Crystals of infectious West Nile virus were obtained and diffracted at best to about 25 Å resolution. Preliminary analysis of the diffraction pattern suggested tight hexagonal packing of the intact virus.
Keywords: West Nile virus, flaviviruses
Abstract
West Nile virus, a human pathogen, is closely related to other medically important flaviviruses of global impact such as dengue virus. The infectious virus was purified from cell culture using polyethylene glycol (PEG) precipitation and density-gradient centrifugation. Thin amorphously shaped crystals of the lipid-enveloped virus were grown in quartz capillaries equilibrated by vapor diffusion. Crystal diffraction extended at best to a resolution of about 25 Å using synchrotron radiation. A preliminary analysis of the diffraction images indicated that the crystals had unit-cell parameters a ≃ b ≃ 480 Å, γ = 120°, suggesting a tight hexagonal packing of one virus particle per unit cell.
1. Introduction
West Nile virus (WNV), a member of the Flaviviridae family, is closely related to other arthropod-borne and medically relevant viruses such as dengue, tick-borne encephalitis, Japanese encephalitis and yellow fever viruses. The positive-strand RNA genome encodes three structural proteins: the capsid protein (C), the precursor membrane protein (prM, which is proteolytically cleaved into pr and M during virus maturation) and the envelope protein (E). Mature West Nile virions are roughly spherical, with a diameter of about 495 Å. Their nucleocapsid core, consisting of RNA (∼11 kb) and C protein (∼12 kDa), is encapsidated by a host-derived lipid bilayer and an outer protein shell composed of the membrane-anchored glycoproteins M (∼8 kDa) and E (∼54 kDa).
Three-dimensional near-atomic resolution structures are available for the C protein (Ma et al., 2004 ▶; Dokland et al., 2004 ▶), the pr peptide (∼11 kDa; Li et al., 2008 ▶) and the soluble ectodomain of the E glycoprotein (Rey et al., 1995 ▶; Modis et al., 2003 ▶; Zhang et al., 2004 ▶; Nybakken et al., 2006 ▶). The highly basic largely α-helical capsid protein interacts with the viral RNA genome to form the nucleocapsid core, but the structure of the assembled core is still unknown (Zhang et al., 2007 ▶). The ectodomain of E has three domains, DI, DII and DIII, with a flexible ‘hinge’ between DI and DII, and contains cellular receptor-binding and major antigenic sites. The pr peptide covers the fusion loop at the distal end of DII in immature flavivirions, preventing premature fusion with the host-cell membranes.
The pseudo-atomic resolution structure of a mature flavivirus has been obtained by fitting X-ray coordinates into a cryo-electron microscopy (cryo-EM) density map of dengue virus. The outer surface of the flavivirus particle is formed by an icosahedral scaffold of 90 E homodimers (Kuhn et al., 2002 ▶). Each E monomer is anchored into the lipid envelope by two transmembrane helices. The partially α-helical ‘stem’ region connecting the E ectodomain to its transmembrane anchor lies essentially flat against the viral membrane (Kuhn et al., 2002 ▶; Zhang et al., 2003 ▶). Buried under the E-protein layer are 180 icosahedrally arranged monomers of the membrane-anchored M protein (Zhang et al., 2003 ▶). However, the atomic structures of the M protein and of the E stem/transmembrane region are not yet known.
Flaviviruses are enveloped viruses that enter host cells by receptor-mediated endocytosis. After replication and assembly, immature virions undergo a protease-dependent and pH-dependent maturation in the trans-Golgi network. Major reshuffling of the viral proteins and their domains occurs within the particles during these processes (Kuhn et al., 2002 ▶; Modis et al., 2004 ▶; Stiasny & Heinz, 2006 ▶; Yu et al., 2008 ▶; Kaufmann et al., 2009 ▶). A near-atomic resolution structure of the complete virion will provide detailed information regarding the interactions between all viral components and might thereby help in understanding the structural rearrangement during entry and maturation. Here, we report the crystallization of WNV and the preliminary analysis of the crystals during our attempt to obtain a near-atomic resolution structure of a whole flavivirus.
2. Materials and methods
2.1. WNV propagation and purification
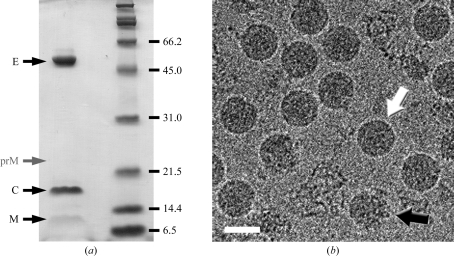
WNV was propagated under Biosafety Level 3 containment in Vero cells and was purified by density-gradient centrifugation as described previously (Kaufmann et al., 2009 ▶). For crystallization, the virus was concentrated to 2–15 mg ml−1 E protein in NTE buffer (12 mM Tris–HCl pH 8.0, 120 mM NaCl, 1 mM EDTA) using Amicon centrifugal filters (100 kDa molecular-weight cutoff; Millipore, Billerica, Massachusetts, USA). The protein concentration and purity and the quality of the final virus preparations were estimated by SDS gel electrophoresis with Coomassie Blue staining and cryo-EM (Fig. 1 ▶). As some degradation of the virus sample was observed within 24 h of purification, crystallization experiments had to be set up immediately after virus preparation.
Figure 1.
WNV sample preparation. (a) Coomassie-stained protein gel showing the protein components (E, C and M) of mature virions. The approximate running position of prM is indicated. Occasionally, some prM was present in mature virus preparations, probably as a result of imperfect maturation processing. (b) Cryo-EM image of WNV. A few broken or partially immature particles (an example is marked by a black arrow) were repeatedly encountered in preparations of smooth mature virus (white arrow), most likely owing to purification stress and incomplete intracellular maturation, respectively. The scale bar represents 500 Å.
2.2. Crystallization and X-ray data collection
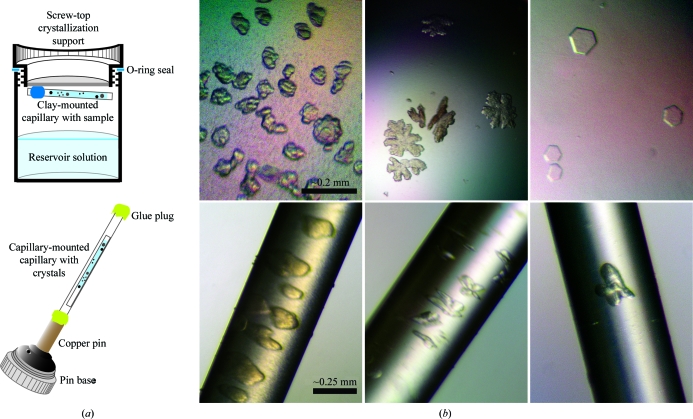
Screening for crystallization conditions was performed using a home-made grid screen (PEG concentration and polymer size versus NaCl and/or glycerol) and with commercially available kits such as PEG/Ion Screen (Hampton Research, Laguna Niguel, California, USA), Crystal Screen 2 (Hampton Research) and the Cryo I sparse-matrix screen (Emerald BioSystems, Bainbridge Island, Washington, USA). Crystal Screen 2, various salts (MgCl2, CaCl2, KCl) and alcohols (2-propanol, 1,6-hexanediol) were subsequently tested as additives in the grid screen. Because of biosafety considerations, crystallization experiments with infectious virus were carried out using screw-in O-ring-sealed EasyXtal crystallization supports (Qiagen, Valencia, California, USA). Initial screens were set up by mixing 1–3 µl virus solution (2–15 mg ml−1 E protein) in a 1:1 ratio with reservoir solution for hanging-drop vapor-diffusion equilibration at 283 and 293 K. Crystal optimization screens were also set up using open, thin-walled quartz capillaries (∼8 mm in length, ∼0.5–0.7 mm in diameter) that were mounted with clay on the crystallization support over 500 µl reservoir solution to equilibrate by vapor diffusion (Fig. 2 ▶ a). The virus sample was mixed in a 1:1 ratio with reservoir solution and drawn into the capillary tube using capillary action. Crystal growth in capillaries was chosen to keep physical damage during crystal handling to a minimum, as described for bacteriophage PRD1 (Cockburn et al., 2003 ▶). Some crystal droplets/capillaries were treated with glutaraldehyde, introducing the cross-linking agent by vapor diffusion (Lusty, 1999 ▶).
Figure 2.
WNV crystallization. (a) Schematic illustration of the crystallization setup in capillaries and the mounting of these capillaries for data collection. (b) Crystals of WNV grown in hanging drops (upper panels) or capillaries (lower panels).
For data collection, crystals from droplets were mounted in a capillary via CryoLoop (Hampton) or MicroMesh (MiTeGen LLC, Ithaca, New York, USA) mounts and sealed tightly using Devcon 5-Minute Epoxy adhesive (ITW Devcon, Danvers, Massachusetts, USA). In compliance with biosafety requirements, crystal-containing capillaries were mounted within a second capillary that was in turn mounted on a CrystalCap Copper pin (Hampton Research; Fig. 2 ▶ a). X-ray diffraction data were collected under Biosafety Level 3 containment, as specified by the Centers for Disease Control and Prevention and the Argonne National Laboratory Institutional Biosafety Committee, on the BioCars beamline 14BM-C (Advanced Photon Source, Argonne National Laboratory, Argonne, Illinois, USA). Single crystals were exposed to the X-ray beam (λ = 0.9002 Å) one by one moving along the capillary. Exposure times varied from 45 to 120 s per image and images were recorded on an ADSC Quantum-315 CCD detector. The crystal-to-detector distance was set to between 700 and 1000 mm. A small beamstop permitted the recording of low-resolution data to about 100 Å. As the buffer-suspended crystals frequently detached from the capillary wall during movement of the goniometer head, oscillation was limited to 0–0.1° per frame. The capillary was cooled to 283 K during data collection by placing it in close proximity to a cold nitrogen-gas stream.
2.3. Results and discussion
WNV crystals of diverse morphology were observed within 12 h of crystallization setup and grew to maximum dimensions of about 0.2 mm (Fig. 2 ▶ b). Most often crystals were obtained with indented edges or snowflake-like appearance, but occasionally perfect-looking hexagonal plates were observed. All crystals seemed very fragile; the physical appearance of the crystals deteriorated within a few days, with their edges growing visually less distinct. Gentle manipulation often resulted in immediate disintegration of the crystals, in particular for the hexagonal plates. The latter crystals could not be reproduced in capillaries. Generally, no or only a few diffraction spots were observed for the crystals mounted from hanging drops. The best-diffracting crystals were obtained in capillaries. In capillaries, crystals grew with their maximum dimensions tangential to the crystal wall, while the third dimension generally remained small (Fig. 2 ▶ b). The best crystals at 283 K were obtained with a virus-sample concentration of ∼4 mg ml−1 E, a precipitant concentration of 2.25–2.75%(w/v) PEG 20 000 or 5.5–6%(w/v) methoxy-PEG 5000, NTE buffer at pH 8 and 5%(v/v) glycerol as an additive. At 293 K, the optimal concentration of virus was ∼6–14 mg ml−1 E using 6.5–7%(w/v) methoxy-PEG 5000 as a precipitant. The addition of 75–150 mM NaCl or the treatment of the crystals with glutaraldehyde had no discernable effect on crystal morphology or the diffraction pattern.
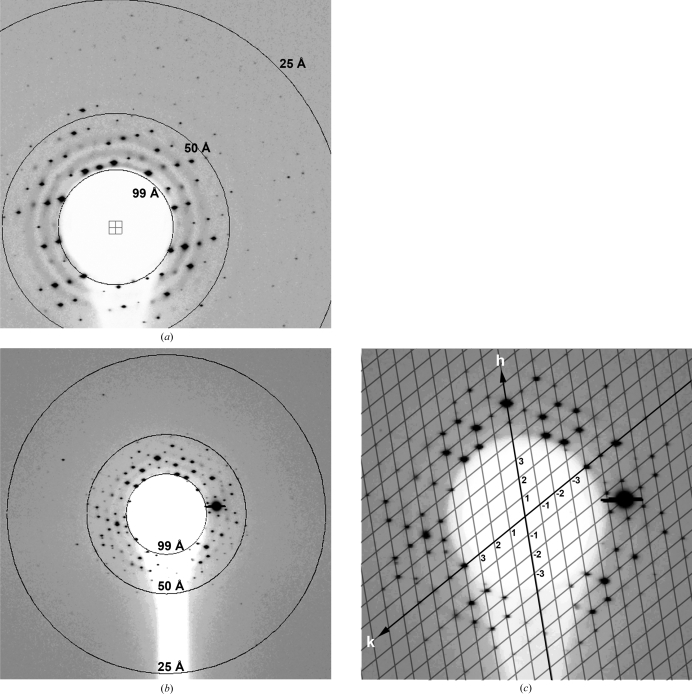
A total of 109 diffraction images with single lattices were collected from about 600 crystals. The crystals showed severe radiation damage after just 1–2 exposures. About a dozen images with a clean single-crystal lattice and sharp diffraction spots were selected for detailed analysis. The diffraction patterns extended at best to about 25 Å resolution (Figs. 3 ▶ a and 3 ▶ b) and contained a maximum of 400 reflections per image. The crystals displayed a hexagonal diffraction pattern when illuminated by the X-ray beam directed perpendicular to their preferred growth direction (Fig. 3 ▶ b). The central part of the diffraction images exhibited an approximately centrosymmetric pattern of reflections belonging to a single reciprocal-lattice plane that passes through the origin of reciprocal space (Fig. 3 ▶ c). The crystal unit-cell parameters in the direction tangential to the capillary side were determined from several images to be a ≃ b ≃ 480 Å, γ = 120°. These unit-cell parameters suggested tight hexagonal packing of mature WNV virions. Given these parameters, the crystals are likely to contain a single whole virus particle per unit cell. Identification of the remaining unit-cell parameters (c, α and β) was not possible because the crystal orientation towards the beam could not be precisely controlled owing to the double capillary-mounting setup and crystal mobility in the liquid. Even the best available diffraction images with the highest resolution data showed reflections from only a few reciprocal planes in the direction perpendicular to the beam, thus providing very little information about the positioning of the reflections in three-dimensional reciprocal space. This was most likely the reason for the unsuccessful autoindexing attempts using the software packages MOSFLM (Leslie, 1992 ▶) and HKL-2000 (Otwinowski & Minor, 1997 ▶). However, diffraction spots from several images were integrated using a custom-written program. Equivalent reflections from different images were merged and scaled, resulting in R factors of 25–30% and indicating some consistency between different crystals and the possibility of obtaining a full data set by combining information from individual images.
Figure 3.
X-ray diffraction analysis of capillary-grown WNV crystals. (a) Diffraction image extending to about 25 Å resolution, (b) typical hexagonal diffraction pattern and (c) interpretation of the centrosymmetric pattern in (b), in which the reflections belong to a single reciprocal-lattice plane.
WNV crystallization conditions and diffraction limits were highly reminiscent of those described previously for enveloped viruses in a study on Sindbis virus (Harrison et al., 1992 ▶) and in early crystallization reports on bacteriophage PRD1 (Bamford et al., 2002 ▶). One critical step in the successful crystallization and structural exploration of PRD1 was the development of a rapid chromatography-based purification method resulting in highly pure virus material. In this work, heterogeneity and instability of the WNV sample were observed, which probably contributed significantly to the formation of poorly ordered crystals with a limited resolution range of diffraction. Initial attempts to improve the WNV purification method were hindered by small virus yields. Nevertheless, the authors suggest that further crystallization trials should focus on improving virus-sample quality to obtain better quality crystals.
Acknowledgments
We are thankful to the staff of the Biosafety Level 3 facility at the BioCARS beamline, especially Vukica Šrajer. We are thankful to Susan L. Hafenstein for her support during data collection and to Sheryl L. Kelly for her help in the preparation of this manuscript. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science under Contract No. DE-AC02-06CH11357. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources under grant No. RR007707. This work was supported by NIH grants RO1-AI76331 to MGR and PO1-AI055672 to RJK and MGR.
References
- Bamford, J. K., Cockburn, J. J., Diprose, J., Grimes, J. M., Sutton, G., Stuart, D. I. & Bamford, D. H. (2002). J. Struct. Biol.139, 103–112. [DOI] [PubMed]
- Cockburn, J. J. B., Bamford, J. K. H., Grimes, J. M., Bamford, D. H. & Stuart, D. I. (2003). Acta Cryst. D59, 538–540. [DOI] [PubMed]
- Dokland, T., Walsh, M., Mackenzie, J. M., Khromykh, A. A., Ee, K. H. & Wang, S. (2004). Structure, 12, 1157–1163. [DOI] [PMC free article] [PubMed]
- Harrison, S. C., Strong, R. K., Schlesinger, S. & Schlesinger, M. J. (1992). J. Mol. Biol.226, 277–280. [DOI] [PubMed]
- Kaufmann, B., Chipman, P. R., Holdaway, H. A., Johnson, S., Fremont, D. H., Kuhn, R. J., Diamond, M. S. & Rossmann, M. G. (2009). PLoS Pathog.5, e1000672. [DOI] [PMC free article] [PubMed]
- Kuhn, R. J., Zhang, W., Rossmann, M. G., Pletnev, S. V., Corver, J., Lenches, E., Jones, C. T., Mukhopadhyay, S., Chipman, P. R., Strauss, E. G., Baker, T. S. & Strauss, J. H. (2002). Cell, 108, 717–725. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Li, L., Lok, S. M., Yu, I. M., Zhang, Y., Kuhn, R. J., Chen, J. & Rossmann, M. G. (2008). Science, 319, 1830–1834. [DOI] [PubMed]
- Lusty, C. J. (1999). J. Appl. Cryst.32, 106–112.
- Ma, L., Jones, C. T., Groesch, T. D., Kuhn, R. J. & Post, C. B. (2004). Proc. Natl Acad. Sci. USA, 101, 3414–3419. [DOI] [PMC free article] [PubMed]
- Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. (2003). Proc. Natl Acad. Sci. USA, 100, 6986–6991. [DOI] [PMC free article] [PubMed]
- Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. (2004). Nature (London), 427, 313–319. [DOI] [PubMed]
- Nybakken, G. E., Nelson, C. A., Chen, B. R., Diamond, M. S. & Fremont, D. H. (2006). J. Virol.80, 11467–11474. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rey, F. A., Heinz, F. X., Mandl, C., Kunz, C. & Harrison, S. C. (1995). Nature (London), 375, 291–298. [DOI] [PubMed]
- Stiasny, K. & Heinz, F. X. (2006). J. Gen. Virol.87, 2755–2766. [DOI] [PubMed]
- Yu, I. M., Zhang, W., Holdaway, H. A., Li, L., Kostyuchenko, V. A., Chipman, P. R., Kuhn, R. J., Rossmann, M. G. & Chen, J. (2008). Science, 319, 1834–1837. [DOI] [PubMed]
- Zhang, W., Chipman, P. R., Corver, J., Johnson, P. R., Zhang, Y., Mukhopadhyay, S., Baker, T. S., Strauss, J. H., Rossmann, M. G. & Kuhn, R. J. (2003). Nature Struct. Biol.10, 907–912. [DOI] [PMC free article] [PubMed]
- Zhang, Y., Kostyuchenko, V. A. & Rossmann, M. G. (2007). J. Struct. Biol.157, 356–364. [DOI] [PMC free article] [PubMed]
- Zhang, Y., Zhang, W., Ogata, S., Clements, D., Strauss, J. H., Baker, T. S., Kuhn, R. J. & Rossmann, M. G. (2004). Structure, 12, 1607–1618. [DOI] [PMC free article] [PubMed]