The endonuclease/exonuclease/phosphatase-homology domain of phosphodiesterase PDE12 has been crystallized. The crystals diffracted to 2.5 Å resolution.
Keywords: phosphodiesterases, PDE12, endonuclease/exonuclease/phosphatase-homology domain
Abstract
Phosphodiesterase PDE12 is a medically important esterase-family member that hydrolyzes 2′–5′-linked oligoadenylates (2-5A), which are involved in the regulation of biological processes related to the antiviral and antitumour activity that can be induced by interferons. Here, cloning, purification and crystallization of the C-terminal endonuclease/exonuclease/phosphatase-homology domain of human PDE12 is reported. The crystals belonged to space group P3121 or P3221, with unit-cell parameters a = b = 111.3, c = 192.4 Å, and diffracted to 2.5 Å resolution. Assuming the presence of three molecules in the asymmetric unit, the solvent content was estimated to be about 44.0%.
1. Introduction
Vertebrates are equipped with adaptive and innate immune systems that respond to attacks by viruses. One of the major pathways pertaining to the antiviral and antitumour activity induced by interferons (IFNs) involves 2′,5′-oligoadenylate synthetase (2′,5′-OAS), phosphodiesterase 12 (PDE12) and RNase L (Player & Torrence, 1998 ▶; Kerr & Brown, 1978 ▶). The system is triggered by the induction of 2′,5′-OAS by IFNs and subsequent activation by viral double-stranded RNA (dsRNA). The activated 2′,5′-OAS catalyzes the de novo synthesis of 5′-triphosphorylated 2′,5′-phosphodiester-linked oligoadenylates [(pp)p(A2′p5′)nA] (2-5A), which bind RNase L. 2-5A-bound RNase L is in turn activated and degrades viral and cellular single-stranded RNAs in infected cells (Rebouillat & Hovanessian, 1999 ▶; Schmidt et al., 1978 ▶; Zhou et al., 1993 ▶).
PDE12, an enzyme that belongs to the endonuclease/exonuclease/phosphatase family, hydrolyzes 2-5A to ATP and AMP and is therefore also referred to as 2′,5′-phosphodiesterase (2′,5′-PDE; Itkes et al., 1984 ▶; Alster et al., 1986 ▶; Torrence et al., 1987 ▶; Johnston & Hearl, 1987 ▶; Kubota et al., 2004 ▶). It plays a role as a negative regulator in the 2-5A system by reducing 2-5A levels in cells. PDE12 displays relatively poor (less than 30%) identity to other members of the endonuclease/exonuclease/phosphatase family: AP endonuclease, DNase I, sphingomyelinase, inositol-1,4,5-trisphosphate phosphatases such as synaptojanin and deadenylases such as nocturnin. An unrooted tree that was constructed from 18 related human genes shows three major families: the inositol phosphatase family, the DNase family and the PDE12 family, which contains members of unknown function (Kubota et al., 2004 ▶). Recently, it has been shown that the PDE12 inhibitor A-74528 isolated from Streptomyces sp. could suppress viral replication with an IC50 value of 34 µg ml−1 (Fujita et al., 2005 ▶). Therefore, this enzyme plays a key role in regulation of the 2-5A system and could be a potential novel target for antiviral and antitumour treatments. Here, we report the cloning, overexpression, crystallization and preliminary X-ray crystallographic analysis of human PDE12 as a first step towards structure determination.
2. Materials and methods
2.1. Cloning and expression of PDE12
PDE12 (NP_808881.3) was cloned from a human leukocyte cDNA library and amplified by the polymerase chain reaction. The PCR product was digested with the restriction endonucleases FspI and BsrGI and then cloned into SfoI/Acc65I-digested pFastBac vector (Invitrogen). The resulting construct expressed full-length protein (residues 1–609) and was co-transfected with a modified baculovirus genome into Spodoptera frugiperda (sf9) insect cells. The recombinant baculovirus was plaque-purified, amplified and overexpressed. Since some degradation of the full-length protein was observed by polyacrylamide gel electrophoresis, limited protease digestion was performed in order to generate a stable domain. As a result, limited elastase digestion yielded a stable 51 kDa domain. The 51 kDa domain was verified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI–TOF MS; JMS Elite, PerSeptiv Inc.) and N-terminal sequence analysis (M4952, Applied Biosystems). These analyses showed that the 51 kDa domain was produced by an N-terminal truncation (Δ1–154) and contained a C-terminal fragment encompassing the endonuclease/exonuclease/phosphatase-homology (EEPH) domain. Residues 155–609 that contained the 51 kDa domain were cloned with the N-terminal tag MSYYHHHHHHDYDIPTTENLYFQGA. Transformed cells were grown in sf900II medium (Invitrogen); after 72 h infection they were harvested and resuspended in lysis buffer containing 50 mM Tris–HCl pH 8.0, 50 mM NaCl and 5 mM β-mercaptoethanol (β-ME).
2.2. Purification of PDE12
Cells were disrupted by sonication and centrifuged at 45 000g for 60 min at 277 K. The resulting supernatant was loaded onto a chromatography column (Bio-Rad) packed with an equal volume of Q-Sepharose (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 5 mM β-ME and 20 mM imidazole. Following protein loading, the column was washed with the same buffer until the absorbance at 280 nm of the effluent returned to baseline. The protein was eluted with the same buffer containing 400 mM NaCl.
The eluted protein was applied onto a nickel–nitrilotriacetic acid (Ni–NTA) column, washed with buffer containing 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 5 mM β-ME and 20 mM imidazole and then eluted with the same buffer containing 200 mM imidazole. The eluted protein was applied onto a HiTrap Q-Sepharose column (GE Healthcare), washed with 50 mM Tris–HCl pH 8.0, 100 mM NaCl and 2 mM dithiothreitol (DTT) and then eluted using an NaCl concentration gradient (100–500 mM).
The His6-tagged protein was cleaved overnight at 277 K using AcTEV protease (Invitrogen). Finally, the cleaved protein was concentrated to 5 ml, loaded onto a Superdex 200 HR 16/60 column (GE Healthcare) pre-equilibrated with buffer containing 50 mM Tris–HCl pH 8.0, 100 mM NaCl and 2 mM DTT and eluted at a flow rate of 1.0 ml min−1. The purified PDE12 (155–609), referred to as PDE12 in the following, was concentrated to 40 mg ml−1 using Centricon-30 filters (Amicon) in preparation for crystallization. The protein concentration was determined by measuring the absorbance at 280 nm and using the molar absorption coefficient of PDE12. Aliquots of the purified protein were frozen at 193 K prior to crystallization experiments.
2.3. Crystallization and X-ray data collection
Initial crystallization screening was performed by the sitting-drop vapour-diffusion method using the commercially available crystallization screening kits Crystal Screen I, Crystal Screen II, Index (Hampton Research), PEGs and PEGs II (Qiagen K.K.) and a Hydra II Plus One crystallization robot (Matrix Technology) at 293 K. Each droplet, which was prepared by mixing 0.2 µl protein solution with an equal volume of reservoir solution, was equilibrated against 50 µl reservoir solution in a 96-well Intelli-Plate (Art Robbins Instruments).
Initial diffraction tests of the crystals obtained were performed using a Rigaku FR-E X-ray generator equipped with a Rigaku R-AXIS VII detector at 100 K. Diffraction data sets were collected on the BL41XU beamline at SPring-8 (Hyogo, Japan). For data collection, the crystals were cryoprotected by soaking them in mother liquor with 10%(v/v) glycerol and were then flash-cooled in liquid nitrogen. A complete data set was collected to 2.5 Å resolution and was integrated and scaled using DENZO and SCALEPACK as implemented in HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
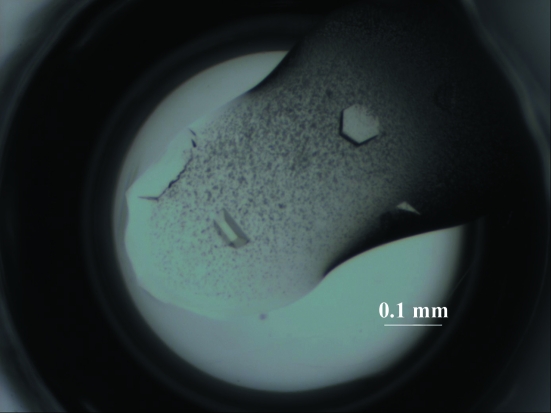
The insect cells produced PDE12 with an additional 25 residues, including six histidine-tag residues, as a soluble protein. The tag-cleaved protein obtained by treatment with TEV protease was also found to be soluble and was purified for crystallization. Two different crystal forms, needle-shaped and hexagonal plate-shaped, were obtained in the same drop under several conditions which contained 0.1 M Tris–HCl pH 8.5, 20%(w/v) polyethylene glycol 3350 (PEG 3350) and 0.2 M of various salts: potassium fluoride or chloride, ammonium chloride, sodium or potassium thiocyanate, potassium nitrate or potassium formate. Hexagonal plate-shaped crystals grew slowly to dimensions that were suitable for X-ray diffraction. The crystallization conditions were optimized using the hanging-drop vapour-diffusion method with droplets that were prepared by mixing 1.0 µl protein solution with 1.0 µl reservoir solution and equilibrated against 500 µl reservoir solution. Larger crystals were obtained using PEG solutions containing potassium chloride or sodium thiocyanate. Diffraction tests revealed that crystals from solutions containing potassium chloride diffracted poorly, whereas crystals from solutions containing sodium thiocyanate diffracted to higher resolution. The best crystals were grown from a solution containing 20 mg ml−1 (0.4 mM) protein, 75 mM Tris–HCl pH 8.0, 9%(w/v) PEG 3350, 1 mM DTT, 50 mM sodium chloride and 90 mM potassium chloride or 90 mM sodium thiocyanate equilibrated against 18%(w/v) PEG 3350 in 100 mM Tris–HCl pH 8.0 containing 180 mM potassium chloride or 180 mM sodium thiocyanate (Fig. 1 ▶). Systematic absences suggested that the PDE12 crystals belonged to space group P3121 or P3221, with unit-cell parameters a = b = 111.3, c = 192.4 Å. The calculated unit-cell volume per unit of molecular mass (V M) was 2.38 Å3 Da−1, with a solvent content of 44.0% by volume (Matthews, 1968 ▶), when the unit cell was assumed to contain three molecules. Data-collection statistics are summarized in Table 1 ▶.
Figure 1.
Hexagonal plate-shaped crystals of human PDE12.
Table 1. Crystal information and data-collection statistics.
Values in parentheses are for the outer resolution shell.
| Beamline | SPring-8, BL41XU |
| Detector | ADSC Quantum 315 |
| Wavelength (Å) | 1.00 |
| Beam size (µm) | 100 |
| Temperature (K) | 100 |
| Space group | P3121 or P3221 |
| Unit-cell parameters (Å) | a = b = 111.3, c = 192.4 |
| Resolution (Å) | 50.0–2.5 (2.58–2.5) |
| No. of reflections (total/unique) | 454742/48923 |
| Completeness (>0σ) (%) | 92.6 (44.3) |
| Mosaicity | 0.906 |
| 〈I/σ(I)〉 | 18.1 (0.73) |
| Rmerge† (%) | 9.1 (35.2) |
| Average redundancy | 9.3 (8.3) |
R
merge = 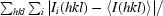
 , where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where I
i(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
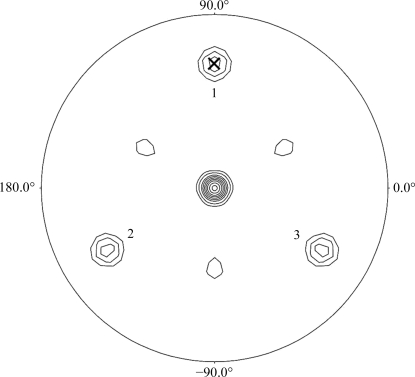
Calculation of the self-rotation function using the program POLARRFN (Collaborative Computational Project, Number 4, 1994 ▶) was performed with structure-factor amplitudes to a maximum resolution of 3.0 Å and revealed clear noncrystallographic symmetry (NCS) peaks on the κ = 120° section (Fig. 2 ▶), suggesting a threefold axis with ω = 60.1° (the inclination to the z axis within the yz plane), ϕ = 90.0° (within the xy plane), κ = 120°. Structure determination using multi-wavelength anomalous dispersion methods is currently in progress.
Figure 2.
κ = 120° section of the self-rotation function calculated using POLARRFN. The integration radius was 30 Å. The peak (ω = 60.1°, ϕ = 90°) is marked by a cross and its symmetry-related peaks are numbered. Contouring starts at the 1σ level with an interval of 1σ.
Acknowledgments
We would like to thank J. Tsukamoto for technical support in performing the MALDI–TOF MS analysis and N-terminal sequence analysis of the proteins and peptides. We are also grateful to the staff at SPring-8 for help with data collection on synchrotron beamline BL41XU. This work was supported by a Grant-in-Aid for Scientific Research (A) and for Scientific Research on Priority Areas to TH from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- Alster, D., Brozda, D., Kitade, Y., Wong, A., Charubala, R., Pfleiderer, W. & Torrence, P. F. (1986). Biochem. Biophys. Res. Commun.141, 555–561. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Fujita, Y., Kasuya, A., Matsushita, Y., Suga, M., Kizuka, M., Iijima, Y. & Ogita, T. (2005). Bioorg. Med. Chem. Lett.15, 4317–4321. [DOI] [PubMed]
- Itkes, A. V., Kartasheva, O. N., Kafiani, C. A. & Severin, E. S. (1984). FEBS Lett.176, 65–68. [DOI] [PubMed]
- Johnston, M. I. & Hearl, W. G. (1987). J. Biol. Chem.262, 8377–8382. [PubMed]
- Kerr, I. M. & Brown, R. E. (1978). Proc. Natl Acad. Sci. USA, 75, 256–260. [DOI] [PMC free article] [PubMed]
- Kubota, K., Nakahara, K., Ohtsuka, T., Yoshida, S., Kawaguchi, J., Fujita, Y., Ozeki, Y., Hara, A., Yoshimura, C., Furukawa, H., Haruyama, H., Ichikawa, K., Yamashita, M., Matsuoka, T. & Iijima, Y. (2004). J. Biol. Chem.279, 37832–37841. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Player, M. R. & Torrence, P. F. (1998). Pharmacol. Ther.78, 55–113. [DOI] [PMC free article] [PubMed]
- Rebouillat, D. & Hovanessian, A. G. (1999). J. Interferon Cytokine Res.19, 295–308. [DOI] [PubMed]
- Schmidt, A., Zilberstein, A., Shulman, L., Federman, P., Berissi, H. & Revel, M. (1978). FEBS Lett.95, 257–264. [DOI] [PubMed]
- Torrence, P. F., Alster, D., Huss, S., Gosselin, G. & Imbach, J.-L. (1987). FEBS Lett.212, 267–270. [DOI] [PubMed]
- Zhou, A., Hassel, B. A. & Silverman, R. H. (1993). Cell, 72, 753–765. [DOI] [PubMed]