deletion of the MsrA gene in mice on lifespan needs further examination [i.e., there are reports of no effect of MsrA knockout (39) and a reduction of ∼40% (32)], overexpression of bovine MsrA, predominantly in the nervous system, extended lifespan of fruit flies by ∼70% (38). Using yeast cells as a model, it was shown that overexpression of either MsrA or MsrB can increase lifespan (25). However, whereas MsrA was effective under regular growth conditions, MsrB could only extend lifespan under calorie restriction conditions in yeast.
Selenium deficiency was previously shown to reduce the expression of MsrB1 in mice (33), but it would be important to examine this regulation under multiple selenium diets. It is also of interest to elucidate if selenium, through methionine sulfoxide reduction and other antioxidant systems, has a role in regulating lifespan in mammals. Although selenium was previously implicated in the aging process, an experiment specifically designed to examine the role of this trace element on lifespan in rodents or other mammals has not been performed (or has not been reported).
To examine regulation of methionine sulfoxide reductase function, we carried out a study that utilized several mouse models, including wild-type mice (Balb/c mice, a commonly used mouse line), and TGFα and TGFα/i6A- transgenic mice. TGFα mice express TGFα, a transgene that promotes carcinogenesis and is capable of modulating cellular redox status (14). TGFα/i6A- express both TGFα and a mutant form of Sec tRNA lacking a modification at a codon flanking the anticodon. The i6A- transgene leads to a dramatic reduction in selenoprotein expression, particularly of stress-related selenoproteins, such as GPx1, whereas several selenoproteins, such as TR3, are little affected (34). The above listed mouse models are inbred strains in which individual animals are genetically very similar, and small sample sizes could be used to represent the population. However, the use of inbred strains may also have disadvantages, particularly with regard to high incidence of strain-specific pathologies. To better relate our findings to human health, we also included an additional outbred control group, a four-way cross HET mice. This outbred model incorporates genetic diversity and has been used in the National Institute of Aging Interventions Testing Program, for example, demonstrating the effect of rapamycin on lifespan extension (18). The outbred and inbred mice were subjected to selenium diets, calorie restriction, and/or aging to examine regulation of methionine sulfoxide reduction at three levels: (i) selenoprotein expression, (ii) MsrA and MsrB1 expression, and (iii) MsrA and MsrB activities.
Materials and Methods
Materials
Chemicals were from Sigma, St. Louis, MO. Anti-GPx1 antibodies were from Genetex, Irvine, CA. Anti-MsrB1 (22), anti-TR3 (45), and anti-MsrA (47) antibodies were previously described. Selenium-defined rodent diets were based on selenium-deficient Torula yeast and were obtained from Harland TekLad, Madison, WI. We used Se-deficient ((-)Se, <0.02 ppm Se), 0.1 ppm Se, 0.15 ppm Se, 0.4 ppm Se, and 2.25 ppm Se diets, which were obtained by supplementing the Se-deficient diet with indicated amounts of selenium in the form of sodium selenite, as described (35). In addition, we used a common rodent diet from Harland TekLad and two independent batches of Purina version of NIH 31 rodent diet with 4% fat (designated NIH 1 and NIH 2). The actual amount of selenium in each diet was determined fluorometrically using 2,3-diaminonaphtharene (DAN) (48) in Oscar E. Olson Biochemistry Analytical Services Laboratory, South Dakota State University, SD.
Strains of mice
Care and treatment of experimental animals were approved by the Animal Care and Use Committee at the University of Nebraska-Lincoln (UNL). Balb/c mice were kindly provided by the UNL animal facility at Manter Hall, and TGFα/+ mice by Dr. Glenn Merlino (National Cancer Institute). TGFα/+ mice on a CD1 background were bred with either wild-type FVB mice or i6A- mice on an FVB background (34) to obtain TGFα/+ and TGFα/i6a- mice on a mixed CD1/FVB background. The HET mice were a four-way cross population initially defined by Dr. R. Miller, called UM-HET3. They were bred from mothers that are CByB6F1/J (JAX stock #100009), and mated with fathers that are C3D2F1/J (JAX stock #100004). From each breeding pair, the initial litters were discarded, and the second and subsequent litters were used for the experiments.
Selenium diets and aging
Following weaning, Balb/c mice were subjected to selenium diets and maintained on these diets for 10 or 30 months. Each of the age groups consisted of three subgroups that received either selenium-deficient ((-)Se, <0.02 ppm Se), 0.1 ppm Se, or 0.4 ppm Se diets. This experiment was carried out for both male and female mice.
Combination of selenium diets and calorie restriction
Weaned Balb/c mice were first maintained on one of four selenium diets, including (-)Se, 0.15 ppm Se, 0.4 ppm Se, and 2.25 ppm Se diets, for 6 weeks to adjust their Se status. Animal weight and amount of consumed food were monitored daily during the last 2 weeks of this procedure, and only the mice with weight deviating not more then 5% from the median weight (calculated separately for males and females) were subjected to the calorie restriction study. During calorie restriction, male mice received 2.3 g and female mice 2.5 g of diet at 9 AM daily. This amount corresponded to 65% of the food consumed by animals having free access to food. The calorie restriction study lasted 6 weeks. At the end of the study, the mice were 4 months old.
TGFa/+ and TGFa/i6a
TGFα/+ and TGFα/i6A- mice were each divided into three groups at 4 weeks of age and subjected to the following selenium levels: (-)Se, 0.1 ppm Se, and 0.4 ppm Se.
HET mice
Four-way cross, 5–7-month HET female mice were divided into six groups and subjected to the following selenium diets for 6 weeks: (-)Se, 0.1 ppm Se, 0.4 ppm Se, two independent batches of NIH diets, and Harlan Teklad standard rodent chow.
Sample preparation
At the end of each experiment, animals were sacrificed, and their livers were dissected and subjected to selenoprotein expression and activity assays. Tissues were homogenized in PBS containing protease inhibitors (Roche, Basel, Switzerland), and the homogenates were normalized with regard to protein concentration. Western blotting analyses and MsrA and MsrB activity assays were then carried out with these samples.
Protein expression analysis
Tissue homogenates were separated on SDS-PAGE gels (10 μg of protein was loaded), and the proteins were transferred onto PVDF membranes and probed with the antibodies indicated. Secondary HRP-linked anti-rabbit antibodies were from GE HealthCare, Little Chalfont, United Kingdom. Nanogram grade ELC substrate for MsrA, GPx1, and TR3 detection was from GE HealthCare, and picogram grade substrate for MsrB1 detection from Sigma.
Msr activity assays
MsrA and MsrB activities were measured in homogenates as previously described (23, 24). Briefly, 200 μg of protein from liver extracts were used in each assay. The reaction was carried out at 37°C for 30 min in the presence of DTT, and either 200 μM dabsyl-methionine-S-sulfoxide (MsrA assay) or 200 μM dabsyl-methionine-R-sulfoxide (MsrB assay) were added to the reaction mixture. To assay for total Msr activity, we used 400 μM of mixed (R,S) methionine sulfoxide. The dabsyl-methionine product was separated from the substrate using an HPLC procedure and peak areas were quantified.
Statistics
Statistical analysis was performed using a Student's t-test or Analysis of Variance (ANOVA). P values of <0.05 were considered significant.
Results
Expression profile of MsrB1, MsrA, GPx1, and TR3 in mice
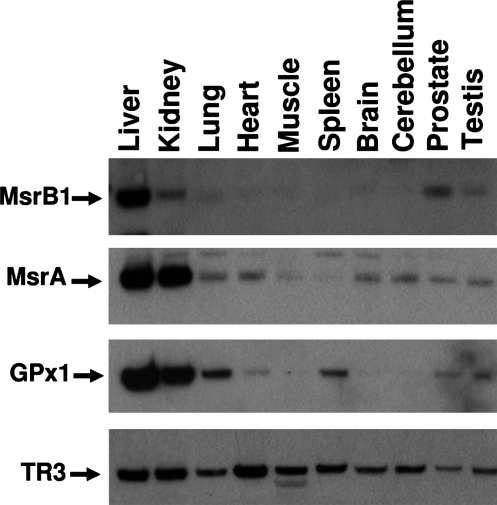
The expression profile of MsrB1 was characterized in mouse tissues by Western blotting and compared with those of MsrA, GPx1, and mitochondrial thioredoxin reductase (TR3, also known as TxnRd2 and TrxR2). As shown in Fig. 1, the highest MsrB1 levels were observed in liver, followed by kidney and prostate. Liver is known to be rich in selenoproteins, with GPx1 being the most abundant selenoprotein in this organ (30, 51). Interestingly, MsrA also showed a high expression level in liver (Fig. 1). Thus, two tested stress-related selenoproteins, MsrB1 and GPx1, as well as MsrA, were highly expressed in this organ. In contrast, TR3 was expressed at similar levels in various mouse tissues (Fig. 1). Based on these data, liver was chosen for further experiments that examined regulation of MsrB1 and MsrA expression and their activities by dietary factors.
FIG. 1.
Expression profiles of selenoproteins and MsrA in mouse organs and tissues. Equal amounts (10 μg of protein per lane) from indicated tissues of a 10-month-old male Balb/c mouse were loaded on SDS-PAGE gels, transferred onto PVDF membranes, and probed with anti-MsrB1, MsrA, GPx1, and TR3 antibodies.
Selenium diets and a strategy to examine regulation of MsrB1 and MsrA expression by dietary selenium
To examine dietary control of methionine sulfoxide reduction in mouse liver, we used the following selenium diets: (-)Se (Se-deficient), 0.1 ppm Se, 0.15 ppm Se, 0.4 ppm Se, 2.25 ppm Se, and three control diets (two batches of NIH 31 Purina diet and Harlan Teklad rodent chow). The Se diets were based on the Se-deficient diet, which was supplemented with defined amounts of selenium as sodium selenite. Expected (based on the amount of Se actually added to the diet) and measured selenium concentrations agreed well for all selenium diets (Supplemental Table 1; see www.liebertonline.com/ars). The Se-deficient diet had ∼0.02 ppm Se. It is noteworthy that both NIH and Harlan Teklad diets contained ∼0.4 ppm of selenium, which corresponded to one of our experimental diets (i.e., 0.4 ppm Se diet). Various mouse models described below were subjected from weaning for indicated periods of time to these diets, and tissue samples were collected.
The following strategy was employed to examine regulation of methionine sulfoxide reductases in mice. For each data point, three liver samples from three different animals were assayed by Western blotting for expression of methionine sulfoxide reductases MsrB1 and MsrA, and selenoproteins GPx1 and TR3 (the latter two were used as controls). In addition, MsrB and MsrA activities were measured as shown in the figures (generally, more samples were used for direct activity assays than for Westerns). We found no statistically significant gender differences in MsrB or MsrA activities in Balb/c mice (data not shown).
Expression of MsrB1 is highly regulated by dietary selenium
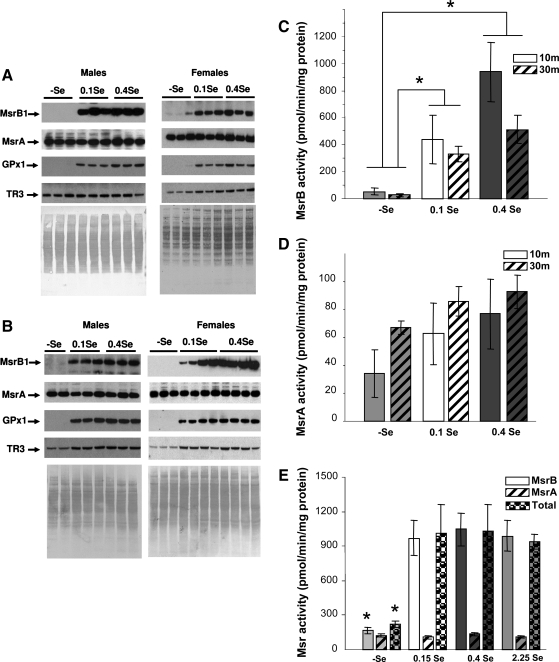
To characterize regulation of Msr function by age, Balb/c mice were maintained on (-)Se, 0.1 ppm, or 0.4 ppm Se diets for 10 or 30 months. As shown in Fig. 2A and B, in both 10- and 30-month-old animals, expression of MsrB1 was dramatically reduced by Se deficiency. This pattern was observed for both male and female mice and mimicked that of GPx1 regulation by dietary selenium. In contrast, little difference in expression levels was observed for TR3, although this protein was slightly reduced on the (-)Se diet. GPx1 is known to be a selenoprotein that is highly responsive to the level of selenium in the diet. Our data place MsrB1 in the same category of selenium-responsive proteins (51).
FIG. 2.
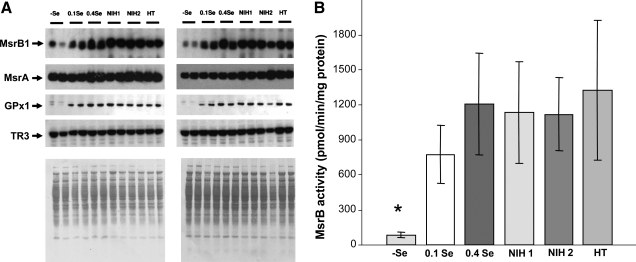
Regulation of MsrB1 and MsrA expression and MsrB and MsrA activities in Balb/c mice by dietary selenium and aging. (A and B) Western blot analyses of liver samples from mice subjected to (-)Se, 0.1 ppm Se, and 0.4 ppm Se diets are shown for 10-month- (A) and 30-month-old (B) animals. The proteins probed are indicated on the left and protein loading is shown by Amido Black staining on bottom images. MsrB1 expression in female mice fed 0.1 ppm Se showed higher variability than in other samples. Also, in general, we observed variability from animal to animal, which is not related to activity and Western analyses. Specific MsrB (C) and MsrA (D) activities in the same set of samples (n = 36, two-way ANOVA interaction, strong evidence for the effect of diet on MsrB1 (pMsrB1 < 0.007) and suggestive evidence for the effect on MsrA (pMsrA < 0.06) activities). (E) Specific MsrB, MsrA, and total Msr activities in liver lysates in 5-month-old mice subjected to indicated selenium diets (n = 16, one-way ANOVA, p < 0.005). Total Msr activity represents a separate measurement with mixed methionine (R,S) sulfoxide substrate rather than the sum of MsrA and MsrB activities.
MsrB1 is the major methionine sulfoxide reductase in mouse liver
Direct assays of MsrB and MsrA activities (Fig. 2C and D) revealed that MsrB activity is approximately fivefold higher than MsrA activity. MsrB activity in both 10- and 30-month-old animal groups was responsive to dietary treatments with selenium. Moreover, in addition to the dramatic reduction in MsrB activity in Se deficiency, we observed a slight, but statistically significant difference, between 0.1 and 0.4 ppm dietary groups. Thus, it appears that regulation of MsrB is similar to that reported for liver GPx1 (2, 6, 29, 43).
To further examine regulation of MsrB activity by supranutritional levels of selenium, we carried out an additional experiment, wherein young (3-month-old) Balb/c mice (four animals per group) received (-)Se, 0.15 Se ppm, 0.4 Se ppm, and 2.25 Se ppm diets for 2 months. As shown in Fig. 2E, MsrB activity was saturated at 0.15 ppm Se. Thus, between 0.1 and 0.15 ppm, Se as sodium selenite was required for maximal MsrB activity. These data further indicate that liver MsrB activity is one of the functions that are most easily regulated by dietary selenium, making MsrB1 a potentially highly useful marker of selenium status in mammals. Indeed, the level at which MsrB1 activity and expression are saturated is similar to (or even slightly higher than) the “plateau break point” previously observed for GPx1 (50).
Since (a) MsrB1 is the only selenoprotein Msr in mammals, (b) MsrB activity is highly regulated by dietary selenium, and (c) MsrA activity is much lower than that of MsrB, we suggest that MsrB1 is the major Msr in mouse liver. It should be noted that the high MsrB activity observed in our study differs from that in a previous report that showed that MsrA and MsrB activities are approximately equal in mouse liver (32). It is not clear whether these differences relate to the differences in assays, sample preparation, mouse strains, or other issues. To further examine the major contribution of MsrB1 to liver Msr activity, we measured total Msr activity as well as MsrA and MsrB activities in the animal group shown in Fig. 2E, and found that the total Msr activity was regulated by dietary selenium similarly to the MsrB activity. These data further support the idea that MsrB1 largely accounts for total Msr activity in mouse liver and that nutritional levels of selenium in the diet affect the overall Msr activity in this organ.
MsrA activity in mice maintained on selenium diets
MsrA activity of young (5-month-old) Balb/c mice placed on the (-)Se diet was slightly reduced compared to those in mice in Se-supplemented groups (Fig. 2E). A small decrease in MsrA activity in mice on the (-)Se diet was also apparent in a 30-month-old group (Fig. 2D), although there was no difference in expression observed by Western blot analyses (Fig. 2B). We also observed Se-dependent changes in MsrA expression and activity in mice which overexpress a mutant form of tRNASec (see below). Since the effect of Se on MsrA activity was not dramatic (as opposed to the effect on total Msr and MsrB activities), further studies will be required to ascertain these observations. For example, it is possible that this effect is due to small impurities of the MsrA substrate. Alternatively, the small effect of selenium deficiency on MsrA activity could be indirect, due to partial inactivation of MsrA following oxidative stress resulting from reduced expression of antioxidant selenoproteins.
MsrB activity is decreased with age, whereas MsrA activity is not
Analysis of liver samples from 10- and 30-month-old animals showed that MsrB activity was reduced with age (Fig. 2C). In the case of 0.4 ppm Se diet, the activity was reduced as much as twofold (p < 0.007). A decrease in MsrB activity was consistent with the reduction in MsrB1 expression levels as shown in Fig. 3B. In contrast, MsrA activity was statistically unchanged, but was characterized by a trend towards increased activity in 30-month-old animals in all three dietary groups (Fig. 2D). Previous research has shown that total Msr activity was reduced with age in rats (37). Our current data suggest that the reduction in MsrB1 activity is responsible for the decrease in total Msr activity with age.
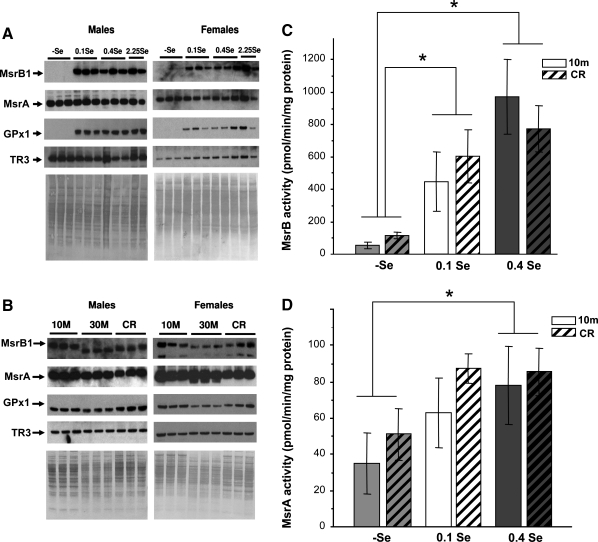
FIG. 3.
Regulation of MsrB1 and MsrA expression and MsrB and MsrA activities in Balb/c mice by a combination of dietary selenium and calorie restriction. (A) Western blot analyses in liver samples, with the proteins probed indicated on the left. These are calorie restriction (CR) samples (compare with Fig. 2A where regular food samples are probed). (B) Comparison of MsrB1, MsrA, GPx1, and TR3 expression in livers of 10-month-, 30 month-old, and CR mice. These mice were fed 0.4 ppm Se diet. The proteins probed are indicated on the left and protein loading is shown by Amido Black staining on bottom images of A and B. Specific activities of MsrB1 (C) and MsrA (D) in 10-month-old and CR animal groups. There was an effect of the diet on both MsrA and MsrB1 activity (n = 43, two-way ANOVA interaction, pMsrA < 0.06 and pMsrB1 < 0.05); however, the effect of CR was not significant (pMsrA > 0.4 and pMsrB1 > 0.12).
Regulation of MsrB1, MsrA, GPx1, and TR3 expression and MsrB and MsrA activities by calorie restriction
Calorie restriction (CR) is a dietary regimen known to extend lifespan in a variety of organisms, including yeast, rotifers, spiders, worms, fish, mice, and rats (27, 40, 49), and this treatment even works in primates (11). Despite extensive characterization in various organisms, the molecular basis for the CR-dependent extension in lifespan is not fully understood. Aging is a multifactorial process, and one of the leading theories posits that it is caused by cumulative oxidative damage by reactive oxygen species (17). Being antioxidants that repair oxidatively damaged proteins, MsrA and MsrB have also been suggested to regulate lifespan. Therefore, it was of interest to examine how CR affects expression of these proteins and how selenium regulates expression of MsrA and MsrB1 under CR conditions. To elucidate this regulation, Balb/c mice were subjected to Se diets in combination with CR. In this experiment, we initially maintained weaned mice on (-)Se, 0.15 ppm Se, 0.4 ppm Se, and 2.25 ppm Se diets for 6 weeks, and then subjected them for an additional 6 weeks to the same diets except that the food was restricted to 65% of the amount the mice normally consumed. Rationale for using 0.15 ppm Se diet in this experiment was that it approximately corresponded to the 0.1 ppm Se group with regard to consumed Se (due to reduced food consumption in the CR group). However, as MsrB1 expression is essentially saturated at 0.15 ppm Se, the difference in food consumption was not expected to affect MsrB1 in CR mice receiving 0.4 ppm Se diet.
Dietary selenium regulated selenoprotein expression in CR animals similarly to that in 10-month-old control mice (Fig. 3A). We found little difference in the expression of MsrA and TR3 in mice on different Se diets, whereas GPx1 and MsrB1 levels were dramatically reduced by Se deficiency under CR conditions. CR itself did not affect expression levels of the three selenoproteins or of MsrA (Fig. 3B, e.g., compare 10-month-old and CR groups). Analyses of Msr activities in these samples again revealed a dramatic reduction in MsrB1 activity and little or no reduction in MsrA activity in mice subjected to (-)Se diet. Interestingly, CR generally stimulated MsrA and MsrB activities (Fig. 3C and D), although a statistically significant difference was found only for MsrB activity on the (-)Se diet. This regulation should be verified in further studies.
Regulation of MsrA and MsrB1 in mice overexpressing mutant Sec tRNA
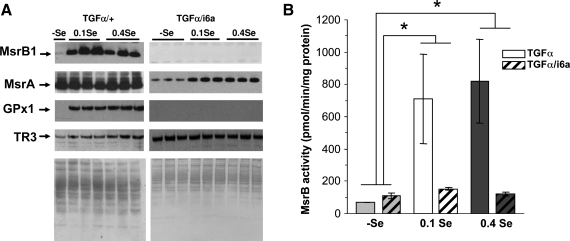
To further characterize regulation of MsrA and MsrB by Se diets, we examined mice overexpressing either TGFα or both TGFα and the i6A- mutant form of Sec tRNA. As shown in Fig. 4A, Se deficiency in TGFα transgenic mice resulted in essentially undetectable levels of MsrB1 and GPx1, but did not affect TR3 and MsrA levels (these mice are known to differentially regulate stress-related selenoproteins, such as GPx1 and TRs (35)). However, double transgenic TGFα/i6A- mice had low levels of MsrB1 and GPx1 at all dietary Se levels, while again fully preserving TR3 levels. In the case of MsrA, the double transgenic mice had an apparent decrease in the expression level of this enzyme when the (-)Se diet was compared with the 0.1 or 0.4 Se ppm diets (p < 0.01). These data suggest that the overall Se status in mouse liver could regulate the expression of MsrA, a non-selenoprotein.
FIG. 4.
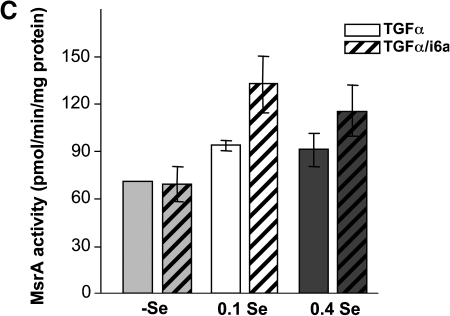
Regulation of MsrB1 and MsrA expression and MsrB and MsrA activities by dietary selenium in TGFα and TGFα/i6A- transgenic mice. (A) Western blot analysis of proteins from TGFα (left panel) and TGFα/i6A- (right panel) mice. The bottom image shows protein staining with Amido Black as a control of protein loading. Analysis of MsrB (B) and MsrA (C) activities in transgenic animals (n = 16, two-way ANOVA). There was an effect of selenium diet on MsrB activity, p < 0.04. However, no evidence was found against the null hypothesis for MsrA activity (p MsrA > 0.28). There was also no significant difference in MsrA and MsrB activities in TGFα and TGFα/i6A- transgenic mice (pMsrA > 0.26, pMsrB1 > 0.23).
Analysis of MsrB (Fig. 4B) and MsrA (Fig. 4C) activities agreed with the Western blot data. TGFα/i6A- mice had very low MsrB activity in all three dietary groups. Since no regulation of MsrB activity by dietary selenium was observed, it is likely that the residual MsrB activity was due to non-selenium MsrB isozymes, MsrB2 and MsrB3. Interestingly, MsrA activity was higher in TGFα/i6A- than in TGFα mice.
Regulation in outbred mice
The studies described above were limited to inbred mouse lines. To extend the findings to the heterogeneous animal population, we subjected a four-way cross HET mice, a model that incorporates genetic diversity (18), to selenium dietary treatments. In addition to the selenium diets, in this experiment we included two batches of the same NIH diet and a common Harlan Teklad rodent diet.
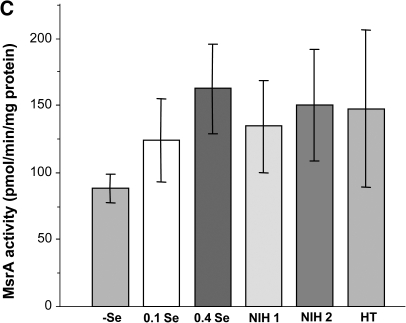
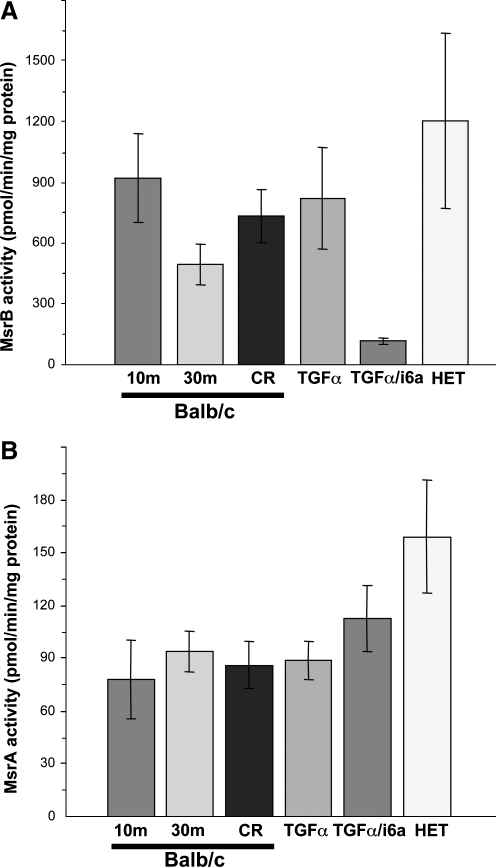
Regulation of selenoprotein expression by dietary selenium in outbred mice was similar to that in Balb/c mice, as MsrB1 and GPx1 levels were reduced and TR3 and MsrA levels were not affected by Se deficiency (Fig. 5A). Consistent with the detected levels of Se in NIH and Harlan Teklad diets (Supplemental Table 1), selenoprotein expression was fully saturated by these diets. Further activity assays found similar MsrA and MsrB activities in 0.4 ppm, NIH 1, NIH 2, and Harlan Teklad dietary groups (Fig. 5B and C). MsrB activity was slightly reduced in the 0.1 ppm Se diet and significantly decreased in Se deficiency (p < 0.005). MsrA activity was slightly higher in Se-supplemented mice compared to animals on the (-)Se diet (p > 0.1). Taken together, these data suggest that the findings on expression and dietary regulation of Msr proteins in Balb/c mice are likely to be similar in a wide variety of mouse models. We compared MsrA and MsrB activities in all 0.4 ppm Se groups of mice used in this study. As shown in Fig. 6, both MsrA and to some degree MsrB activities were suppressed in inbred models.
FIG. 5.
Regulation of MsrB1 and MsrA expression and MsrB and MsrA activities in HET outbred mice. (A) Female mice were subjected to six indicated diets (four animals per diet). Their liver samples were analyzed by Western blotting, as indicated. The proteins probed are indicated on the left and protein loading is shown by Amido Black staining on bottom image. Analysis of MsrB (B) and MsrA (C) activities (n = 24, one-way ANOVA), pMsrB1 < 0.005 and pMsrA > 0.1.
FIG. 6.
Comparison of MsrB and MsrA activities in different mouse strains. 10-month-old Balb/c (10m), 30 month-old Balb/c (30m), calorie restriction (CR) group, TGFα transgenic, TGFα/i6A- double transgenic, and HET outbred mice were maintained on a 0.4 ppm Se diet, and MsrB (A) and MsrA (B) activities determined as shown.
Discussion
In this study, we found that MsrB1 is the major Msr in mouse liver. MsrA is also abundant in this organ (as well as in kidney), whereas these enzymes are expressed at low levels in several other organs. In this regard, it would be interesting to determine the contribution of MsrB2 and MsrB3 in the context of low MsrB1 and MsrA activities. For example, MsrB2 can protect leukemia cells against oxidative damage (9).
Previous research suggested that, being a selenoprotein, MsrB1 is regulated by dietary selenium (33, 35, 36). Interestingly, we found that this protein was highly responsive to the level of selenium in the diet. Its regulation closely resembled that of another stress-related selenoprotein, GPx1, at both expression and activity levels. Approximately 0.15 ppm selenium in the diet was required to saturate MsrB1 activity and expression, making this protein one of the most susceptible to minor changes in Se levels in the diet. Our study also revealed that manipulations in dietary Se can be used to dramatically change total Msr activity in mouse liver. This feature may be used in future studies that examine the role of Msr system in disease.
We also observed that MsrB1, but not MsrA, activity was reduced with age, and that this regulation was independent of the Se status in the diet. Thus, MsrB1 accounts for the reduced methionine sulfoxide reduction in aging liver. Since aging is also accompanied by changes in trace element metabolism, supplementation of the diet with Se may be important in the aging population. We also found that regulation of MsrB1 and GPx1 by dietary Se was preserved under the conditions of CR, yet this dietary regimen itself had little influence on these enzymes. The data suggest that MsrB1 is not the basis for CR-dependent increase in lifespan, but these two factors might work synergistically in the extension of lifespan. If so, this would be similar to the finding in yeast, wherein a combination of CR and high levels of MsrB expression was particularly effective in extending lifespan (25).
Overall, our findings demonstrate that the system responsible for methionine sulfoxide reduction in mammals can be regulated at multiple levels by a simple dietary factor, selenium, as well as the age of animals. Our data provide insights into the strategies to regulate and possibly upregulate this repair process in diseased and aging organisms.
Supplementary Material
Abbreviations Used
- GPx
glutathione peroxidase
- Msr
methionine sulfoxide reductase
- MsrA
methionine-S-sulfoxide reductase
- MsrB
methionine-R-sulfoxide reductase
- Se
selenium
- Sec
selenocysteine
- TR
thioredoxin reductase
Acknowledgments
We would like to thank Vyacheslav Labunskyy, Mikalai Malinovski, Anton Turanov, and Yukiho Shinogawa for their help with processing of mouse samples, and Alexey Lobanov for help with statistical analysis. This work was supported by National Institutes of Health AG021518 and in part by GM065204, CA080946 and GM061603 to VNG, by the Korean Science and Engineering Foundation via the Aging-Associated Vascular Disease Research Center at Yeungnam University (R13-2005-005-01004-0) to HYK, and by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research to DLH.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Baker RD. Baker SS. LaRosa K. Whitney C. Newburger PE. Selenium regulation of glutathione peroxidase in human hepatoma cell line Hep3B. Arch Biochem Biophys. 1993;304:53–57. doi: 10.1006/abbi.1993.1320. [DOI] [PubMed] [Google Scholar]
- 2.Bermano G. Nicol F. Dyer JA. Sunde RA. Beckett GJ. Arthur JR. Hesketh JE. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J. 1995;311:425–430. doi: 10.1042/bj3110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry MJ. Banu L. Chen YY. Mandel SJ. Kieffer JD. Harney JW. Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3' untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 4.Berry MJ. Banu L. Harney JW. Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brot N. Weissbach H. Biochemistry and physiological role of methionine sulfoxide residues in proteins. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 6.Burk RF. Hill KE. Regulation of selenoproteins. Annu Rev Nutr. 1993;13:65–81. doi: 10.1146/annurev.nu.13.070193.000433. [DOI] [PubMed] [Google Scholar]
- 7.Burk RF. Selenium, an antioxidant nutrient. Nutr Clin Care. 2002;5:75–79. doi: 10.1046/j.1523-5408.2002.00006.x. [DOI] [PubMed] [Google Scholar]
- 8.Burk R. Hill KE. Selenoprotein P: An extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215–235. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- 9.Cabreiro F. Picot CR. Perichon M. Castel J. Friguet B. Petropoulos I. Overexpression of mitochondrial methionine sulfoxide reductase B2 protects leukemia cells from oxidative stress-induced cell death and protein damage. J Biol Chem. 2008;283:16673–16681. doi: 10.1074/jbc.M708580200. [DOI] [PubMed] [Google Scholar]
- 10.Chavatte L. Brown BA. Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 11.Colman RJ. Anderson RM. Johnson SC. Kastman EK. Kosmatka KJ. Beasley TM. Allison DB. Cruzen C. Simmons HA. Kemnitz JW. Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copeland PR. Fletcher JE. Carlson BA. Hatfield DL. Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diwadkar–Navsariwala V. Diamond AM. The link between selenium and chemoprevention: A case for selenoproteins. J Nutr. 2004;134:2899–2902. doi: 10.1093/jn/134.11.2899. [DOI] [PubMed] [Google Scholar]
- 14.Factor VM. Kiss A. Woitach JT. Wirth PJ. Thorgeirsson SS. Disruption of redox homeostasis in the transforming growth factor-alpha/c-myc transgenic mouse model of accelerated hepatocarcinogenesis. J Biol Chem. 1998;273:15846–15853. doi: 10.1074/jbc.273.25.15846. [DOI] [PubMed] [Google Scholar]
- 15.Fagegaltier D. Hubert N. Yamada K. Mizutani T. Carbon P. Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flohe L. Andreesen JR. Brigelius-Flohe R. Maiorino M. Ursini F. Selenium, the element of the moon, in life on earth. IUBMB Life. 2000;49:411–420. doi: 10.1080/152165400410263. [DOI] [PubMed] [Google Scholar]
- 17.Harman D. Free radicals in aging. Mol Cell Biochem. 1988;84:155–161. doi: 10.1007/BF00421050. [DOI] [PubMed] [Google Scholar]
- 18.Harrison DE. Strong R. Sharp ZD. Nelson JF. Astle CM. Flurkey K. Nadon NL. Wilkinson JE. Frenkel K. Carter CS. Pahor M. Javors MA. Fernandez E. Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatfield DL. Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmgren A. Thioredoxin and glutaredoxin: Small multi-functional redox proteins with active-site disulphide bonds. Biochem Soc Trans. 1988;16:95–96. doi: 10.1042/bst0160095. [DOI] [PubMed] [Google Scholar]
- 21.Hubert N. Walczak R. Sturchler C. Myslinski E. Schuster C. Westhof E. Carbon P. Krol A. RNAs mediating cotranslational insertion of selenocysteine in eukaryotic selenoproteins. Biochimie. 1996;78:590–596. doi: 10.1016/s0300-9084(96)80005-8. [DOI] [PubMed] [Google Scholar]
- 22.Kim HY. Gladyshev VN. Methionine sulfoxide reduction in mammals: Characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HY. Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HY. Fomenko DE. Yoon YE. Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koc A. Gasch AP. Rutherford JC. Kim HY. Gladyshev VN. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc Natl Acad Sci USA. 2004;101:7999–8004. doi: 10.1073/pnas.0307929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohrle J. Oertel M. Gross M. Selenium supply regulates thyroid function, thyroid hormone synthesis and metabolism by altering the expression of the selenoenzymes Type I 5'-deiodinase and glutathione peroxidase. Thyroidology. 1992;4:17–21. [PubMed] [Google Scholar]
- 27.Koubova J. Guarente L. How does calorie restriction work? Genes Dev. 2003;17:313–321. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- 28.Kryukov GV. Kumar RA. Koc A. Sun Z. Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei XG. Evenson JK. Thompson KM. Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 30.Lei XG. Glutathione peroxidase-1 gene knockout on body antioxidant defense in mice. Biofactors. 2001;14:93–99. doi: 10.1002/biof.5520140113. [DOI] [PubMed] [Google Scholar]
- 31.Low SC. Grundner–Culemann E. Harney JW. Berry MJ. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000;19:6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moskovitz J. Bar–Noy S. Williams WM. Requena J. Berlett BS. Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moskovitz J. Stadtman ER. Selenium-deficient diet enhances protein oxidation and affects methionine sulfoxide reductase (MsrB) protein level in certain mouse tissues. Proc Natl Acad Sci USA. 2003;100:7486–7490. doi: 10.1073/pnas.1332607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moustafa ME. Carlson BA. El–Saadani MA. Kryukov GV. Sun QA. Harney JW. Hill KE. Combs GF. Feigenbaum L. Mansur DB. Burk RF. Berry MJ. Diamond AM. Lee BJ. Gladyshev VN. Hatfield DL. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novoselov SV. Calvisi DF. Labunskyy VM. Factor VM. Carlson BA. Fomenko DE. Moustafa ME. Hatfield DL. Gladyshev VN. Selenoprotein deficiency and high levels of selenium compounds can effectively inhibit hepatocarcinogenesis in transgenic mice. Oncogene. 2005;24:8003–8011. doi: 10.1038/sj.onc.1208940. [DOI] [PubMed] [Google Scholar]
- 36.Oien DB. Moskovitz J. Selenium and the methionine sulfoxide reductase system. Molecules. 2009;14:2337–2344. doi: 10.3390/molecules14072337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petropoulos I. Mary J. Perichon M. Friguet B. Rat peptide methionine sulphoxide reductase: Cloning of the cDNA, and down-regulation of gene expression and enzyme activity during aging. Biochem J. 2001;355:819–825. doi: 10.1042/bj3550819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan H. Tang XD. Chen ML. Joiner ML. Sun G. Brot N. Weissbach H. Heinemann SH. Iverson L. Wu CF. Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmon AB. Pérez VI. Bokov A. Jernigan A. Kim G. Zhao H. Levine RL. Richardson A. Lack of methionine sulfoxide reductase A in mice increases sensitivity to oxidative stress but does not diminish life span. FASEB J. 2009;23:3601–3608. doi: 10.1096/fj.08-127415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohal RS. Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadtman TC. Discoveries of vitamin B12 and selenium enzymes. Annu Rev Biochem. 2002;71:1–16. doi: 10.1146/annurev.biochem.71.083101.134224. [DOI] [PubMed] [Google Scholar]
- 42.Stadtman ER. Van Remmen H. Richardson A. Wehr NB. Levine RL. Methionine oxidation and aging. Biochim Biophys Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Sunde RA. Regulation of glutathione peroxidase-1 expression. In: Hatfield OL, editor; Berry MJ, editor; Gladyshev VN, editor. Selenium: Its Molecular Biology and Role in Human Health. Second. New York: Springer; 2006. pp. 149–160. [Google Scholar]
- 44.Tujebajeva RM. Copeland PR. Xu XM. Carlson BA. Harney JW. Driscoll DM. Hatfield DL. Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turanov AA. Su D. Gladyshev VN. Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J Biol Chem. 2006;281:22953–22963. doi: 10.1074/jbc.M604326200. [DOI] [PubMed] [Google Scholar]
- 46.Vogt W. Oxidation of methionyl residues in proteins: Tools, targets, and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 47.Vougier S. Mary J. Friguet B. Subcellular localization of methionine sulphoxide reductase A (MsrA): Evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem J. 2003;373:531–537. doi: 10.1042/BJ20030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watkinson JH. Fluorometric determination of selenium in biological material with 2,3-diaminonaphthalene. Anal Chem. 1966;38:92–97. doi: 10.1021/ac60233a025. [DOI] [PubMed] [Google Scholar]
- 49.Weindruch R. Naylor PH. Goldstein AL. Walford RL. Influences of aging and dietary restriction on serum thymosin alpha 1 levels in mice. J Gerontol. 1988;43:B40–42. doi: 10.1093/geronj/43.2.b40. [DOI] [PubMed] [Google Scholar]
- 50.Weiss SL. Evenson JK. Thompson KM. Sunde RA. The selenium requirement for glutathione peroxidase mRNA level is half of the selenium requirement for glutathione peroxidase activity in female rats. J Nutr. 1996;126:2260–2267. doi: 10.1093/jn/126.9.2260. [DOI] [PubMed] [Google Scholar]
- 51.Weiss Sachdev S. Sunde RA. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and −4 in rat liver. Biochem J. 2001;357:851–858. doi: 10.1042/0264-6021:3570851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.