The first crystal structure of phosphoglucose isomerase from M. tuberculosis H37Rv is reported.
Keywords: Mycobacterium tuberculosis, PGI, isomerases, 5-phosphoarabinonate
Abstract
Phosphoglucose isomerase (PGI) plays a key role in both glycolysis and gluconeogenesis inside the cell, whereas outside the cell it exhibits cytokine properties. PGI is also known to act as an autocrine motility factor, a neuroleukin agent and a differentiation and maturation mediator. Here, the first crystal structure of PGI from Mycobacterium tuberculosis H37Rv (Mtb) is reported. The structure was refined at 2.25 Å resolution and revealed the presence of one molecule in the asymmetric unit with two globular domains. As known previously, the active site of Mtb PGI contains conserved residues including Glu356, Glu216 and His387 (where His387 is from the neighbouring molecule). The crystal structure of Mtb PGI was observed to be rather more similar to human PGI than other nonbacterial PGIs, with only a few differences being detected in the loops, arm and hook regions of the human and Mtb PGIs, suggesting that the M. tuberculosis enzyme uses the same enzyme mechanism.
1. Introduction
Mycobacterium tuberculosis, which infects one-third of the human population, remains the leading cause of mortality among bacterial pathogens. It is responsible for approximately two million fatalities and nine million new cases annually (Murray & Salomon, 1998 ▶). Globally, tuberculosis has shown a resurgence owing to (i) the deadly synergy of the bacterium with HIV and (ii) the emergence of multidrug-resistant strains (MDR), including extensively drug-resistant strains (XDR-TB) that are virtually untreatable using the available chemotherapies (Janowski et al., 2009 ▶). The pathogen is capable of surviving and replicating in the macrophage for a long period of time, which is an important factor in the prevalence of the disease. Consequently, there is an increased necessity to understand the detailed mechanisms employed by the pathogen for its persistence in the macrophage, as understanding these mechanisms may offer new targets for the development of highly effective therapeutic agents. Traditionally, antibiotic agents have largely been targeted at processes that are essential for the growth of the pathogen under optimal conditions. However, enzymatic pathways that are involved in growth and survival under nutritionally restrictive conditions represent attractive alternative targets for the development of novel drugs. M. tuberculosis harvests most of its energy from glycolysis when the bacterium is persisting in a ‘quiescent’ state inside macrophages. Therefore, the key enzymes of this pathway are good targets for the development of new inhibitors and drugs.
Phosphoglucose isomerase is a ubiquitously present enzyme that catalyzes the second step of glycolysis, namely the interconversion of glucose-6-phosphate (G6P) and fructose-6-phosphate (F6P). As in most organisms, PGI from M. tuberculosis is a homodimeric enzyme, with each monomer comprising 553 amino-acid residues. PGI has an analogous structure in both eukaryotic and prokaryotic species, with the most conserved residues located in and around the active site. Apart from its enzymatic functions, PGI also has a number of other ‘moonlighting’ functions: it acts as an autocrine motility factor (AMF) and neuroleukin agent (NLK), a serine proteinase inhibitor and a differentiation and maturation mediator (DMM) (Cao et al., 2000 ▶; Chaput et al., 1988 ▶; Faik et al., 1988 ▶; Watanabe et al., 1991 ▶).
X-ray crystal structures of PGIs with and without substrates and inhibitors have been reported from a number of species (Chou et al., 2000 ▶; Cordeiro et al., 2004 ▶; Davies & Muirhead, 2002 ▶, 2003 ▶; Jeffery et al., 2001 ▶; Lin et al., 2009 ▶; Meng et al., 1999 ▶; Solomons et al., 2004 ▶; Sun et al., 1999 ▶; Tanaka et al., 2006 ▶; Yamamoto et al., 2008 ▶). These studies have provided considerable insight into the structure and catalytic mechanisms of the enzyme. Comparisons of the structures with and without bound ligands have revealed a multistep catalytic reaction mechanism and the residues involved in enzyme catalysis (Meng et al., 1999 ▶; Solomons et al., 2004 ▶; Sun et al., 1999 ▶). Despite their similar overall fold, there are significant structural differences between PGIs, mainly in the large domain of the enzyme (Chou et al., 2000 ▶; Cordeiro et al., 2004 ▶; Davies & Muirhead, 2002 ▶, 2003 ▶; Jeffery et al., 2000 ▶, 2001 ▶; Solomons et al., 2004 ▶; Sun et al., 1999 ▶). Subtle differences can also be found in the conformation of the small domain of different PGIs (Chou et al., 2000 ▶). PGI, which is an essential enzyme for the survival of M. tuberculosis (Mtb) in macrophages, can be targeted for selective inhibition of the pathogen (Mathur et al., 2005 ▶; Sassetti et al., 2003 ▶). Structural characterization of Mtb PGI will help to identify differences that may exist between Mtb PGI and that of its human host.
2. Experimental procedures
2.1. Expression and purification of native and mutant PGIs of M. tuberculosis H37Rv
The expression and purification of native Mtb PGI was performed using the methods described previously by Mathur et al. (2007 ▶). Single amino-acid substitution mutations were made on PGI using a QuikChange Site-Directed mutagenesis kit as per the manufacturer’s instructions, using the Mtb gene cloned into pET22b(+) as template (Mathur et al., 2005 ▶). The putative mutant transformants were analyzed and the mutations were confirmed by automated DNA sequencing (Applied Biosystems Model 393). The efficiency of the kit was found to be 80%. The Thr211Ala and Gly157Tyr mutant PGIs were purified from the soluble fraction using one-step Ni2+–NTA affinity chromatography. The yield of purified protein was 25–30 mg protein per litre of culture.
The enzyme activity was determined spectrophotometrically essentially as described by Mathur et al. (2005 ▶), using a reaction mixture containing 0.1 mM Tris–HCl pH 7.6, 2 mM EDTA, 0.5 mM β-NADP+, 1 mM fructose-6-phosphate, 1 U glucose-6-phosphate dehydrogenase and an aliquot of purified recombinant enzyme in a final reaction volume of 1 ml. The change in absorption at 340 nM was followed as a measure of the reduction of the β-NADP+ intermediate by glucose-6-phosphate dehydrogenase. One unit of enzyme activity is defined as 1 µmol of β-NADP+ reduced per minute under the experimental conditions.
2.2. Circular-dichroism (CD) spectroscopy of the recombinant and mutant PGIs
Circular-dichroism measurements of the recombinant proteins were performed at various temperatures on a JASCO J710 spectropolarimeter (Easton, Missouri, USA) using a 0.1 cm path-length cell. The temperature was controlled using a PTC-348W temperature controller. The protein samples were dissolved to a concentration of 0.15 mg ml−1 in 5 mM Tris–phosphate buffer pH 7.6. The spectra in the far ultraviolet (190–320 nm) were recorded at a scanning speed of 20 nm min−1. Ten spectra were accumulated in each case and averaged, followed by baseline correction by subtraction of the buffer spectrum. The mean residue ellipticity was calculated and expressed in units of deg cm2 dmol−1.
2.3. Crystallization of Mtb PGI
Crystals of Mtb PGI were grown using the hanging-drop vapour-diffusion method. Equal volumes (1 µl each) of protein solution (10 mg ml−1) and crystallization buffer were mixed on a cover slip, which was subsequently suspended over a reservoir containing 500 µl crystallization buffer. The crystals were optimized by micro-seeding using reservoir containing 1.2 M ammonium sulfate pH 8.5 (0.1 M HEPES) and 5% glycerol at 283 K. The diffraction improved from 2.8 Å resolution (as reported previously by Mathur et al., 2007 ▶) to 2.25 Å resolution. It was observed that aging of the protein (∼2 weeks at 277 K) in combination with small changes in the reservoir solution and the seeding techniques improved the diffraction quality. The best crystals grew in about 10 d and were harvested by transfer to a cryoprotectant solution consisting of 15% ethylene glycol in the mother-liquor solution, suspended in a nylon loop (Hampton Research, USA) and flash-cooled in liquid nitrogen.
2.4. Data collection and structure determination and refinement
Details of diffraction data collection are given in Table 1 ▶. The data were processed and scaled using the XDS program package (Kabsch, 1993 ▶). The structure was solved by the molecular-replacement method using the program Phaser (Read, 2001 ▶), with the structure of human phosphoglucose isomerase (PDB code 1iat; Read et al., 2001 ▶) as a search model. Two flexible loops (residues 104–117, 414–422, 438–464; B factor = 57 Å2) and the C-terminal region (amino acid residues 512–554; B factor = 47 Å2) of the search model were truncated for the phase solution. Cycles of adjustment of the model were performed using the graphical program Coot (Emsley & Cowtan, 2004 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell. The structure was determined using data derived from a single crystal.
| Diffraction data statistics | |
| X-ray source | Synchrotron† |
| Detector | ADSC Q105 CCD |
| Wavelength (Å) | 0.97942 |
| Temperature (K) | 100 |
| Crystal oscillation (°) | 1.0 |
| Crystal information | |
| Space group | I212121 |
| Unit-cell parameters (Å) | a = 109.7, b = 118.5, c = 137.6 |
| Resolution (Å) | 30.00–2.25 |
| Rmeas‡ (%) | 8.2 |
| I/σ(I) | 17.52 |
| Completeness (%) | 99.4 |
| Redundancy | 7.2 |
| Refinement | |
| Resolution (Å) | 2.25 (2.30–2.25) |
| No. of reflections | 42594 |
| Rwork/Rfree§ | 18.2/22.3 |
| No. of atoms | 4492 |
| Protein | 4191 |
| Water | 291 |
| Ligand (sulfate) | 10 |
| B factors (average B value; Å2) | |
| Protein | 45.94 |
| Water | 53.85 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.15 |
Beamline ID29 at the European Synchrotron Radiation Facility, Grenoble, France.
R meas is the redundancy-independent R factor (intensities).
3% of the total reflections were excluded for cross-validation.
Alternate rounds of computational refinement were performed using PHENIX (Adams et al., 2002 ▶). The first electron-density map showed interpretable density for the major fold of the PGI molecule. Most of the amino-acid residues were clearly visible in the electron-density map, with the exception of the side chains of the surface residues Lys248 and Arg147, which were disordered. Therefore, the side-chain occupancies of these atoms were kept at zero. In the later stages, water molecules were modelled using the graphical program Coot (Emsley & Cowtan, 2004 ▶) and included in the refinement cycles. The geometry of the final model was checked using PROCHECK (Laskowski et al., 1993 ▶). Molecular diagrams were drawn using PyMOL (http://pymol.sourceforge.net).
3. Results and discussion
3.1. Crystallization, data collection and structure determination
Crystals of the Mtb PGI protein that diffracted to 2.25 Å resolution were obtained using the hanging-drop vapour-diffusion technique and were optimized using the microseeding technique. The crystals of Mtb PGI belonged to space group I212121, with one molecule in the asymmetric unit. In this crystal form, the two molecules of the dimer were related by a crystallographic twofold axis parallel to a. The unit-cell parameters were a = 109.7, b = 118.5, c = 137.6 Å, α = β = γ = 90°. The structure of Mtb PGI was solved by the molecular-replacement method using the coordinates of truncated human PGI (see §2) as a search model (PDB code 1iat; Read et al., 2001 ▶). Manual fitting of the model following the refinement cycles eventually led to a final R work of 18.2% and an R free of 22.3% with good stereochemistry (Table 1 ▶). In the final model no residues were in disallowed regions of Ramachandran space (95.2% preferred, 100% allowed) as analysed using PROCHECK (Laskowski et al., 1993 ▶). The protein folds in a similar fashion as previously reported for rabbit and human PGI (Berrisford et al., 2006 ▶). The model showed good electron density for amino-acid residues 6–554, including two residues, Leu and Glu, which were introduced at the C-terminus of the mature protein as a result of the convenient cloning of the PGI gene in the expression vector. The N-terminal residues 1–5 were not built because of the poor electron-density map in this region.
3.2. Overall structure of Mtb PGI
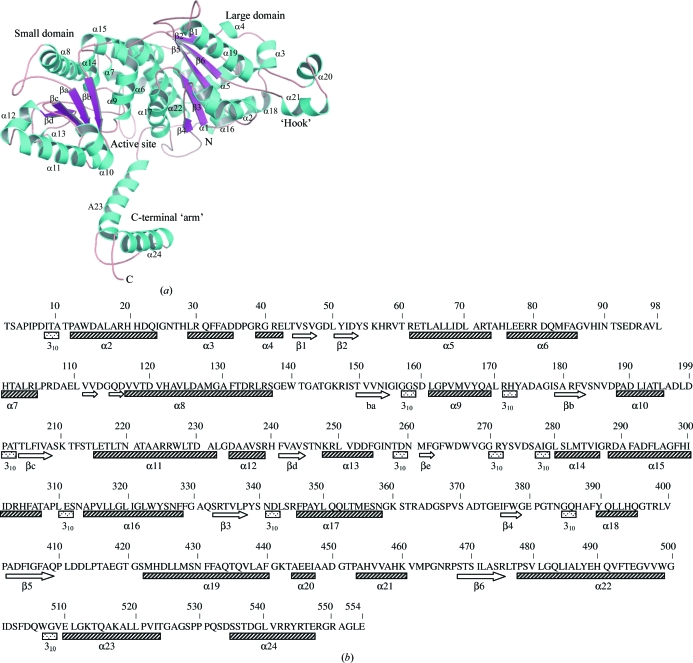
The 2.25 Å resolution crystal structure of the Mtb PGI monomer shows that the molecule is comprised of two globular domains (namely large and small; Fig. 1 ▶ a). The secondary-structure assignments are shown in Fig. 1 ▶(b). Three-dimensional structures of PGI (native as well as in complex with substrates or inhibitors) from different sources are known (Table 2 ▶). In all cases, both domains of PGI show α/β folding motifs containing a β-sheeted central core flanked by α-helices. A long protruding loop extending from the large domain into the C-terminal region gives an ellipsoidal shape to the molecule. This loop region (residues 441–464) consists of two small α-helices (α20 and α21).
Figure 1.
(a) Cartoon diagram of the overall three-dimensional structure of PGI from M. tuberculosis H37Rv. The small and large domains and the active site are indicated. Helices are shown in cyan and strands are shown in magenta. The N- and C-termini are labelled N and C, respectively. (b) The assignment of the secondary-structure elements of Mtb PGI H37Rv is indicated below the amino-acid sequence. α-Helices are displayed as hatched boxes, β-strands are shown as arrows and 310-helices are shown as dotted boxes.
Table 2. Structures related to Mtb H37Rv identified in the PDB.
| PDB code | Source | Resolution (Å) | No. of subunits in ASU | Ligand† | Sequence identity to Mtb (%) | Reference |
|---|---|---|---|---|---|---|
| 1g98 | Oryctolagus cuniculus | 1.9 | 2 | PA5 | 46 | Jeffery et al. (2001 ▶) |
| 1gzd | Sus scrofa | 2.5 | 1 | SO42− | 49 | Davies & Muirhead (2002 ▶) |
| 1gzv | S. scrofa | 3.5 | 1 | PA5 | 46 | Davies & Muirhead (2002 ▶) |
| 1hm5 | O. cuniculus | 1.8 | 2 | No ligand | 45 | Arsenieva & Jeffery (2002 ▶) |
| 1iat | Homo sapiens | 1.6 | 1 | BME, SO42− | 46 | Read et al. (2001 ▶) |
| 1jlh | H. sapiens | 2.1 | 4 | 48 | Cordeiro et al. (2003 ▶) | |
| 1koj | O. cuniculus | 1.9 | 2 | 5-Phospho-D-arabinohydroxamic acid | 45 | Arsenieva et al. (2002 ▶) |
| 1n8t | O. cuniculus | 2.5 | 2 | 48 | Davies & Muirhead (2003 ▶) | |
| 1q50 | Leishmania mexicana | 2.6 | 1 | 48 | Cordeiro et al. (2004 ▶) | |
| 1qxj | Pyrococcus furiosus | 1.8 | 2 | Ni2+ | 3 | Swan et al. (2003 ▶) |
| 1qxr | P. furiosus | 1.7 | 2 | PA5, Ni2+ | 3 | Swan et al. (2003 ▶) |
| 1qy4 | P. furiosus | 1.8 | 2 | Ni2+, 6-phosphogluconic acid | 3 | Swan et al. (2003 ▶) |
| 1t10 | L. mexicana mexicana | 2.3 | 1 | F6P | 48 | Cordeiro et al. (2004 ▶) |
| 1tzb | Pyrobaculum aerophilum strain IM2 | 1.2 | 2 | GOL, SO42− | 2 | Swan et al. (2004 ▶) |
| 1tzc | P. aerophilum strain IM2 | 1.5 | 2 | GOL, PA5, SO42− | 6 | Swan et al. (2004 ▶) |
| 1u0e | Mus musculus | 1.6 | 2 | GOL, BME, SO42− | 47 | Solomons et al. (2004 ▶) |
| 1u0f | M. musculus | 1.6 | 2 | α-D-G6Q, G6Q, SO42−, BME, GOL | 47 | Solomons et al. (2004 ▶) |
| 1u0g | M. musculus | 1.7 | 2 | SO42−, erythose-4-phosphate, BME, GOL | 47 | Solomons et al. (2004 ▶) |
| 1x7n | Pyrococcus furiosus | 1.9 | 1 | PA5, Mn2+ | 3 | Berrisford et al. (2004 ▶) |
| 1x82 | P. furiosus | 1.5 | 1 | PA5 | 3 | Berrisford et al. (2004 ▶) |
| 1x8e | P. furiosus | 2.8 | 2 | 3 | Berrisford et al. (2004 ▶) | |
| 1x9h | Pyrobaculum aerophilum | 1.5 | 2 | GOL, F6P, SO42− | 6 | Swan et al. (2004 ▶) |
| 1x9i | P. aerophilum | 1.2 | 2 | GOL, G6Q | 6 | Swan et al. (2004 ▶) |
| 1xtb | O. cuniculus | 2.0 | 2 | D-Sorbitol-6-phosphate | 49 | Lee & Jeffery (2005 ▶) |
| 2cer | Sulfolobus solfataricus | 2.3 | 2 | ACT, (5R,6R,7S,8S)-5-(hydroxymethyl)-2-(2-phenylethyl)-1,5,6,7,8,8a-hexahydroimidazo[1,2-a]pyridine-6,7,8-triol | 5 | Gloster et al. (2006 ▶) |
| 2cet | Thermotoga maritima | 2.0 | 2 | Ca2+, (5R,6R,7S,8S)-5-(hydroxymethyl)-2-(2-phenylethyl)-1,5,6,7,8,8a-hexahydroimidazo[1,2-a]pyridine-6,7,8-triol, ACT | 1 | Gloster et al. (2006 ▶) |
| 2cvp | M. musculus | 1.8 | 1 | GOL, ACT | 47 | Tanaka et al. (2006 ▶) |
| 2cxn | M. musculus | 1.4 | 1 | GOL, phosphate ion | 47 | Tanaka et al. (2006 ▶) |
| 2cxo | M. musculus | 1.8 | 1 | GOL, D-4-phosphoerythronic acid | 47 | Tanaka et al. (2006 ▶) |
| 2cxp | M. musculus | 1.7 | 1 | GOL, arabinose-5-phosphate | 47 | Tanaka et al. (2006 ▶) |
| 2cxq | M. musculus | 1.5 | 1 | GOL, D-sorbitol-6-phosphate | 47 | Tanaka et al. (2006 ▶) |
| 2cxr | M. musculus | 1.7 | 1 | GOL, 6-phosphogluconic acid | 47 | Tanaka et al. (2006 ▶) |
| 2cxs | M. musculus | 1.5 | 1 | GOL, F6P | 47 | Tanaka et al. (2006 ▶) |
| 2cxt | M. musculus | 1.5 | 1 | GOL, F6P | 47 | Tanaka et al. (2006 ▶) |
| 2cxu | M. musculus | 1.6 | 1 | GOL, phosphate ion | 47 | Tanaka et al. (2006 ▶) |
| 2gc0 | Pyrococcus furiosus | 2.0 | 2 | 5-Phospho-D-arabinohydroxamic acid, Zn2+ | 3 | Berrisford et al. (2006 ▶) |
| 2gc1 | P. furiosus | 2.0 | 2 | Zn+2, D-sorbitol-6-phosphate | 3 | Berrisford et al. (2006 ▶) |
| 2gc2 | P. furiosus | 2.1 | 2 | D-F6P (open form), Zn2+ | 3 | Berrisford et al. (2006 ▶) |
| 2gc3 | P. furiosus | 2.1 | 2 | α-D-Mannose-6-phosphate, Zn2+ | 3 | Berrisford et al. (2006 ▶) |
| 2o2c | Trypanosoma brucei brucei | 1.6 | 1 | GOL, G6Q | 16 | Arsenieva et al. (2002 ▶) |
| 2o2d | T. brucei brucei | 1.9 | 1 | GOL, citric acid | 19 | Arsenieva et al. (2002 ▶) |
| 2q8n | Thermotoga maritima MSB8 | 1.8 | 1 | Nonaethylene glycol, Cl−, SO42− | 21 | Joint Center for Structural Genomics (unpublished) |
| 3cv0 | Trypanosoma brucei | 2.0 | 1 | 1,2-Ethanediol | 7 | Sampathkumar et al. (2008 ▶) |
| 3cvl | T. brucei | 2.2 | 1 | 7 | Sampathkumar et al. (2008 ▶) | |
| 3ff1 | Staphylococcus aureus subsp. aureus COL | 1.6 | 1 | Na+, G6Q | 16 | S. M. Anderson, J. S. Brunzelle, O. Onopriyenko, S. Peterson, W. F. Anderson & A. Savchenko (unpublished) |
| 3hjb | Vibrio cholerae | 1.5 | 1 | Ca2+, Cl−, tetraethylene glycol, Na+, di(hydroxyethyl)ether | 51 | Center for Structural Genomics of Infectious Diseases (unpublished) |
| 3ifs | Bacillus anthracis | 2.0 | 1 | SO42−, Cl−, triethylene glycol, Li+ | 19 | Center for Structural Genomics of Infectious Diseases (unpublished) |
| 2wu8 | Mycobacterium tuberculosis H37Rv | 1 | SO42− | This work |
PA5, 5-phosphoarabinonic acid; F6P, fructose-6-phosphate; GOL, glycerol; BME, β-mercaptoethanol; G6Q, glucose-6-phosphate; ACT, acetate ion.
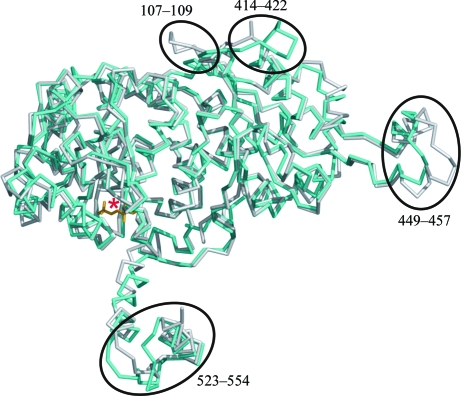
The large domain contains a six-stranded mixed parallel/antiparallel β-sheet surrounded by four α-helices on each side. The small domain comprises a five-stranded parallel β-sheet encircled on each side by loops and three α-helices in a similar fashion to the large domain. The structure has a 50-residue extension (Fig. 1 ▶ a) at the C-terminus that extends away from the small and large domains but wraps around the other monomer in the dimer. This extended region is usually termed the ‘arm’. Both the hook and the arm make several contacts with the neighbouring monomer. These interactions allow PGI to remain highly stable under denaturing conditions (Blackburn & Noltmann, 1972 ▶; Dyson & Noltmann, 1969 ▶). The overall topology of the monomer of phosphoglucose isomerase from M. tuberculosis H37Rv was similar to other reported structures of PGI (Arsenieva et al., 2002 ▶; Davies & Muirhead, 2003 ▶; Lee et al., 2001 ▶; Read et al., 2001 ▶; Shaw & Muirhead, 1977 ▶). The structure of Mtb PGI can be best superimposed on that of native human PGI (PDB code 1iat; Read et al., 2001 ▶) with a root-mean-square deviation (r.m.s.d.) value of 0.8 Å for the main-chain Cα atoms (Fig. 2 ▶), excluding both terminal helices and the loop (residues 441–464).
Figure 2.
Superposition of the overall structures of Mtb and human PGI in complex with the inhibitor PAB. The Mtb PGI structure superposed well with native human PGI (PDB code 1iat), resulting in an r.m.s.d. value of 0.8 Å for 497 Cα pairs. The Mtb and human PGIs are shown in cyan and grey, respectively. The red asterisk represents the transition-state analogue inhibitor PAB (5-phosphoarabinonate; yellow) in Mtb PGI. The regions of major differences in the ‘hook’ (residues 449–457), loop (residues 107–109 and 414–422) and ‘arm’ (residues 523–554) regions are circled.
3.3. Active site of Mtb PGI
Analysis of the crystal structure revealed that the active-site geometry retains the same arrangement as has been described for PGIs from other species. The crystal structures of PGI in complex with substrate and inhibitors as reported in Table 2 ▶ provide details of the active-site geometry. The active site is comprised of several loops which connect the secondary structures of the small domain, i.e. the connections βa–α9, βb–α10, βc–α11 and βe–α14. On the other side, the active site is formed by the C-terminal region of α17, the N-terminal region of α23 and the middle section of α19. The contribution of the α18 helix and a 310-helix by the neighbouring molecule indicates that the active site of one monomer requires elements from an adjacent monomer. The conserved active-site residues included the side-chain hydroxyl groups of Ser159, Ser209, Thr211 and Thr214. Other conserved residues included Lys210, Glu216, Gln352, Glu356, Gln506, Glu510 and Lys517. His387 from the adjacent molecule was crucial for the formation of the active-site pocket by two subunits of PGI as described by Jeffery et al. (2000 ▶) and references therein. In the case of this crystal form of Mtb PGI, the His387 residue from the other monomer of the crystallographic dimer contributes to the active site. Glu216 O∊2 interacts via a water molecule (bond distance of 2.72 Å) on one side, whereas Glu216 O∊1 interacts with His387 (bond distance of 2.64 Å) from the neighbouring molecule. Arg271 forms hydrogen bonds to both Gln506 (bond distance of 2.77 Å) and Glu356 (bond distance of 2.81 Å).
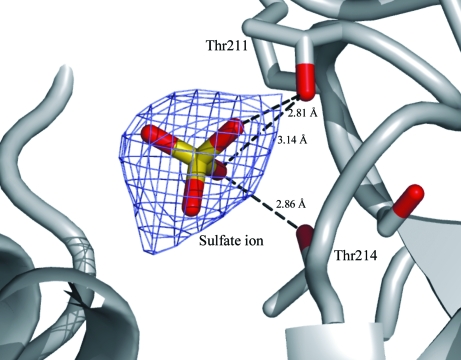
In this native enzyme structure, a sulfate ion from the crystallization precipitant (ammonium sulfate) was bound in the active site, indicating the substrate-binding site of the enzyme. A superposition with the inhibitor-bound structure of human PGI shows the superposed sulfate ions in the cavity. The position of the sulfate ion indicated potential hydrogen-bonding interactions between the substrate and the active-site residues (Fig. 3 ▶); it made interactions with Thr211 (2.81 Å) and Thr214 (2.86 Å). Lys513 lies in the close vicinity of the active-site sulfate ion.
Figure 3.
Electron-density map of the bound sulfate ion in the active-site cavity. The potential hydrogen bonds to the surrounding threonine residues of the substrate-binding sites are shown as dashed lines. The numbers represent the bond distances (in Å).
The substrate-binding site of PGI is highly conserved in various species (Achari et al., 1981 ▶; Mo et al., 1975 ▶; Muramatsu & Noso, 1971 ▶). The substrate-binding site of Mtb PGI has a deep binding pocket located between the large domain, the small domain and the C-terminus. The two conserved regions of PGI that form the substrate-binding site, GGRxxxxG and DQxGVExxK, correspond to amino-acid residues 269–279 of the small domain and residues 505–513 of the large domain of Mtb PGI, respectively (Bairoch et al., 1996 ▶; Sun et al., 1999 ▶). These two conserved regions were located within the cleft area in our structure, indicating the same overall evolutionary origin and catalytic mechanism as for all other PGIs.
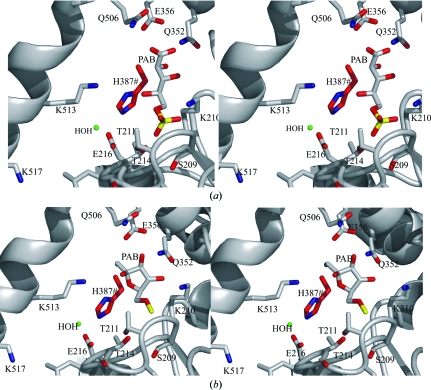
We performed superposition of the substrate d-fructose 6-phosphate (F6P) and the inhibitor 5-phosphoarabinonate (PAB) (Figs. 4 ▶ a and 4 ▶ b) using mammalian PGI (PDB codes 1hox and 1nuh, respectively; Lee et al., 2001 ▶; Davies et al., 2003 ▶) structures as templates. PGI catalysis proceeds using both G6P (d-glucose 6-phosphate) and F6P as substrate (Achari et al., 1981 ▶). PAB is a competitive inhibitor of PGI that is believed to mimic the cis-enediolate intermediate of the catalytic reaction (Chirgwin & Noltmann, 1975 ▶). The complexes of Mtb PGI with F6P and with PAB superposed well onto the structures of rabbit PGI complexed with F6P (r.m.s. deviation of 0.6 Å for 488 Cα pairs; PDB code 1hox; Lee et al., 2001 ▶) and human PGI complexed with PAB (r.m.s. deviation of 0.9 Å for 500 Cα pairs; PDB code 1nuh; Davies et al., 2003 ▶), especially in the active-site region. The phosphate groups of the both F6P and the PAB inhibitor are oriented by the conserved residues Ser159, Thr211, Thr214, the main-chain amide of Lys210 and Thr217 (via a water molecule). The atomic positions of the substrate analogue PAB and the substrate d-fructose 6-phosphate are within hydrogen-bonding distance limits. Most of the interactions of the PAB molecule in the substrate-binding site were also conserved, as described previously (Arsenieva et al., 2002 ▶; Arsenieva & Jeffery, 2002 ▶; Davies & Muirhead, 2003 ▶; Jeffery et al., 2000 ▶).
Figure 4.
Putative models of ligand complexes. (a) Stereo diagram showing F6P substrate in the substrate-binding site of Mtb PGI. (b) The inhibitor PAB binds well in the substrate-binding cavity. The residues in the close vicinity that are involved in several interactions for both ligands (F6P and PAB) are shown. The water molecule interacting with Glu216 is shown in green. The His residue (His387) involved in catalysis is from the neighbouring molecule and is shown in red (see §3.3 for details).
3.4. Comparison with other PGIs
Sequence alignment of Mtb PGI with PGIs from different species revealed low sequence identity to bacterial/archeal PGIs (4–10%), but high sequence identity was observed to mammalian PGIs (48–49%). The overall topology of PGI from M. tuberculosis is more similar to those of mammalian PGIs (r.m.s. deviations in the range 0.6–1.0 Å from the Cα atoms of published structures of mammalian PGIs) than those of bacterial PGIs. The main-chain atoms of Mtb PGI can be superimposed onto the native human PGI structure with an r.m.s. deviation of 0.8 Å for 497 Cα pairs (Fig. 2 ▶; PDB code 1iat; Read et al., 2001 ▶), indicating that the two enzymes fold in a highly similar manner. The r.m.s. deviation of 1.6 Å for 381 Cα pairs on superposition with the bacterial homologue Bacillus stearothermophilus PGI (PDB code 2pgi; Sun et al., 1999 ▶) was much larger. The overall sequence and structural features of the bacterial and mammalian PGIs in the N-terminal region are quite different (Sun et al., 1999 ▶). However, Mtb PGI and human PGI fold in a similar manner and exhibit similar catalytic properties (Mathur et al., 2005 ▶), suggesting that the evolutionary origin of Mtb PGI may be a mammalian PGI.
Although Mtb PGI is more similar to mammalian PGIs, a few regions of the large domain, especially in the hook and the C-terminal arm regions, differ from mammalian PGIs (Fig. 2 ▶). The major differences are that the loop before helix α19 (Arg416–Gly423) was larger than in mammalian, archeal and other PGIs, the ‘hook’ region (Ala449–Val457) was found to be smaller than in mammalian PGIs and in other PGIs, including B. stearothermophilus PGI, and the turn between β-sheets, α1 and α2 was four residues smaller than that present in the mammalian PGI 1iat. There were also some sequence-specific differences such as the amino-acid residue at position 40, which is always a large aromatic residue (Phe) in other PGI structures but was evolutionarily adapted to a glycine residue in Mtb PGI. The small residue provides enough space for the loop between helices α23 and α24 of the neighbouring molecule to form a stable dimer. The Ala residue at position 74 differed from other PGI structures known to date, in which an arginine was present instead. In Mtb PGI the alanine residue contributes to the hydrophobic core region in the small domain, unlike the other PGI structures, e.g. human PGI, in which the long side chain of Arg was flipped out of the hydrophobic cavity in the solvent channel (Read et al., 2001 ▶). The amino acid at position 275 that lies between the two 310-helices in Mtb PGI was again evolutionarily changed from a hydrophobic Trp residue to a rather polar Asp residue. It was positioned close to helices α7, α15 and α22.
The C-terminal region was quite flexible and has much larger deviations when compared with other PGI structures from various species (data not shown). The arm and the loop regions of Mtb PGI showed significant differences from other bacterial PGIs (e.g. PGI from B. stearothermophilus; PDB code 2pgi; Sun et al., 1999 ▶). The N-terminal region from residues 7 to 51, which includes a helix, deviated (r.m.s. deviation of ∼2.2 Å) most from B. stearothermophilus PGI. The loop regions of Mtb PGI (Gly88–Ala98, Pro107–Asp122) were larger than those of 2pgi and deviated significantly from the PGIs of other species. The loop region in the C-terminus (residues Gly549–Glu554) points in a totally different direction compared with other PGIs. The overall comparison after superposition of Mtb PGI onto PGIs from different sources showed a highly conserved active site for all PGIs and significant differences in the large domain, particularly when compared with the bacterial homologue.
3.5. Site-directed mutagenesis of Mtb PGI
On the basis of the sequence homology of phosphoglucose isomerase across various species, two amino-acid residues in the active site (Gly157 and Thr211) that have remained conserved in phylogenetically diverse species were identified for site-directed mutagenesis in order to evaluate their role in the catalytic activity of PGI. Expression of these mutant proteins was seen in the soluble fractions after induction and was decreased compared with that of wild-type phosphoglucose isomerase. The expression of all mutants was confirmed by Western blotting (data not shown).
The Gly157Tyr mutation resulted in a complete loss of activity of Mtb PGI. According to homology-modelling studies, Gly157 is part of the highly conserved GIGGS motif inside the active-site cavity. Glycine residues do not usually form catalytic residues. The small size of this residue was believed to create space for substrate binding and therefore this residue was believed to be involved in maintaining the conformation of the catalytic pocket. Replacement of Gly157 by the bulky group of tyrosine probably results in steric hindrance and may prevent the binding and/or docking of substrate. Alternatively, the removal of the glycine residue may reduce the conformational flexibility of the active-site loop.
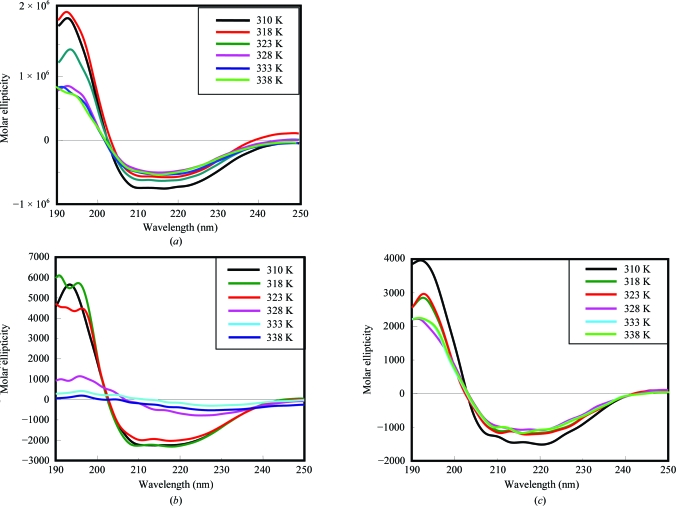
Thr211 is part of the active-site cavity. The mutant Thr211Ala showed decreased activity on the descending part of the bell-shaped curve (the region of the curve where the activity of the enzyme starts to decrease after reaching a peak as a function of pH). This could be because the mutation of the hydrophilic side chain of Thr to the smaller and hydrophobic Ala side chain results in weaker interaction with the substrate. Enzyme-kinetic studies of the Thr211Ala mutant indeed revealed a higher K m (0.42 mM for the substrate fructose-6-phosphate) compared with that of the wild-type protein (0.27 mM). CD spectra of the mutant taken at increasing temperatures showed dramatic differences betwen the spectra of wild-type and mutant Thr211Ala, with the loss of secondary structure being much more pronounced in the mutant (Fig. 5 ▶).
Figure 5.
CD spectra of (a) wild-type PGI, (b) T211A mutant PGI and (c) G157Y mutant PGI in the far-UV region at different temperatures. See §3.5 for further details.
The three-dimensional native structure of phosphoglucose isomerase from M. tuberculosis H37Rv has been determined. Our X-ray experiment has provided information about the role of the residues that differ in size, location and conformation. Based on the superposition of F6P and the competitive inhibitor (PAB) in the active site, a putative substrate-binding site for Mtb PGI has been described. The structure also provided insight into the similarities and differences that exist between the overall structures of the native human and Mtb PGIs. The significant differences in the loops (residues 104–117 and 414–422), the ‘hook’ (residues 441–464) and the ‘arm’ (residues 523–554) between the human and Mtb PGIs identified in the present study can be exploited for the design of effective and suitable inhibitors against the pathogen. The complete high-resolution structure of PGI presented here offers a starting point for further crystallographic, biochemical, genetic and neurobiological studies.
Supplementary Material
PDB reference: phosphoglucose isomerase, 2wu8
Acknowledgments
Data collection was performed on beamline ID29 at the European Synchrotron Radiation Facility, Grenoble, France; we thank the beamline staff for assistance. The authors thank Dr Manfred Weiss for discussion and comments on the manuscript. KA is funded by a FEBS longterm fellowship and a Marie Curie reintegration grant (ERG). The Council of Scientific and Industrial Research, New Delhi, India is acknowledged for a research fellowship to AA. Financial support from the Department of Biotechnology, India is acknowledged.
References
- Achari, A., Marshall, S. E., Muirhead, H., Palmieri, R. H. & Noltmann, E. A. (1981). Philos. Trans. R. Soc. Lond. B Biol. Sci.293, 145–157. [DOI] [PubMed]
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Arsenieva, D., Hardre, R., Salmon, L. & Jeffery, C. J. (2002). Proc. Natl Acad. Sci. USA, 99, 5872–5877. [DOI] [PMC free article] [PubMed]
- Arsenieva, D. & Jeffery, C. J. (2002). J. Mol. Biol.323, 77–84. [DOI] [PubMed]
- Bairoch, A., Bucher, P. & Hofmann, K. (1996). Nucleic Acids Res.24, 189–196. [DOI] [PMC free article] [PubMed]
- Berrisford, J. M., Akerboom, J., Brouns, S., Sedelnikova, S. E., Turnbull, A. P., van der Oost, J., Salmon, L., Hardre, R., Murray, I. A., Blackburn, G. M., Rice, D. W. & Baker, P. J. (2004). J. Mol. Biol.343, 649–657. [DOI] [PubMed]
- Berrisford, J. M., Hounslow, A. M., Akerboom, J., Hagen, W. R., Brouns, S. J., van der Oost, J., Murray, I. A., Blackburn, G. M., Waltho, J. P., Rice, D. W. & Baker, P. J. (2006). J. Mol. Biol.358, 1353–1366. [DOI] [PubMed]
- Blackburn, M. N. & Noltmann, E. A. (1972). J. Biol. Chem.247, 5668–5674. [PubMed]
- Cao, M. J., Osatomi, K., Matsuda, R., Ohkubo, M., Hara, K. & Ishihara, T. (2000). Biochem. Biophys. Res. Commun.272, 485–489. [DOI] [PubMed]
- Chaput, M., Claes, V., Portetelle, D., Cludts, I., Cravador, A., Burny, A., Gras, H. & Tartar, A. (1988). Nature (London), 332, 454–455. [DOI] [PubMed]
- Chirgwin, J. M. & Noltmann, E. A. (1975). J. Biol. Chem.250, 7272–7276. [PubMed]
- Chou, C.-C., Sun, Y.-J., Meng, M. & Hsiao, C.-D. (2000). J. Biol. Chem.275, 23154–23160. [DOI] [PubMed]
- Cordeiro, A. T., Godoi, P. H., Silva, C. H., Garratt, R. C., Oliva, G. & Thiemann, O. H. (2003). Biochim. Biophys. Acta, 1645, 117–122. [DOI] [PubMed]
- Cordeiro, A. T., Hardré, R., Michels, P. A. M., Salmon, L., Delboni, L. F. & Thiemann, O. H. (2004). Acta Cryst. D60, 915–919. [DOI] [PubMed]
- Davies, C. & Muirhead, H. (2002). Proteins, 49, 577–579. [DOI] [PubMed]
- Davies, C. & Muirhead, H. (2003). Acta Cryst. D59, 453–465. [DOI] [PubMed]
- Davies, C., Muirhead, H. & Chirgwin, J. (2003). Acta Cryst. D59, 1111–1113. [DOI] [PubMed]
- Dyson, J. E. & Noltmann, E. A. (1969). Biochemistry, 8, 3533–3543. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Faik, P., Walker, J. I., Redmill, A. A. & Morgan, M. J. (1988). Nature (London), 332, 455–457. [DOI] [PubMed]
- Gloster, T. M., Roberts, S., Perugino, G., Rossi, M., Moracci, M., Panday, N., Terinek, M., Vasella, A. & Davies, G. J. (2006). Biochemistry, 45, 11879–11884. [DOI] [PubMed]
- Janowski, R., Panjikar, S., Eddine, A. N., Kaufmann, S. H. & Weiss, M. S. (2009). J. Struct. Funct. Genomics, 10, 137–150. [DOI] [PMC free article] [PubMed]
- Jeffery, C. J., Bahnson, B. J., Chien, W., Ringe, D. & Petsko, G. A. (2000). Biochemistry, 39, 955–964. [DOI] [PubMed]
- Jeffery, C. J., Hardre, R. & Salmon, L. (2001). Biochemistry, 40, 1560–1566. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Lee, J. H., Chang, K. Z., Patel, V. & Jeffery, C. J. (2001). Biochemistry, 40, 7799–7805. [DOI] [PubMed]
- Lee, J. H. & Jeffery, C. J. (2005). Protein Sci.14, 727–734. [DOI] [PMC free article] [PubMed]
- Lin, H.-Y., Kao, Y.-H., Chen, S.-T. & Meng, M. (2009). Biochim. Biophys. Acta, 1794, 315–323. [DOI] [PubMed]
- Mathur, D., Ahsan, Z., Tiwari, M. & Garg, L. C. (2005). Biochem. Biophys. Res. Commun.337, 626–632. [DOI] [PubMed]
- Mathur, D., Anand, K., Mathur, D., Jagadish, N., Suri, A. & Garg, L. C. (2007). Acta Cryst. F63, 353–355. [DOI] [PMC free article] [PubMed]
- Meng, M., Chane, T.-L., Sun, Y.-J. & Hsiao, C.-D. (1999). Protein Sci.8, 2438–2443. [DOI] [PMC free article] [PubMed]
- Mo, Y., Young, C. D. & Gracy, R. W. (1975). J. Biol. Chem.250, 6747–6755. [PubMed]
- Muramatsu, N. & Noso, Y. (1971). Arch. Biochem. Biophys.144, 245–252. [DOI] [PubMed]
- Murray, C. J. & Salomon, J. A. (1998). Proc. Natl Acad. Sci. USA, 95, 13881–13886. [DOI] [PMC free article] [PubMed]
- Read, J., Pearce, J., Li, X., Muirhead, H., Chirgwin, J. & Davies, C. (2001). J. Mol. Biol.309, 447–463. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Sampathkumar, P., Roach, C., Michels, P. A. & Hol, W. G. (2008). J. Mol. Biol.381, 867–880. [DOI] [PubMed]
- Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2003). Mol. Microbiol.48, 77–84. [DOI] [PubMed]
- Shaw, P. J. & Muirhead, H. (1977). J. Mol. Biol.109, 475–485. [DOI] [PubMed]
- Solomons, J. T. G., Zimmerly, E. M., Burns, S., Krishnamurthy, N., Swan, M. K., Krings, S., Muirhead, H., Chirgwin, J. & Davies, C. (2004). J. Mol. Biol.342, 847–860. [DOI] [PubMed]
- Sun, Y.-J., Chou, C.-C., Chen, W.-S., Wu, R.-T., Meng, M. & Hsiao, C.-D. (1999). Proc. Natl Acad. Sci. USA, 96, 5412–5417. [DOI] [PMC free article] [PubMed]
- Swan, M. K., Hansen, T., Schonheit, P. & Davies, C. (2003). Protein Pept. Lett.10, 517–520. [DOI] [PubMed]
- Swan, M. K., Hansen, T., Schonheit, P. & Davies, C. (2004). J. Biol. Chem.279, 39838–39845. [DOI] [PubMed]
- Tanaka, N., Haga, A., Naba, N., Shiraiwa, K., Kusakabe, Y., Hashimoto, K., Funasaka, T., Nagase, H., Raz, A. & Nakamura, K. T. (2006). J. Mol. Biol.356, 312–324. [DOI] [PubMed]
- Watanabe, H., Carmi, P., Hogan, V., Raz, T., Silletti, S., Nabi, I. R. & Raz, A. (1991). J. Biol. Chem.266, 13442–13448. [PubMed]
- Yamamoto, H., Miwa, H. & Kunishima, N. (2008). J. Mol. Biol.382, 747–762. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: phosphoglucose isomerase, 2wu8
PDB reference: phosphoglucose isomerase, 2wu8