Crystals of C-terminally His-tagged ThyX from H. pylori belonged to space group C2, with unit-cell parameters a = 221.92, b = 49.43, c = 143.02 Å, β = 98.84°, and diffracted to beyond 2.5 Å resolution.
Keywords: flavin-dependent thymidylate synthases, Helicobacter pylori, ThyX
Abstract
The ThyX enzymes that have recently been identified in various bacteria, including some important human pathogens such as Helicobacter pylori and Mycobacterium tuberculosis, are flavin-dependent thymidylate synthases that function in the place of classic thymidylate synthase enzymes in the biosynthesis of dTMP, which is one of the building blocks of DNA. They are promising targets for the development of novel antibiotics because they utilize catalytic mechanisms that are distinct from those of the classic thymidylate synthases found in most organisms, including humans. In this study, H. pylori ThyX was purified and crystallized in complex with flavin adenine dinucleotide (FAD) and a diffraction data set was collected to 2.5 Å resolution. The crystals belonged to space group C2, with unit-cell parameters a = 221.92, b = 49.43, c = 143.02 Å, β = 98.84°.
1. Introduction
In most prokaryotes and nearly all eukaryotes, including humans, dTMP, which is one of the building blocks of DNA, is synthesized by the classic thymidylate synthases (TSs) by reductive methylation of dUMP. Classic TSs are homodimers that have one active site per dimer. They utilize N 5,N 10-methylenetetrahydrofolate (CH2H4folate or CH2THF) as both a methylene-group donor and as a source of reductive power and produce dihydrofolate (H2folate) (Carreras & Santi, 1995 ▶). The product H2folate is recycled by reduction to 5,6,7,8-tetrahydrofolate (H4folate) by dihydrofolate reductase and subsequent methylenation to CH2H4folate. Organisms that are dependent on classic TSs for dTMP biosynthesis universally possess dihydrofolate reductase.
In recent years, it has been found that some microorganisms, including some important human pathogens such as Helicobacter pylori and Mycobacterium tuberculosis (Mathews et al., 2003 ▶), lack both the classic TS and dihydrofolate reductase. They instead depend on an enzyme encoded by the thyX gene for the production of dTMP (Myllykallio et al., 2002 ▶). The ThyX enzymes do not have any sequence homology to classic TSs and have distinct structures and enzymatic mechanisms. They exist and function as homotetramers with four active sites per tetramer. They use FAD as a coenzyme and receive hydride moieties from reduced nicotinamides or other reductants; they use CH2H4folate as a methylene donor and produce H4folate instead of H2folate.
Because of the facts that the ThyX-dependent dTMP-biosynthesis pathway does not exist in humans and other mammals and that ThyX-dependent microorganisms do not possess the classic TS-dependent dTMP-biosynthesis pathway (Leduc et al., 2004 ▶; Agrawal et al., 2004 ▶; Graziani et al., 2006 ▶; Koehn et al., 2009 ▶), ThyX enzymes have emerged as a promising target for the development of novel antibiotics.
In order to better understand the structure–function relationship of ThyX enzymes and as part of longstanding efforts to identify potential drug targets in H. pylori, we have embarked on crystallographic analysis of H. pylori ThyX (HpThyX). We have purified HpThyX and crystallized the protein in a complexed form with FAD. Here, we report the crystallization, diffraction data collection and preliminary crystallographic studies of the HpThyX complex.
2. Materials and methods
2.1. Cloning, expression and purification
The HpThyX gene was amplified from the genome of H. pylori 26695 (conserved in our laboratory) and cloned into an expression vector derived from the pET-30a(+) plasmid (Novagen) and placed between NdeI and XhoI restriction sites. A six-histidine tag (LEHHHHHH) was engineered into the C-terminus of the protein. The insert was sequenced and found to be in complete agreement with the expected sequence.
Escherichia coli BL21 (DE3) pLysS cells (Invitrogen) transformed with the pET30a-ThyX plasmid were grown at 310 K in 2 l Luria–Bertani medium supplemented with kanamycin (50 µg ml−1) until the optical density at 600 nm (OD600) reached 0.6; isopropyl β-d-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 1 mM to induce expression of the recombinant protein. Cell growth continued at 310 K for 4 h. The cells were harvested by centrifugation and the bacterial pellets were resuspended in lysis buffer (20 mM phosphate buffer pH 8.0, 300 mM NaCl and 10 mM imidazole) and disrupted by ultrasonication. Insoluble cellular material was removed by centrifugation. The supernatant was purified on a nickel–nitrilotriacetic acid agarose (Ni–NTA; Novagen) column previously equilibrated with lysis buffer. The column was washed with wash buffer (20 mM phosphate buffer pH 8.0, 300 mM NaCl and 20 mM imidazole). His-tagged HpThyX was then eluted using 250 mM imidazole. The eluted protein was concentrated to about 1 ml by ultrafiltration (Millipore) and loaded onto a Superdex 200 HiLoad 16/60 column (GE Healthcare) previously equilibrated with 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM DTT, 5% glycerol. The peak corresponding to the HpThyX tetramer was collected; the protein was concentrated to about 10 mg ml−1 and flash-frozen at 203 K. Although FAD was not added during the cell-culture and purification steps, the protein preparation displayed a characteristic yellow colour on the chromatographic column during and after elution from the column, which was obviously from bound FAD moieties; this was verified by visible absorption spectroscopy, which showed a signification characteristic absorption peak at 454 nm during purification.
2.2. Crystallization
The concentration of the HpThyX protein in the buffer from the last column as used in crystallization trials was approximately 10 mg ml−1 as determined from the absorbance at 280 nm. The initial crystallization condition was determined by the hanging-drop vapour-diffusion method using Hampton Research Crystal Screen kits. Each drop was formed by mixing equal volumes (1 µl) of protein solution and reservoir solution and was allowed to equilibrate against 500 µl reservoir solution at 293 K. Conditions giving positive hits were optimized by varying the type and concentration of precipitant, salts, buffers and organic compounds, by varying the pH and by the use of additives (Additive Screen HT; Hampton Research HR2-138).
2.3. Data collection and processing
Diffraction data from HpThyX crystals were collected at a wavelength of 1.0 Å on beamline BL17A at Photon Factory, KEK, Japan using an ADSC Quantum 270 CCD detector. For cryoprotection, crystals were soaked in reservoir solution containing glycerol at a concentration of 10–15%(v/v) for 1 min before being mounted on nylon cryoloops and flash-frozen in a liquid-nitrogen stream at 95 K. A total of 180 frames of data were collected with an oscillation angle of 0.6° and an exposure time of 4 s for each image. MOSFLM (v.7.0.4; Leslie, 2006 ▶) and SCALA (v.6.0) from the CCP4 program suite (v.6.0.2; Collaborative Computational Project, Number 4, 1994 ▶) were used for processing, reduction and scaling of the diffraction data.
3. Results
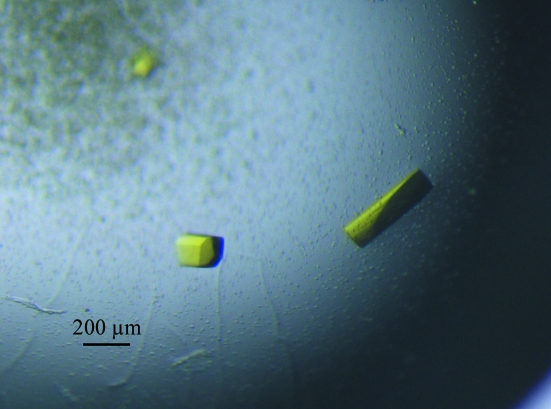
Crystals were initially obtained using Hampton Research Index screen conditions No. 5 (0.1 M HEPES pH 7.5, 2.0 M ammonium sulfate) and No. 19 (1.4 M sodium/potassium phosphate pH 8.2). The optimized crystallization condition consisted of 2.4 M ammonium sulfate, 0.15–0.3 M sodium potassium tartrate, 5% glycerol and 100 mM HEPES pH 6.8–7.0 and used hanging-drop vapour diffusion by mixing equal volumes of protein solution and reservoir solution at 277 K. Crystals appeared within 1–3 d and grew to dimensions of about 0.1 × 0.2 × 0.4 mm in a few days (Fig. 1 ▶).
Figure 1.
HpThyX crystals grown by the hanging-drop method in 2.4 M ammonium sulfate, 0.15 M sodium potassium tartrate, 5% glycerol and 100 mM HEPES pH 7.0. Typical crystal dimensions are 0.1 × 0.1 × 0.3 mm.
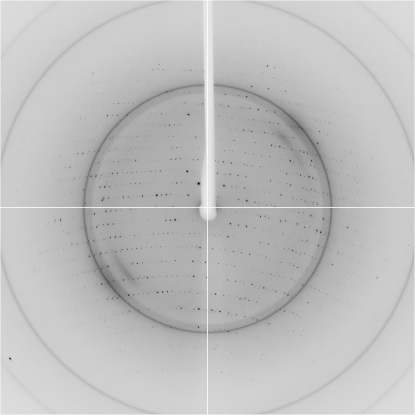
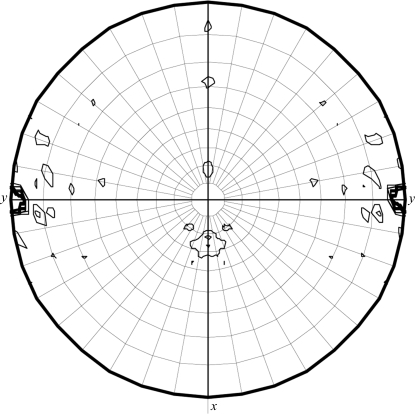
The data-collection statistics for the native crystal are given in Table 1 ▶. The crystal diffracted to a resolution of 2.5 Å and belonged to space group C2, with unit-cell parameters a = 222.92, b = 49.43, c = 143.02 Å, β = 98.84° (Fig. 2 ▶). Calculations showed that there were probably four, five or six HpThyX monomers in the asymmetric unit, with corresponding Matthews coefficients of 3.88, 3.10 or 2.58 Å3 Da−1 and solvent contents of 68.28, 60.35 or 52.42%, respectively. Self-rotation functions calculated by MOLREP in the CCP4 program suite (v.6.0.2; Collaborative Computational Project, Number 4, 1994 ▶) at various resolutions all had strong threefold or sixfold rotation axes along b (Fig. 3 ▶), indicating that there are probably six monomers in the asymmetric unit. This is intriguing as all characterized ThyX proteins exist and function as tetramers. Research is in progress to determine the three-dimensional structure and the oligomerization state of HpThyX.
Table 1. Data-collection statistics for a native HpThyX crystal.
Values in parentheses are for the highest resolution shell.
| Space group | C2 |
| Unit-cell parameters | |
| a (Å) | 221.92 |
| b (Å) | 49.43 |
| c (Å) | 143.02 |
| α (°) | 90 |
| β (°) | 98.84 |
| γ (°) | 90 |
| Wavelength (Å) | 1.00000 |
| Resolution (Å) | 55.63–2.50 (2.64–2.50) |
| No. of reflections | 53927 |
| Rmerge† (%) | 10.7(39.3) |
| Multiplicity | 7.5 |
| Mean I/σ(I) | 16.6 (4.3) |
| Completeness (%) | 96.1 (89.0) |
R
merge = 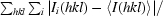
 , where 〈I(hkl)〉 is the mean of the observations I
i(hkl) of reflection hkl.
, where 〈I(hkl)〉 is the mean of the observations I
i(hkl) of reflection hkl.
Figure 2.
A typical diffraction pattern of an HpThyX crystal.
Figure 3.
Stereographic plot of the χ = 60° section of the self-rotation function. The radius of integration was 55 Å and the upper resolution limit was 5 Å. The contour lines were drawn at the 1σ level starting from 6σ.
Acknowledgments
The X-ray diffraction data were collected at Photon Factory, KEK, Japan. The authors thank Dr Y. Han of the Core Facilities, Institute of Biophysics for technical support. This work was supported by funding from the National Science Foundation of China (30771992) and the National 863 Program (2006AA02A208).
References
- Agrawal, N., Lesley, S. A., Kuhn, P. & Kohen, A. (2004). Biochemistry, 43, 10295–10301. [DOI] [PubMed]
- Carreras, C. W. & Santi, D. V. (1995). Annu. Rev. Biochem.64, 721–762. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Graziani, S., Bernauer, J., Skouloubris, S., Graille, M., Zhou, C.-Z., Marchand, C., Decottignies, P., van Tilbeurgh, H., Myllykallio, H. & Liebl, U. (2006). J. Biol. Chem.281, 24048–24057. [DOI] [PubMed]
- Koehn, E. M., Fleischmann, T., Conrad, J. A., Palfey, B. A., Lesley, S. A., Mathews, I. I. & Kohen, A. (2009). Nature (London), 458, 919–923. [DOI] [PMC free article] [PubMed]
- Leduc, D., Graziani, S., Meslet, C. L., Sodolescu, A. L. & Myllykallio, H. (2004). Biochem. Soc. Trans.32, 231–235. [DOI] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Mathews, I. I., Deacon, A. M., Canaves, J. M., McMullan, D., Lesley, S. A., Agarwalla, S. & Kuhn, P. (2003). Structure, 11, 677–690. [DOI] [PubMed]
- Myllykallio, H., Lipowski, G., Leduc, D., Filee, J., Forterre, P. & Liebl, U. (2002). Science, 297, 105–107. [DOI] [PubMed]