Abstract
Selenium is an essential trace element in mammals. The major biological form of this micronutrient is the amino acid selenocysteine, which is present in the active sites of selenoenzymes. Seven of 25 mammalian selenoproteins have been identified as residents of the endoplasmic reticulum, including the 15-kDa selenoprotein, type 2 iodothyronine deiodinase and selenoproteins K, M, N, S, and T. Most of these proteins are poorly characterized. However, recent studies implicate some of them in quality control of protein folding in the ER, retrotranslocation of misfolded proteins from the ER to the cytosol, metabolism of the thyroid hormone, and regulation of calcium homeostasis. In addition, some of these proteins are involved in regulation of glucose metabolism and inflammation. This review discusses evolution and structure–function relations of the ER-resident selenoproteins and summarizes recent findings on these proteins, which reveal the emerging important role of selenium and selenoproteins in ER function. Antioxid. Redox Signal. 12, 839–849.
Introduction
Selenocysteine (Sec, one-letter code is U) is the 21st amino acid in the genetic code; it is found in diverse proteins, known as selenoproteins, in bacteria, archaea, and eukaryotes. One major advantage of selenoproteins as enzymes is their higher catalytic activity, due to the presence of the Sec residue, in comparison to cysteine (Cys)-containing mutants (28) and often, but not always, to their natural Cys-containing counterparts. Twenty-five selenoprotein genes have been found in humans, whereas rodents have 24 such genes (34). Mammalian selenoenzymes of known function catalyze various thiol-dependent reactions and are involved in antioxidant defense, intracellular redox homeostasis, and thyroid hormone metabolism. More specifically, these proteins (a) directly eliminate reactive oxygen species (ROS) by catalyzing the reduction of hydrogen peroxide and phospholipid hydroperoxides [e.g., the glutathione peroxidases (GPxs) (72), selenoprotein H (SelH) (59) and the N-terminal domain of selenoprotein P (SelP) (70)]; (b) remove the consequences of ROS action by catalyzing the reduction of oxidized cysteine and methionine residues in proteins [e.g., the thioredoxin reductases (TRs) (3) and methionine sulfoxide reductases (Msrs) (30)]; and (c) monodeiodinate the thyroid prohormone thyroxine and convert it into the active hormone triiodothyronine, as well as inactivate this hormone by further deiodination [i.e., the iodothyronine deiodinases (DIs) (36)].
Physiological roles of GPxs, TRs, Msrs, and DIs (i.e., the metabolic pathways that include these proteins, their effectors, targets beyond enzymatic substrates, phenotypes associated with impairment of their activities, and knockdown/knockout of their genes) were recently reviewed (3, 6, 20, 24, 30, 36, 50, 63, 68, 72). Selenoproteins with partially characterized biologic functions [i.e., SelH, SelI, SelM, SelN, SelS, SelT, SelW, and the 15-kDa selenoprotein (Sep15)] or unknown functions (i.e., SelK, SelO, SelV) have been less well studied. However, most of them are likely redox enzymes, as their active sites have selenocysteine, which thus far has always been associated with redox functions. Sec is a highly reactive residue with lower pKa than that of Cys (5.2 vs. 8.3) and is a better nucleophile at physiologic pH. In addition, many of them possess a thioredoxin (Trx)-like fold [i.e., SelH, SelT, SelV, SelW (13), SelM and Sep15 (15)]. The Trx fold, described as a two-layer α/β/α sandwich with a βαβββα secondary structure pattern and a conserved CxxC (two Cys separated by two residues) or CxxS/T (one Cys replaced with serine or threonine) active-site motifs, is a characteristic feature of many oxidoreductases (e.g., Trxs, glutaredoxins, and protein disulfide isomerases) involved in regulation of various redox processes (51).
In this review, we focus on mammalian selenoproteins that are located in the endoplasmic reticulum (ER). To date, seven ER-resident selenoproteins have been identified (Table 1). The ER-resident selenoproteins include type 2 DI (D2), SelK, SelS, Sep15, SelM, SelN and SelT. Five of these seven proteins were identified through bioinformatics approaches before obtaining experimental data, including SelK (34), SelS (34), SelM (32), SelN (42), and SelT (35). Two other selenoproteins, D2 and Sep15, were initially identified by using experimental procedures (68, 21). Although no data describe their catalytic activity and direct targets (except for D2), recent data suggested their roles in the quality control of protein folding in the ER (40), retrotranslocation of misfolded proteins from the ER to the cytosol (43, 76) and regulation of calcium homeostasis (23, 29). Here, we highlight the known biologic functions and evolution of ER-resident selenoproteins, beginning with a brief description of eukaryotic selenoprotein synthesis. Then each of the ER selenoproteins and their roles in the ER function are discussed in detail. Finally, we highlight recent comparative evolutionary analyses of these ER selenoproteins.
Table 1.
Human Selenoproteins
| Selenoproteins | Accession number | Subcellular location |
|---|---|---|
| Deiodinase 1 (D1) | NP_000783.2 | Plasma membrane |
| Deiodinase 2 (D2) | NP_054644.1 | ER membrane |
| Deiodinase 3 (D3) | NP_001353.3 | Plasma membrane |
| Glutathione peroxidase 1 (GPx1) | CAA68491.1 | Cytosol, mitochondria |
| Glutathione peroxidase 2 (GPx2) | NP_002074.2 | Cytosol |
| Glutathione peroxidase 3 (GPx3) | NP_002075.2 | Secreted |
| Glutathione peroxidase 4 (GPx4) | NP_002076.2 | Cytosol, mitochondria, nucleus |
| Glutathione peroxidase 6 (GPx6) | NP_874360.1 | Unknown |
| Methionine sulfoxide reductase (MsrB1, SelR, SelX) | NP_057416.1 | Cytosol, nucleus |
| Selenoprotein H (SelH) | NP_734467.1 | Nucleus |
| Selenoprotein I (SelI) | NP_277040.1 | Unknown |
| Selenoprotein K (SelK) | NP_067060.2 | ER and plasma membrane |
| Selenoprotein M (SelM) | NP_536355.1 | ER lumen |
| Selenoprotein N (SelN) | NP_996809.1 | ER membrane |
| Selenoprotein O (SelO) | NP_113642.1 | Unknown |
| Selenoprotein P (SelP) | NP_001087195.1 | Secreted |
| Selenoprotein S (SelS) | NP_060915.2 | ER and plasma membrane |
| Selenoprotein T (SelT) | NP_057359.2 | ER membrane |
| Selenoprotein V (SelV) | NP_874363.1 | Unknown |
| Selenoprotein W (SelW) | NP_003000.1 | Cytosol |
| 15-kDa selenoprotein (Sep15) | NP_004252.2 | ER lumen |
| Selenophosphate synthase 2 (SPS2) | NP_036380.2 | Cytosol |
| Thioredoxin reductase 1 (TR1, TrxR1, TxnRd1) | NP_001087240.1 | Cytosol, nucleus |
| Thioredoxin glutathione reductase (TGR, TR2, TrxnRd3) | XP_001130163.1 | Cytosol |
| Thioredoxin reductase 3 (TR3, TxnRd2) | NP_006431.2 | Mitochondria |
Eukaryotic Selenoprotein Synthesis in Brief
Eukaryotic cells have developed a complex machinery for selenoprotein synthesis that has been thoroughly described in a recently published book (24) and many reviews (e.g., 2, 22, 25, 61, 65). The Sec-incorporation machinery has been conserved during evolution in eukaryotes, but many of these organisms lack it (and therefore, also lack selenoproteins). Sec is encoded by TGA, a codon that usually serves a translation-termination function. Decoding TGA for Sec insertion requires (a) Sec tRNA, tRNA[Ser]Sec; (b) a mRNA stem–loop structure located in the 3'-UTR of eukaryotic selenoproteins genes, called the Sec insertion sequence (SECIS) element; and (c) Sec-decoding protein factors: Sec-specific elongation factor eEFSec, SECIS-binding protein 2 (SBP2), L30 ribosome protein (rpL30), the 43-kDa RNA-binding protein (SECp43), and SECIS-interacting nucleolin (see 2, 22, 24, 25, 61, 65, and references therein). The free Sec pool in cells is extremely low [Sec, as well as selenide, reacts with oxygen, leading to ROS formation (37)], and this free amino acid is not used for specific Sec insertion into proteins. Instead, Sec is synthesized on its tRNA[Ser]Sec, and several enzymes are required for its synthesis, including phosphoseryl-tRNA[Ser]Sec kinase (PSTK), pyridoxal-phosphate–dependent Sec synthase (SecS), and selenophosphate synthetase 2 (SPS2) (2, 8, 24, 61, 65, 75).
Selenoproteins in the Endoplasmic Reticulum
The ER is responsible for various functions in eukaryotic cells, including synthesis of secretory and membrane proteins; transport of these proteins (e.g., transmembrane receptors and integral proteins); membrane trafficking; synthesis of phospholipids, steroids, glycogen, and other molecules; and sequestration of calcium ions and their regulated release into cytosol. At the same time, the ER lumen is a major site for protein folding, disulfide bond formation, N-linked glycosylation, and glycophosphatidylinositol-anchor addition in eukaryotic cells (14). Polypeptides co-translationally inserted into the ER lumen achieve a properly folded mature structure with the aid of ER-resident factors, such as chaperones (e.g., BiP, calnexin, calreticulin, and glucose-regulated protein GRP94) and thiol-disulfide oxidoreductases (e.g., ERp57 and protein disulfide isomerase) (14). Improperly folded proteins are recognized by the ER quality-control machinery and exported from the ER to the cytosol in the process termed retrotranslocation or ER-associated protein degradation (ERAD) (54). As discussed in this review, ER-resident selenoproteins are involved in some of these processes.
Type 2 iodothyronine deiodinase
D2 is a member of a well-described deiodinase family (for reviews, see 4, 5, 20, 36, 68). This protein catalyzes the activation of thyroid prohormone thyroxine (T4) to the biologically active form, 3, 5, 3'-triiodothyronine (T3). D2, the major T4-activating deiodinase, could also catalyze monodeiodination of reverse-T3. T3 effectively binds to the nuclear thyroid hormone receptor that regulates transcription of T3-dependent genes. Various developmental and metabolic processes are affected by this regulation.
Human D2 monomer (31 kDa, 272 aa) is a single-spanning membrane protein, oriented with a short N-terminal region (∼20 aa) in the ER lumen and a catalytic globular domain in the cytosol (4, 36) (a schematic representation of D2-domain arrangement is shown in Fig. 1). The hydrophobic nature of the N-terminus suggests that this part of the D2 molecule is an uncleaved sequence combining both signal and retention functions (4). The entire deiodinase family (D1 to D3) has the same domain organization with homologous sequences in the area surrounding the active site containing an SxxU motif (4, 7, 36). The catalytic domain of these proteins has two parts of the Trx fold (βαβ and ββα), separated by α-l-iduronidase (IDUA)-like sequence (7). The IDUA-like sequence is critical for iodothyronine binding in the active center (7). A unique feature of D2 cDNA in the deiodinase family is a presence of two in-frame TGA codons. The second TGA codon is known as a place for Sec insertion (64); however, this Sec (Fig. 1) and the remaining seven C-terminal amino acids are neither conserved nor critical for deiodination (64), and the specific function of this second Sec is unknown.
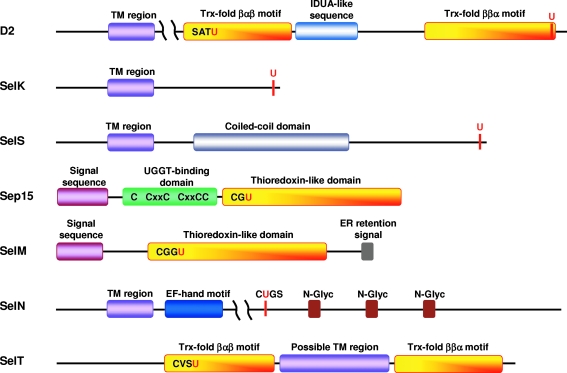
FIG. 1.
Domain organization of ER-resident mammalian selenoproteins. D2, SelK, SelS, and SelN are single-spanning membrane proteins containing an N-terminal transmembrane (TM) region (violet). The TM region begins 20 to 25 amino acids downstream from the initiator Met residue for D2, SelK, SelS, and SelN. A possible TM region of SelT also is shown in violet. Thioredoxin-like fold is known for D2 [βαβ motif (42 aa) and ββα motif (41 aa)], Sep15 (72 aa), SelM (72 aa), and SelT (141 aa) (yellow). The N-terminal signal peptide detected for Sep15 (32 aa) and SelM (24 aa) is shown in pink. Other annotated domains or predicted elements include the IDUA-like sequence in D2 (blue), the coiled-coil domain in SelS (colored in gray), UGGT-binding domain in Sep15 (green), the ER retention signal in SelM (grey), and EF-hand motif (dark blue) and three predicted N-glycosylated sites (residues 482, 504, and 530) in SelN (brown). Location of Sec (U) in all selenoproteins is shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
D2 is active as an enzyme in a homodimeric form because proper conformation for each active center can be achieved only in the dimeric state (20). The proposed catalytic mechanism for D1 to D3 includes generation of iodinated enzyme (D-SeI) followed by reduction with an unidentified endogenous reductant and release of iodide (36). The proximity of ER-resident D2 to the nucleus (4, 20) gives D2-generated, biologically active T3 easier access to the nuclear thyroid hormone receptor than T3 generated by D1 at the plasma membrane. This may be viewed as a mechanism developed by the cell for efficient transcriptional and translational regulation of T3-dependent genes.
Thyroid hormone produced by D2 is important for cochlear development and hearing, and is particularly important for brain development under conditions of thyroid insufficiency in mice (65). In humans, D2 is an important source of T3 in the anterior pituitary and thyroid glands, skeletal and heart muscle, placental and brown adipose tissue, and is the only 5′-deiodinase in the adult central nervous system (5, 20, 36). Further information about the regulation of D2 synthesis and degradation, posttranslational regulation, catalytic mechanism, D2-regulated thyroid hormone signaling pathways, D2 knockout and transgenic mouse models, and roles of D2 in development and energy homeostasis can be found in the following reviews (4, 5, 20, 36, 68).
Selenoprotein K
Human SelK is a small protein (11 kDa, 94 aa) with no assigned biochemical or biologic functions. SelK is a single-spanning integral protein, containing a short N-terminal sequence (∼20 amino acids) in the ER lumen and Sec-containing C-terminal sequence in the cytosol (Fig. 1). The Sec residue in human SelK is the 92nd amino acid. The topology of SelK was determined by using Drosophila Sec-containing homologue (10). SelK is an ER- (10) and plasma membrane-located protein (34). The cytosolic tail of SelK does not have a pronounced secondary structure and is extremely rich in glycine (16%), proline (15%), and the positively charged amino acids lysine, arginine, and histidine (22%). Such an unstructured, positively charged polypeptide might be an attractive target for negatively charged surfaces of other proteins. However, no SelK-binding partners have been found to date. It appears that the N-terminal transmembrane domain is responsible for SelK targeting to the ER membrane and its retention in the ER, as no N-terminal signal and C-terminal ER-retention sequences were found by analysis of SelK sequences.
Only one published study linked SelK to a possible function in cellular redox homeostasis (45). Overexpression of an N-terminal myc-tagged human SelK in neonatal rat cardiomyocytes reduced ROS production and protected cells from oxidative stress induced by hydrogen peroxide. However, it is not clear from the description of these experiments which form of human SelK (i.e., Sec- or Cys-containing) was used in this study (45). Knockdown of SelK expression in Drosophila embryos and in cultured Drosophila Schneider S2 cells by RNAi did not reveal changes in total antioxidant status of embryos and cells (56). It should be noted that Drosophila selenoproteins are not critical for protection against oxidative stress (29), and the role of mammalian SelK in these processes should be clarified by further experimental approaches.
Northern blot analysis of eight human tissues (heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas) displayed ubiquitous SelK mRNA distribution, with particularly high levels in heart, skeletal muscle, and pancreas (45). A recent study measured SelK mRNA levels in mouse tissues by real-time PCR (27), which again revealed the ubiquitous expression of this protein, which was particularly high in testes and high in intestine, kidney, liver, and spleen, and medium in brain, heart, and lung. Detectable SelK in all tested adult mouse and human tissues (12 in total) suggested a role of this protein in a biologic process common to many if not all cell types.
Selenoprotein S
Human SelS (also designated as SEPS1, Tanis, VIMP, and SELENOS) is a single-spanning membrane protein (21 kDa, 189 aa) with a short luminal segment and a longer cytosolic Sec-containing tail of ∼132 amino acids (Fig. 1). The Sec residue is the 188th amino acid. The topology of SelS was determined by co-precipitation of the recombinant GST-fused SelS cytosolic fragment (amino acids 48 to 187) with the recombinant ATPase p97 or with endogenous rat ATPase p97 (p97 is a cytosolic enzyme and a binding partner of SelS in ERAD; Fig. 2) (76). SelS is an ER (76) and plasma (34) membrane-located protein. The SelS cytosolic tail contains a coiled-coil domain (amino acids 52 to 122; PDB ID: 2Q2F) that is probably responsible for dimerization (oligomerization) of SelS (76) or for binding other proteins. The C-terminal end downstream of the coiled-coil domain does not have a pronounced secondary structure and is rich in glycine (21%), proline (10%), and lysine plus arginine (19%). This unstructured part might be needed for binding an unknown negatively charged target. The function of the C-terminal penultimate Sec is unknown. No N-terminal signal and C-terminal ER retention sequences were found by analysis of SelS sequences.
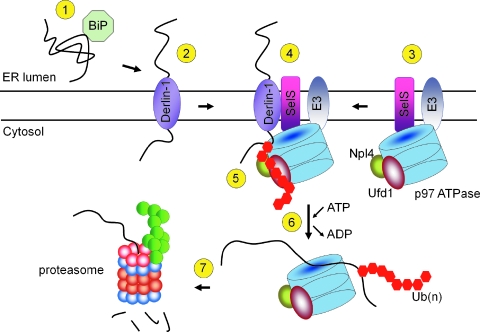
FIG. 2.
ER-associated protein degradation mediated by the Derlin-1 channel. In step 1, unfolded proteins, recognized by the ER chaperone BiP, are targeted to the multispanning Derlin-1. In step 2, retro-translocation is initiated when an unfolded polypeptide emerges in cytosol through the channel postulated to contain Derlin-1 (43, 76). In step 3, which likely occurs simultaneous with step 4, a complex containing SelS, ubiquitin ligase (E3), and p97 ATPase, together with cofactors Ufd1 and Npl4, is assembled. In step 4, the emerging polypeptide is captured in the cytosol by p97 with a complex formed in step 3. In step 5, the polypeptide undergoes polyubiquitination [shown as Ub(n)] by E3 and recognized by both p97 and Ufd1. In step 6, p97 extracts the polypeptide from the ER. Finally, the polyubiquitinated polypeptide is delivered to proteasome for degradation. The figure was adopted from (44, 76, 77). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
SelS was identified as a component of mammalian ERAD machinery (43, 76). The ERAD pathway is responsible for the transport of unfolded and misfolded proteins from the ER to the cytosol, followed by ubiquitin-proteasome system degradation (Fig. 2). In 2004, two groups discovered a new type of retrotranslocation channel that contains Derlin-1, a mammalian homologue of yeast Der1 (43, 76). In this pathway, SelS is proposed to mediate the interaction of cytosolic p97 ATPase and Derlin-1 (43, 76). The C-terminal cytosolic tail of SelS weakly interacts with the cytosolic tail of Derlin-1 (76), indicating that the interactions between SelS and Derlin-1 are mostly mediated by their membrane domains [Derlin-1 contains four transmembrane regions (43)]. Ubiquitin ligases E3 (gp78, Hrd1, SEL1L) identified as p97 binding partners did not interact with SelS (44, 78) (Fig. 2). Subsequently, Derlin-2 and Derlin-3, which are additional components of the ERAD complex, were found (44, 60). Endogenous Derlin-1 and Derlin-2 (44) and Derlin-2 and Derlin-3 (60) form hetero- and oligomers and have been proposed as components of two groups of transmembrane channels (60). SelS also was found as a Derlin-2 binding partner (44, 60), suggesting that it participates in both types of the proposed channels. Derlins contain two Cys residues per molecule; however, their topology is unknown, and a role of SelS in the reduction of disulfides or other forms of oxidized Cys in Derlins is possible. Interestingly, the latter study showed that the interaction between p97 and Derlin-1 was not affected when SelS levels were reduced by shRNA (44). These data indicate that the proposed function of SelS (i.e., binding p97 to Derlins) may be dispensable and that additional partners of SelS in the ERAD complex must be identified to elucidate the function of SelS in retrotranslocation.
The function of SelS in ERAD that plays a critical role in neutralizing ER stress and cell survival may explain the previously observed upregulation of SelS under three diverse conditions: glucose deprivation and treatments with N-glycosylation inhibitor tunicamycin and Ca2+-ATPase blocker thapsigargin (18, 19). All these conditions induce aggregation of improperly folded proteins in the ER due to accumulation of underglycosylated proteins, formation of unspecific disulfide bonds, or Ca2+ depletion in the ER. To overcome ER stress, cells induce expression of ERAD machinery components through transcriptional activation (60). Based on the role of SelS in ERAD, two additional effects of SelS overexpression, (a) protection of Min6 pancreatic cells from oxidative stress (19), and (b) protection of RAW264.7 murine macrophages from ER-induced apoptosis (31), could be clarified. It appears that one consequence of systemic oxidative stress (that results in the ER stress) can be effectively overcome by the retrotranslocation mechanism. Participation of SelS in ERAD may be a better mechanism to protect cells from oxidative stress than the previously proposed antioxidant function of SelS. Interestingly, Kim et al. (31) did not observe a correlation between SelS expression in RAW264.7 cells and apoptotic agents (staurosporine, anti-Fas) that do not cause ER stress, whereas a clear effect was found in the case of pharmacologic ER-stress agents (tunicamycin and thapsigargin). However, SelS-dependent protective mechanisms against ER stress–induced apoptosis are not yet sufficiently understood, and it is possible that other ERAD components than SelS may protect against the ER stress–induced apoptosis.
Before SelS identification as a selenoprotein (34) and as a component of ERAD machinery (44, 76), the TGA codon in SelS was interpreted as a stop signal (74). We further briefly discuss these prior studies, as they are important for understanding the possible function of SelS. One study (74) identified SelS in a type II diabetes animal model Psammomys obesus (Israeli sand rat) and named it Tanis (Hebrew word for “fasting”). SelS was found to be expressed in many rat and mouse tissues (27, 74). Expression of SelS was downregulated in liver, adipose tissue, and skeletal muscle in fed state of diabetic P. obesus in comparison to healthy animals (74). This selenoprotein was markedly increased in fasting diabetic, but not in nondiabetic animals (74). Based on the yeast two-hybrid screen with GST-tagged C-terminal end of SelS (i.e., truncated form) as a bait and transformants from a human liver cDNA library, serum amyloid A1β (SAA1β) protein was found as an SelS-binding partner (74).
SAA proteins are a family of apolipoproteins associated with high-density lipoprotein in plasma. SAA1 is secreted by liver and adipose tissue during the acute phase of inflammation and is thought to be involved in chronic diseases associated with diabetes. SelS was proposed as a receptor for SAA1β (74). The topology of SelS on the plasma membrane is unknown, but interaction of the SelS cytosolic tail with SAA was detected (74), suggesting that the C-terminal Sec-containing end of SelS may be located outside the cell. The function of plasma membrane SelS is unknown.
Other evidence for the role of SelS in mediating inflammation came from the study of the SelS promoter region. A promoter polymorphism, −105G → A, has been shown to increase the proinflammatory cytokine expression (12). Several clinical studies have shown the association of this SelS polymorphism with coronary heart disease and ischemic stroke (1), preeclampsia (57), and gastric cancer (66). However, another case study showed the lack of association between SelS polymorphism and type I diabetes, rheumatoid arthritis, or inflammatory bowel disease (53). Association of SelS and inflammatory response must be clarified in further studies.
15-kDa selenoprotein and selenoprotein M
Human Sep15 (18 kDa, 165 aa, with an ER signal peptide) and SelM (16 kDa, 145 aa, with an ER signal peptide) share 31% sequence identity and form a evolutionarily distinct selenoprotein family within the Trx superfamily (15). Analysis of tissue expression patterns in mammals revealed that Sep15 and SelM have different, but overlapping distributions. Sep15 showed high expression levels in prostate, testes, brain, liver, and kidney, whereas the highest level of SelM expression was observed in the brain (32, 38). Both Sep15 and SelM encode an N-terminal signal sequence that is likely cleaved after translocation into the ER and a single Sec residue within the redox-active CxxU (SelM) or CxU (Sep15) motif (see Fig. 1 for structural elements). In addition, Sep15 contains a distinct Cys-rich domain in the N-terminal part of the protein (39). SelM lacks this Cys-rich domain, but, in contrast to Sep15, it has a C-terminal ER retention signal. The structural studies demonstrated that both Sep15 and SelM have a Trx-like fold, suggesting an oxidoreductase function for these selenoproteins (15).
UDP-glucose:glycoprotein glucosyltransferase (UGGT) has been identified as a binding partner of Sep15 (33). This ER-localized chaperone is involved in the quality control of protein folding in the ER. UGGT functions primarily as a folding sensor and initiates prolonged association of incorrectly folded glycoproteins with calnexin (CNX) and protein disulfide isomerase ERp57, which facilitate protein folding and catalyze thiol-disulfide bond exchange. However, recent studies demonstrated that UGGT may also directly assist folding of a group of CNX substrates. This group of glycoproteins requires UGGT or associated factors or both for correct folding, as the rate of their release from CNX was delayed in UGGT-deficient mouse embryonic fibroblasts (67). Sep15 forms a tight 1:1 complex with UGGT through its N-terminal Cys-rich domain (39). Association of Sep15 with UGGT suggests the possibility that Sep15 may function as a protein disulfide isomerase that targets glycoproteins in this class of CNX substrates (40, 67).
The redox potential of Sep15 was determined for recombinant Drosophila Sep15 (15), which is not a selenoprotein and contains a CxC motif in the active site. This potential was found to be −225 mV and corresponded to the lowest redox potential among known thiol-dependent oxidoreductases that reside in the ER and assist in protein folding (15). Oxidoreductases involved in reduction of disulfide bonds in the ER of eukaryotic cells have not previously been reported. Recently, ER-resident protein ERdj5 (73) was proposed to be a major enzyme with reductase activity in the ER. ERdj5 was found to be responsible for the reduction of disulfide bonds in misfolded proteins targeted for retrotranslocation. However, the redox potential of mouse ERdj5 is approximately the same as that of Drosophila Sep15 (−218 mV vs. −225 mV), and the Cys-containing Sep15 could be considered a strong reductant in the ER (selenodisulfides usually have lower redox potential than their disulfide homologues). A reductant that is able to reduce proteins with such low redox potential as Sep15 and ERdj5 is unknown, as the ER does not have a major reductant such as cytosolic Trx (−270 mV). The question “Which systems reduce selenenylsulfide bonds involving Sec and a resolving Cys in the ER-resident selenoproteins?” could be asked for all selenoproteins with Sec in the ER lumen.
Selenoprotein N
Initially, two isoforms of human SelN (also known as SEPN1 and SepN) were deduced from cDNA analyses (55). Isoform 1 corresponded to the full-length sequence with two in-frame TGA codons, whereas isoform 2 had a segment containing the first TGA codon deleted and therefore had only one in-frame TGA codon. However, subsequent analysis of SelN expression in six various human adult and fetal tissues (liver, brain, heart, diaphragm, skeletal muscle, and stomach) by Western blotting with three different anti-SelN antibodies revealed the presence of only one protein corresponding to isoform 2 (62).
SelN (66 kDa, 590 aa) is an integral ER-membrane protein (62) with a recently determined topology that was reported as unpublished data (41). It is a single-spanning membrane protein (Fig. 1) with a small N-terminus in the cytosol and the major part of the protein, including the predicted active site, in the ER lumen (41). The SelN Sec residue is located 458 amino acids downstream from the initiator Met in a CUGS redox motif (Fig. 1). A similar motif is present in TRs (3, 34). The presence of a transmembrane region in the N-terminal sequence was indirectly and unintentionally confirmed in one of the initial studies on this protein (62), when the first 61 amino acids corresponding to the first exon were removed and localization of the truncated SelN was found to be mostly nuclear. Initially, the N-terminus was considered the ER-targeting and -retention sequence of SelN (62). SelN has a candidate calcium-binding EF-hand motif that was predicted by bioinformatics analyses (62). It was proposed that this motif contributes to the overall structure and does not serve for signaling purposes (41). Deglycosylation assays involving SelN have demonstrated that this selenoprotein is a glycoprotein (62). Five putative N-glycosylated asparagines (residues 126, 189, 482, 504, and 530) in Asn-Xaa-Ser/Thr motifs were predicted by NetNGlyc 1.0, and three of them are shown in Fig. 1.
SelN attracted much attention from the scientific community when several research groups linked mutations in the SelN gene to various muscular disorders now collectively known as SEPN1-related myopathy (for review, see 41). It includes disorders ranging from congenital muscular dystrophy to rigid-spine muscular dystrophy (55), the classic form of multiminicore disease (17), desmin-related myopathy with Mallory body–like inclusions (16), and congenital fiber-type disproportion myopathy (11). Patients with SEPN1-related myopathy are characterized by scoliosis, neck weakness, spinal rigidity, severe respiratory insufficiency, and poor axial muscle strength (11, 16, 17, 54). SelN point mutations identified in patients with SEPN1-related myopathy are scattered throughout the coding region of SelN (41). These mutations lead to premature termination of translation or decreased Sec-insertion efficiency that significantly reduce SelN levels (2, 41).
A recent innovative study (29) linked SelN insufficiency and muscular myopathy at the molecular level. An association of rabbit SelN with the ryanodine receptor (RyR) intracellular calcium-release channel was demonstrated in vivo through immunoprecipitation and colocalization experiments (29). In addition, both SelN and RyR were required for calcium-flux activity in zebrafish embryos (29). It was proposed that SelN and RyR are part of a common complex. Moreover, SelN was required for RyR activity (i.e., a decrease in the RyR activity was detected in SelN-depleted muscles). Thus, SelN may serve a reductase function for a homotetrameric RyR channel that has ∼400 Cys residues and whose activity is known to be subject to redox regulation (29). In this regard, SelN could be viewed as a redox regulator of calcium homeostasis in muscle.
Finally, SelN is a ubiquitously expressed protein, and its function in other tissues is not characterized. SelN expression is increased in mouse fetal tissues (in liver, stomach, heart, and muscle) in comparison with adult tissues (62).
Selenoprotein T
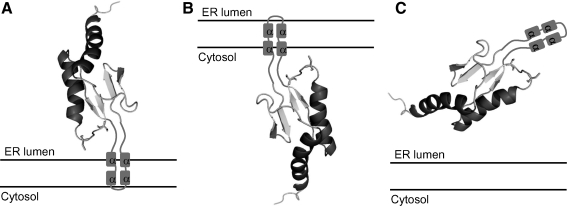
Human SelT is a 22-kDa protein consisting of 195 aa with no assigned function. The proposed Trx-like catalytic domain of the protein has two parts of the Trx fold, βαβ and ββα, separated by an α-helical insertion and a CxxU redox motif in the βαβ motif (Fig. 1) (13). Localization of SelT in the ER was shown in two studies (13, 23), but a GFP-fused N-terminal SelT was localized predominantly in the Golgi, with a possible occurrence in the ER and cytosol (13). The N-terminal signal sequence predicted by SignalP at the ExPASy server with a probability of 100% was tested in two studies (13, 23), which showed that SelT distribution was independent of the presence of its N-terminal sequence. However, a hydrophobic region located in the insertion between two parts of the Trx fold at positions 87 to 102 was found to be required for targeting SelT to the ER (23) (Fig. 1). It is likely that this hydrophobic region may be responsible, not only for targeting SelT to the ER, but also for the integration and maintenance of SelT in this compartment (23). The length of the α-helical insertion (67 aa) separating the two Trx parts is sufficient to pass the ER membrane twice and form the globular Trx-fold domain on either the ER lumen or the cytosolic sides (Fig. 3A and B, respectively). A possibility also exists that SelT is not a transmembrane protein (Fig. 3C). If the hydrophobic regions indeed form the transmembrane domain, SelT would be a double-spanning membrane protein.
FIG. 3.
Three possible variants of SelT topology in the ER. The 3D structure of mouse SelT was modeled with Modeler 9 v6 based on N- and C-terminal sequence similarity to mouse SelW (PDB 2NPB). The α-helical insertion separating two Trx-motifs (13) was removed from the SelT sequence before modeling. It consists of four predicted α-helixes that are depicted by squares. (A, B) SelT, shown as a membrane protein, Sec-containing globular domain may be localized in the ER lumen (A) or cytosol (B). (C) SelT is shown as ER-resident protein, but not an integral ER membrane protein.
SelT mRNA was detected in all tested mouse adult tissues examined with Northern blot analysis, with the highest levels observed in kidney, followed by brain, heart, thymus, and testes (13). Real time PCR analysis revealed the highest SelT mRNA levels in mouse testes (27), in rat testes, and in the anterior lobe of the pituitary (23, 27). In situ hybridization of rat embryo showed that SelT was ubiquitously expressed at the E14 and E21 development stages (23). Widespread distribution observed in early as well as late stages of embryogenesis and in adults suggests a basic and general function of SelT in various tissues of developing and adult animals.
A recent study (23) implicated SelT in the regulation of Ca2+ homeostasis and neuroendocrine secretion in response to a neuropeptide, pituitary adenylate cyclase–activating polypeptide (PACAP). PACAP induces the differentiation of rat pheochromocytoma PC12 cells and regulates the secretion of neuropeptides. Initially, it was found that PACAP and cAMP induced a rapid and long-lasting increase in SelT expression in PC12 cells. Subsequent experiments showed that overexpression of Sec-containing SelT in PC12 cells increased the intracellular Ca2+ concentration, and overexpression of the Sec-to-Ala mutant of SelT prevented the effect of SelT on Ca2+ release, implying that SelT could regulate Ca2+ homeostasis through a redox mechanism (23). Conversely, knockdown of SelT expression by shRNA inhibited the PACAP-induced increase in intracellular Ca2+ and decreased the growth and hormone secretion, suggesting that SelT is involved in the signaling pathway activated by PACAP, perhaps through calcium regulation (23). Neither targets of SelT nor the SelT-linked intracellular Ca2+ channel have been identified.
Evolutionary Trends of Mammalian ER-Resident Selenoproteins
In the past decade, dramatic advances in genomics have led to the generation of complete or almost complete genomic sequences for a large number of organisms from the three domains of life. Comparative genomics, which examines the relations of genome structure and its genes and other functional elements across species, plays an increasingly important role in providing new insights into understanding the evolution of ancestral and modern species, which in turn helps us to understand gene/protein functions, metabolic and signaling pathways, and other biologic processes in living organisms. Recently, several comparative genomic analyses have been carried out to examine selenoproteins and selenium utilization (47, 79, 80). These studies have improved our understanding of the occurrence and evolutionary trends in the use of selenoproteins, including those localized to the ER.
The selenoproteomes (set of selenoproteins) of several recently sequenced organisms, such as Ostreococcus tauri (26 selenoproteins), O. lucimarinus (29 selenoproteins), Dictyostelium discoideum (five selenoproteins), Drosophila pseudoobscura (three selenoproteins), Thalassiosira pseudonana (16 selenoproteins), fish (32 to 37 selenoproteins), and several other organisms with complete genomes were recently described (46, 47). Combined with the previously characterized selenoproteomes of mammals (34), D. melanogaster (9, 52), Caenorhabditis elegans (71), Chlamydomonas reinhardtii (58), Trypanosoma and Leishmania (48), and Plasmodium (49), these studies revealed general patterns in the use of selenoproteins. The size of eukaryotic selenoproteome varies from zero (e.g., plants, fungi and some protists) to more than 30 (fish and algae). Significant differences in the composition of selenoproteomes could be seen even among related organisms. The distribution of ER-resident selenoproteins and their Cys-containing homologues in representative model eukaryotes is shown in Fig. 4.
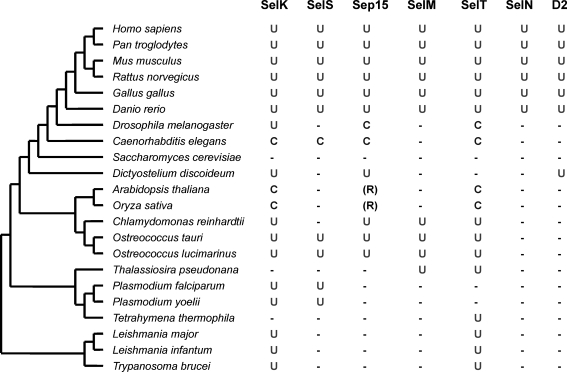
FIG. 4.
Occurrence of ER-resident selenoproteins in eukaryotes. A set of model organisms (from unicellular organisms to humans) was selected to illustrate the distribution of these selenoproteins in eukaryotes. In each organism, the presence of Sec (U)- or Cys (C)-containing forms of each ER-resident selenoprotein family is indicated. Homologues of Sep15 were detected in land plants, Arabidopsis thaliana and Oryza sativa, which contain arginine (R) in place of Sec/Cys.
Among ER selenoproteins, SelK is the most widespread selenoprotein (47). This protein of unknown function is present in nearly all eukaryotes that use Sec, but it is replaced with a Cys-containing homologue in nematodes and several other organisms. Thus, SelK appears to be an ancient selenoprotein that evolved in the early ancestor of eukaryotes. Other ER selenoproteins, including SelT, SelS, Sep15, and SelM, also were detected in both lower eukaryotes (e.g., Ostreococcus, Dictyostelium, and/or Thalassiosira) and in mammals. In contrast, SelN has a more narrow distribution and appears to be specific to vertebrates. Thus, most of the mammalian ER-resident selenoproteins can be traced back to the ancestral, unicellular eukaryotes. Almost all organisms containing D2 (as well as D1 and D3) are vertebrates. However, recent studies have identified the presence of a Sec-containing deiodinase-like protein in D. discoideum (47) and several Sec-containing deiodinase-like proteins in prokaryotes (80) and lower eukaryotes. This may suggest that, despite the functions different from those of human D1-3, members of the deiodinase family may have evolved in the ancestral single-celled organism, but were lost in most eukaryotic clades. This is consistent with a general evolutionary trend of selenoproteins in eukaryotes: core selenoprotein families (like SelK) evolved first, followed by the origin of additional selenoproteins in narrower groups of organisms (like SelN).
Open Questions
The identity, evolution, and function of mammalian selenoproteins have received considerable attention in recent years. However, the unexpected enrichment of these proteins in the ER has largely been overlooked. This compartment is known for its redox processes, the most prominent of which is the disulfide bond–formation system, but how selenoproteins fit into this function is not known. The specific functions of the majority of ER-resident selenoproteins are not known, but recent studies identified unique linkages of individual selenoproteins to particular redox processes in the ER. SelN appears to serve as a redox cofactor for ryanodine receptor, whereas the oxidoreductase Sep15 associates with the protein involved in the quality control of protein folding. D2 is involved in the metabolism of thyroid hormones, also catalyzing a redox reaction. SelS has been linked to retrotranslocation of proteins from the ER for their subsequent degradation. Whether redox processes play a role in this function is not known, but we note that currently no functional information exists regarding the Sec residue in SelS. It would be important to understand the specific function of each of the ER selenoproteins. Such studies will surely lead to a better understanding of the role of selenium in biology and human health.
Abbreviations Used
- CNX
calnexin
- ER
endoplasmic reticulum
- ERAD
ER-associated protein-degradation machinery
- IDUA
α-l-iduronidase
- PACAP
pituitary adenylate cyclase–activating polypeptide
- ROS
reactive oxygen species
- SAA
serum amyloid A
- SelK
selenoprotein K
- SelM
selenoprotein M
- SelN
selenoprotein N
- SelS
selenoprotein S
- SelT
selenoprotein T
- Sep15
the 15-kDa selenoprotein
- Trx
thioredoxin
- UGGT
UDP-glucose:glycoprotein glucosyltransferase
Acknowledgments
We thank Dr. Dmitri Fomenko for providing a modeled SelT structure. This work was supported by NIH grants to V.N.G., and NCI Intramural Research Program and the Center for Cancer Research to D.L.H.
References
- 1.Alanne M. Kristiansson K. Auro K. Silander K. Kuulasmaa K. Peltonen L. Salomaa V. Perola M. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum Genet. 2007;122:355–365. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- 2.Allmang C. Wurth L. Krol A. The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim Biophys Acta. 2009;1790:1415–1423. doi: 10.1016/j.bbagen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Arnér ESJ. Focus on mammalian thioredoxin reductases: important selenoproteins with versatile functions. Biochim Biophys Acta. 2009;R901:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Bianco AC. Larsen PR. Cellular and structural biology of the deiodinases. Thyroid. 2005;15:777–786. doi: 10.1089/thy.2005.15.777. [DOI] [PubMed] [Google Scholar]
- 5.Bianco AC. Salvatore D. Gereben B. Berry MJ. Larsen PR. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- 6.Borchert A. Wang CC. Ufer C. Schiebel H. Savaskan NE. Kuhn H. The role of phospholipid hydroperoxide glutathione peroxidase isoforms in murine embryogenesis. J Biol Chem. 2006;281:19655–19664. doi: 10.1074/jbc.M601195200. [DOI] [PubMed] [Google Scholar]
- 7.Callebaut I. Curcio-Morelli C. Mornon JP. Gereben B. Buettner C. Huang S. Castro B. Fonseca TL. Harney JW. Larsen PR. Bianco AC. The iodothyronine selenodeiodinases are thioredoxin-fold family proteins containing a glycoside hydrolase clan GH-A-like structure. J Biol Chem. 2003;278:36887–36896. doi: 10.1074/jbc.M305725200. [DOI] [PubMed] [Google Scholar]
- 8.Carlson BA. Xu XM. Kryukov GV. Rao M. Berry MJ. Gladyshev VN. Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellano S. Morozova N. Morey M. Berry MJ. Serras F. Corominas M. Guigó R. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CL. Shim MS. Chung J. Yoo HS. Ha JM. Kim JY. Choi J. Zang SL. Hou X. Carlson BA. Hatfield DL. Lee BJ. G-rich, a Drosophila selenoprotein, is a Golgi-resident type III membrane protein. Biochem Biophys Res Commun. 2006;348:1296–1301. doi: 10.1016/j.bbrc.2006.07.203. [DOI] [PubMed] [Google Scholar]
- 11.Clarke NF. Kidson W. Quijano-Roy S. Estournet B. Ferreiro A. Guicheney P. Manson JI. Kornberg AJ. Shield LK. North KN. SEPN1: associated with congenital fiber-type disproportion and insulin resistance. Ann Neurol. 2006;59:546–552. doi: 10.1002/ana.20761. [DOI] [PubMed] [Google Scholar]
- 12.Curran JE. Jowett JB. Elliott KS. Gao Y. Gluschenko K. Wang J. Abel Azim DM. Cai G. Mahaney MC. Comuzzie AG. Dyer TD. Walder KR. Zimmet P. MacCluer JW. Collier GR. Kissebah AH. Blangero J. Genetic variation in selenoprotein S influences inflammatory response. J Nat Genet. 2005;37:1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- 13.Dikiy A. Novoselov SV. Fomenko DE. Sengupta A. Carlson BA. Cerny RL. Ginalski K. Grishin NV. Hatfield DL. Gladyshev VN. SelT, SelW, SelH, and Rdx12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry. 2007;46:6871–6882. doi: 10.1021/bi602462q. [DOI] [PubMed] [Google Scholar]
- 14.Ellgaard L. Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson AD. Labunskyy VM. Fomenko DE. Arac D. Chelliah Y. Amezcua CA. Rizo J. Gladyshev VN. Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- 16.Ferreiro A. Ceuterick-de Groote C. Marks JJ. Goemans N. Schreiber G. Hanefeld F. Fardeau M. Martin JJ. Goebel HH. Richard P. Guicheney P. Bönnemann CG. Desmin-related myopathy with Mallory body-like inclusions is caused by mutations of the selenoprotein N gene. Ann Neurol. 2004;55:676–686. doi: 10.1002/ana.20077. [DOI] [PubMed] [Google Scholar]
- 17.Ferreiro A. Quijano-Roy S. Pichereau C. Moghadaszadeh B. Goemans N. Bönnemann C. Jungbluth H. Straub V. Villanova M. Leroy JP. Romero NB. Martin JJ. Muntoni F. Voit T. Estournet B. Richard P. Fardeau M. Guicheney P. Mutations of the selenoprotein N gene, which is implicated in rigid spine muscular dystrophy, cause the classical phenotype of multiminicore disease: reassessing the nosology of early-onset myopathies. Am J Hum Genet. 2002;71:739–749. doi: 10.1086/342719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fradejas N. Pastor MD. Mora-Lee S. Tranque P. Calvo S. SEPS1 gene is activated during astrocyte ischemia and shows prominent antiapoptotic effects. J Mol Neurosci. 2008;35:259–265. doi: 10.1007/s12031-008-9069-3. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y. Feng HC. Walder K. Bolton K. Sunderland T. Bishara N. Quick M. Kantham L. Collier GR. Regulation of the selenoprotein SelS by glucose deprivation and endoplasmic reticulum stress: SelS is a novel glucose-regulated protein. FEBS Lett. 2004;563:185–190. doi: 10.1016/S0014-5793(04)00296-0. [DOI] [PubMed] [Google Scholar]
- 20.Gereben B. Zavacki AM. Ribich S. Kim BW. Huang SA. Simonides WS. Zeöld A. Bianco AC. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev. 2008;29:898–938. doi: 10.1210/er.2008-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gladyshev VN. Jeang KT. Wootton JC. Hatfield DL. A new human selenium-containing protein. J Biol Chem. 1998;273:8910–8915. doi: 10.1074/jbc.273.15.8910. [DOI] [PubMed] [Google Scholar]
- 22.Gromer S. Eubel JK. Lee L. Jacob J. Human selenoproteins at a glance. Cell Mol Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grumolato L. Ghzili H. Montero-Hadjadje M. Gasman S. Lesage J. Tanguy Y. Galas L. Ait-Ali D. Leprince J. Guérineau NC. Elkahloun AG. Fournier A. Vieau D. Vaudry H. Anouar Y. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+ mobilization and neuroendocrine secretion. FASEB J. 2008;22:1756–1768. doi: 10.1096/fj.06-075820. [DOI] [PubMed] [Google Scholar]
- 24.Hatfield DL, editor; Berry MJ, editor; Gladyshev VN, editor. Selenium: Its Molecular Biology and Role in Human Health. 2nd. New York: Springer; 2006. p. 410. [Google Scholar]
- 25.Hatfield DL. Carlson BA. Xu XM. Mix H. Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog Nucleic Acid Res Mol Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 26.Hirosawa-Takamori M. Chung HR. Jackle H. Conserved selenoprotein synthesis is not critical for oxidative stress defence and the lifespan of Drosophila. EMBO Rep. 2004;5:317–322. doi: 10.1038/sj.embor.7400097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann PR. Höge SC. Li PA. Hoffmann FW. Hashimoto AC. Berry MJ. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson L. Gafvelin G. Arnér ESJ. Selenocysteine in proteins: properties and biotechnological use. Biochim Biophys Acta. 2005;1726:1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Jurynec MJ. Xia R. Mackrill JJ. Gunther D. Crawford T. Flanigan KM. Abramson JJ. Howard MT. Grunwald DJ. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc Natl Acad Sci U S A. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HY. Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3:2080–2089. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim KH. Gao Y. Walder K. Collier GR. Skelton J. Kissebah AH. SEPS1 protects RAW264.7 cells from pharmacological ER stress agent-induced apoptosis. Biochem Biophys Res Commun. 2007;354:127–132. doi: 10.1016/j.bbrc.2006.12.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korotkov KV. Novoselov SV. Hatfield DL. Gladyshev VN. Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol Cell Biol. 2002;22:1402–1411. doi: 10.1128/mcb.22.5.1402-1411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korotkov KV. Kumaraswamy E. Zhou Y. Hatfield DL. Gladyshev VN. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2001;276:15330–15336. doi: 10.1074/jbc.M009861200. [DOI] [PubMed] [Google Scholar]
- 34.Kryukov GV. Castellano S. Novoselov SV. Lobanov AV. Zehtab O. Guigó R. Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 35.Kryukov GV. Kryukov VM. Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 36.Kuiper GGJM. Kester MHA. Peeters RP. Visser TJ. Biochemical mechanisms of thyroid hormone deiodination. Thyroid. 2005;15:787–798. doi: 10.1089/thy.2005.15.787. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S. Björnstedt M. Holmgren A. Selenite is a substrate for calf thymus thioredoxin reductase and thioredoxin and elicits a large non-stoichiometric oxidation of NADPH in the presence of oxygen. Eur J Biochem. 1992;207:435–439. doi: 10.1111/j.1432-1033.1992.tb17068.x. [DOI] [PubMed] [Google Scholar]
- 38.Kumaraswamy E. Malykh A. Korotkov KV. Kozyavkin S. Hu Y. Kwon SY. Moustafa ME. Carlson BA. Berry MJ. Lee BJ. Hatfield DL. Diamond AM. Gladyshev VN. Structure-expression relationships of the 15-kDa selenoprotein gene: possible role of the protein in cancer etiology. J Biol Chem. 2000;275:35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- 39.Labunskyy VM. Ferguson AD. Fomenko DE. Chelliah Y. Hatfield DL. Gladyshev VN. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose:glycoprotein glucosyltransferase. J Biol Chem. 2005;280:37839–37845. doi: 10.1074/jbc.M508685200. [DOI] [PubMed] [Google Scholar]
- 40.Labunskyy VM. Hatfield DL. Gladyshev VN. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59:1–5. doi: 10.1080/15216540601126694. [DOI] [PubMed] [Google Scholar]
- 41.Lescure A. Rederstorff M. Krol A. Guicheney P. Allamand V. Selenoprotein function and muscle disease. Biochim Biophys Acta. 2009;1790:1569–1574. doi: 10.1016/j.bbagen.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Lescure A. Gautheret D. Carbon P. Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J Biol Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 43.Lilley BN. Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 44.Lilley BN. Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C. Qiu F. Zhou H. Peng Y. Hao W. Xu J. Yuan J. Wang S. Qiang B. Xu C. Peng X. Identification and characterization of selenoprotein K: an antioxidant in cardiomyocytes. FEBS Lett. 2006;580:5189–5197. doi: 10.1016/j.febslet.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 46.Lobanov AV. Hatfield DL. Gladyshev VN. Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol. 2008;9:R62. doi: 10.1186/gb-2008-9-3-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lobanov AV. Fomenko DE. Zhang Y. Sengupta A. Hatfield DL. Gladyshev VN. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 2007;8:R198. doi: 10.1186/gb-2007-8-9-r198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobanov AV. Gromer S. Salinas G. Gladyshev VN. Selenium metabolism in Trypanosoma: characterization of selenoproteomes and identification of a Kinetoplastida-specific selenoprotein. Nucleic Acids Res. 2006;34:4012–4024. doi: 10.1093/nar/gkl541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lobanov AV. Delgado C. Rahlfs S. Novoselov SV. Kryukov GV. Gromer S. Hatfield DL. Becker K. Gladyshev VN. The Plasmodium selenoproteome. Nucleic Acids Res. 2006;34:496–505. doi: 10.1093/nar/gkj450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J. Holmgren A. Selenoproteins. J Biol Chem. 2009;284:723–287. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 51.Martin JL. Thioredoxin: a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 52.Martin-Romero FJ. Kryukov GV. Lobanov AV. Carlson BA. Lee BJ. Gladyshev VN. Hatfield DL. Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J Biol Chem. 2001;276:29798–29804. doi: 10.1074/jbc.M100422200. [DOI] [PubMed] [Google Scholar]
- 53.Martínez A. Santiago JL. Varadé J. Márquez A. Lamas JR. Mendoza JL. de la Calle H. Díaz-Rubio M. de la Concha EG. Fernández-Gutiérrez B. Urcelay E. Polymorphisms in the selenoprotein S gene: lack of association with autoimmune inflammatory diseases. BMC Genomics. 2008;9:329. doi: 10.1186/1471-2164-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meusser B. Hirsch C. Jarosch E. Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 55.Moghadaszadeh B. Petit N. Jaillard C. Brockington M. Roy SQ. Merlini L. Romero N. Estournet B. Desguerre I. Chaigne D. Muntoni F. Topaloglu H. Guicheney P. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- 56.Morozova N. Forry EP. Shahid E. Zavacki AM. Harney JW. Kraytsberg Y. Berry MJ. Antioxidant function of a novel selenoprotein in Drosophila melanogaster. Genes Cells. 2003;8:963–971. doi: 10.1046/j.1365-2443.2003.00687.x. [DOI] [PubMed] [Google Scholar]
- 57.Moses EK. Johnson MP. Tømmerdal L. Forsmo S. Curran JE. Abraham LJ. Charlesworth JC. Brennecke SP. Blangero J. Austgulen R. Genetic association of preeclampsia to the inflammatory response gene SEPS1. Am J Obstet Gynecol 198: 336. 2008:e1–e5. doi: 10.1016/j.ajog.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 58.Novoselov SV. Rao M. Onoshko NV. Zhi H. Kryukov GV. Xiang Y. Weeks DP. Hatfield DL. Gladyshev VN. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novoselov SV. Kryukov GV. Xu XM. Carlson BA. Hatfield DL. Gladyshev VN. Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J Biol Chem. 2007;282:11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- 60.Oda Y. Okada T. Yoshida H. Kaufman RJ. Nagata K. Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papp LV. Lu J. Holmgren A. Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 62.Petit N. Lescure A. Rederstorff M. Krol A. Moghadaszadeh B. Wewer UM. Guicheney P. Selenoprotein N: an endoplasmic reticulum glycoprotein with an early developmental expression pattern. Hum Mol Genet. 2003;12:1045–1053. doi: 10.1093/hmg/ddg115. [DOI] [PubMed] [Google Scholar]
- 63.Reeves MA. Hoffmann PR. The human selenoproteome: recent insights into functions and regulation. Cell Mol Life Sci. 2009;66:2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salvatore D. Harney JW. Larsen PR. Mutation of the Secys residue 266 in human type 2 selenodeiodinase alters 75Se incorporation without affecting its biochemical properties. Biochimie. 1999;81:535–538. doi: 10.1016/s0300-9084(99)80106-0. [DOI] [PubMed] [Google Scholar]
- 65.Squires JE. Berry MJ. Eukaryotic selenoprotein synthesis: mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60:232–235. doi: 10.1002/iub.38. [DOI] [PubMed] [Google Scholar]
- 66.Shibata T. Arisawa T. Tahara T. Ohkubo M. Yoshioka D. Maruyama N. Fujita H. Kamiya Y. Nakamura M. Nagasaka M. Iwata M. Takahama K. Watanabe M. Hirata I. Selenoprotein S (SEPS1) gene −105G>A promoter polymorphism influences the susceptibility to gastric cancer in the Japanese population. BMC Gastroenterol. 2009;9:2. doi: 10.1186/1471-230X-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solda T. Galli C. Kaufman RJ. Molinari M. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol Cell. 2007;27:238–249. doi: 10.1016/j.molcel.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 68.St.Germain DL. Galton VA. Hernandez A. Minireview: defining the roles of the iodothyronine deiodinases: current concepts and challenges. Endocrinology. 2009;150:1097–1107. doi: 10.1210/en.2008-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.St.Germain DL. Galton VA. The deiodinase family of selenoproteins. Thyroid. 1997;7:655–68. doi: 10.1089/thy.1997.7.655. [DOI] [PubMed] [Google Scholar]
- 70.Takebe G. Yarimizu J. Saito Y. Hayashi T. Nakamura H. Yodoi J. Nagasawa S. Takahashi K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem. 2002;277:41254–41258. doi: 10.1074/jbc.M202773200. [DOI] [PubMed] [Google Scholar]
- 71.Taskov K. Chapple C. Kryukov GV. Castellano S. Lobanov AV. Korotkov KV. Guigó R. Gladyshev VN. Nematode selenoproteome: the use of the selenocysteine insertion system to decode one codon in an animal genome? Nucleic Acids Res. 2005;33:2227–2238. doi: 10.1093/nar/gki507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toppo S. Flohé L. Ursini F. Vanin S. Maiorino M. Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim Biophys Acta. 2009;1790:1486–1500. doi: 10.1016/j.bbagen.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Ushioda R. Hoseki J. Araki K. Jansen G. Thomas DY. Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 74.Walder K. Kantham L. McMillan JS. Trevaskis J. Kerr L. De Silva A. Sunderland T. Godde N. Gao Y. Bishara N. Windmill K. Tenne-Brown J. Augert G. Zimmet PZ. Collier GR. Tanis: a link between type 2 diabetes and inflammation? Diabetes. 2002;51:1859–1866. doi: 10.2337/diabetes.51.6.1859. [DOI] [PubMed] [Google Scholar]
- 75.Xu XM. Carlson BA. Mix H. Zhang Y. Saira K. Glass RS. Berry MJ. Gladyshev VN. Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye Y. Shibata Y. Yun C. Ron D. Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 77.Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Ye Y. Shibata Y. Kikkert M. van Voorden S. Wiertz E. Rapoport TA. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y. Romero H. Salinas G. Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y. Gladyshev VN. Trends in selenium utilization in marine microbial world revealed through the analysis of the global ocean sampling (GOS) project. PLoS Genet. 2008;4:e1000095. doi: 10.1371/journal.pgen.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]