Abstract
Naturally occurring mutations in the cardiac ryanodine receptor (RyR2) have been linked to certain types of cardiac arrhythmias and sudden death. Two mutation hotspots that lie in the N-terminal and central regions of RyR2 are predicted to interact with one another and to form an important channel regulator switch. To monitor the conformational dynamics involving these regions, we generated a fluorescence resonance energy transfer (FRET) pair. A yellow fluorescent protein (YFP) was inserted into RyR2 after residue Ser437 in the N-terminal region, and a cyan fluorescent protein (CFP) was inserted after residue Ser2367 in the central region, to form a dual YFP- and CFP-labeled RyR2 (RyR2S437-YFP/S2367-CFP). We transfected HEK293 cells with RyR2S437-YFP/S2367-CFP cDNAs, and then examined them by using confocal microscopy and by measuring the FRET signal in live cells. The FRET signals are influenced by modulators of RyR2, by domain peptides that mimic the effects of disease causing RyR2 mutations, and by various drugs. Importantly, FRET signals were also readily detected in cells co-transfected with single CFP (RyR2S437-YFP) and single YFP (RyR2S2367-CFP) labeled RyR2, indicating that the interaction between the N-terminal and central mutation regions is an inter-subunit interaction. Our studies demonstrate that FRET analyses of this CFP- and YFP-labeled RyR2 can be used not only for investigating the conformational dynamics associated with RyR2 channel gating, but potentially, also for identifying drugs that are capable of stabilizing the conformations of RyR2.
Keywords: Ryanodine receptor, Ca2+-release channel, Sudden cardiac death, Ventricular arrhythmias, Inter-subunit interaction
Introduction
Calcium ions are major second messengers in signaling pathways in cells. In the heart, Ca2+ regulates muscle contraction, electrical signals that determine the cardiac rhythm, and cell growth pathways. One of the major Ca2+ channels, responsible for Ca2+ release from intracellular stores, is the type 2 ryanodine receptor (Bers, 2001). Sudden cardiac death is a sudden, unexpected death caused by loss of heart function, and it is the leading cause of natural death in the United States (Rosamond et al., 2008). Sudden cardiac death occurs when the electrical system in the heart malfunctions, leading to a fatal arrhythmia. Most cases of sudden cardiac death cases are associated with cardiovascular abnormalities that are identifiable at autopsy, but some sudden cardiac death can be associated with a structurally normal heart (Tester et al., 2004). Abnormal Ca2+ release through dysfunctional RyR2 has been implicated in some of these cases, and molecular genetic analysis has demonstrated that mutations in the RyR2 gene are linked to about 14% of sudden cardiac death cases in which no heart structural defects are apparent (Tester et al., 2004).
Since the first mutation in the human RyR2 gene was identified in 2001 (Priori et al., 2001), more than 70 naturally occurring mutations in RYR2 have been linked to two autosomal dominant forms of ventricular arrhythmias: catecholaminergic polymorphic ventricular tachycardia (CPVT) and arrhythmogenic right ventricular dysplasia type 2 (ARVD2) (Choi et al., 2004; Laitinen et al., 2001; Priori et al., 2002). CPVT is a genetic arrhythmogenic disorder characterized by stress-induced, bidirectional ventricular tachycardia that can degenerate into cardiac arrest and cause sudden cardiac death. ARVD2 is characterized by partial degeneration of the myocardium of the right ventricle, electrical instability, and sudden death.
The mutations are largely clustered in three regions of the RyR2 sequence: region 1 is near the N-terminus, from residues 77 to 466; region 2 is near the middle of the sequence, spanning residues 1724-2534; and region 3 includes both cytoplasmic- and membrane-associated regions and comprises residues 3778-4959. Intriguingly, the same regions are homologous to three malignant hyperthermia and central core disease mutation regions in the skeletal RyR isoform, RyR1 (Dirksen and Avila, 2005). The conservation of these mutant regions is not coincidental; they almost certainly fall within functional domains that are critical for the regulation of both RyR1 and RyR2, and the molecular mechanisms underlying these RyR-linked skeletal and cardiac muscle diseases are likely to be similar.
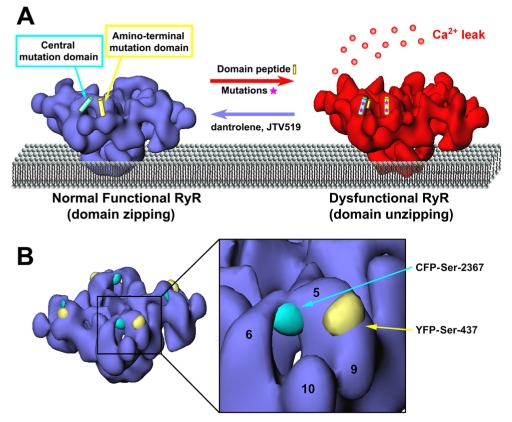
Two mutation hotspots that lie in cytoplasmic regions, the N-terminal region and the central region, are well separated in the primary sequence (by >1200 amino acids). According to the hypothesis of Ikemoto and Yamamoto, these two regions occur in structural domains that physically interact with one another in three-dimensional (3D) space, and changes in the strength of their interaction affects channel gating (Ikemoto and Yamamoto, 2002). According to this hypothesis, the interaction between these domains serves as a regulatory switch for channel gating activity; a tight ‘zipping’ of the interacting domains stabilizes the channel in the closed state. A mutation in either domain weakens such domain-domain interaction, thus increasing the tendency toward ‘unzipping’, which causes activation and leakiness of the Ca2+ release channel (Fig. 1A). In support of the hypothesis, a synthetic peptide, DPc10 (which corresponds to the Gly2460-Pro2495 region of the central mutation of RyR2), apparently interferes with the domain-domain interaction as evidenced by enhanced [3H]ryanodine binding activity, increased sensitivity of RyR2 to activating Ca2+, and induced Ca2+ leakage from the sarcoplasmic reticulum (SR): effects mimicking the typical phenotypes of cardiac diseases (Yamamoto and Ikemoto, 2002). Previous studies pertaining to the interaction between these domains have relied mainly on indirect biochemical assays to detect conformational changes in RyR1 and RyR2 (Yamamoto et al., 2000), and there has been little information describing local or global changes in the 3D architecture of RyR. Our laboratory used 3D cryo-electon microscopy (cryo-EM) to test one prediction of the inter-domain hypothesis, namely, that the N-terminal and central domains are in close proximity to one another. We mapped one central mutation hotspot (Ser2367) in the 3D structure of RyR2 by GFP labeling (Liu et al., 2005). Another site within the N-terminal mutant region (Ser437) has been located by the same method (Wang et al., 2007). GFP inserted in the N-terminal region (Ser437) was mapped to domains 5 and 9, whereas GFP in the central region (Ser2367) mapped to the bridge between domains 5 and 6 (Fig. 1B). These results clearly showed that the two locations are on the opposite sides of domain 5 in the cytoplasmic assembly of RyR, a spatial relationship consistent with the inter-domain zipping-unzipping hypothesis of Ikemoto and colleagues. To further test the inter-domain zipping-unzipping hypothesis, we have generated a FRET pair based on the 3D cryo-EM information, and characterized conformational changes involving these regions. The dynamic conformational changes within the inter-domain switch are revealed directly as changes in FRET efficiencies, thus making FRET particularly suitable for testing anti-arrhythmia drugs that are designed to correct the defective domain-domain interaction in the dysfunctional RyR2. Furthermore, our FRET study has also established that the domain-domain interaction occurs between two neighboring subunits within one tetrameric RyR2 molecule.
Fig. 1.
Design of a FRET pair to test the hypotheses of domain-domain interaction. (A) Hypothetical model showing that an inter-domain zipping-unzipping interaction within RyR affects channel gating. The N-terminal and central regions interact with one another, forming a regulatory switch for channel gating. A mutation in either region weakens the inter-domain interaction (unzipping), and the weakening leads to hyperactivity (leakiness) of the Ca2+ channel. The destabilized interaction can be reversed by therapeutic reagents such as JTV519 (which suppresses the effect of RyR2 mutations that promote sudden death) and dantrolene (which inhibits the effect of RyR1 mutations related to the skeletal muscle diseases malignant hyperthermia). Figure adapted from Yano et al. (Yano et al., 2005) with modifications. (B) A close-up view from an angle that shows the locations of the Ser437-YFP and Ser2367-CFP fluorophore pair, based upon previous 3D cryo-EM of RyR2-GFP chimeras. The distance between CFP and YFP is 50Å (center to center). Functional domains 5, 6, 9 and 10 are indicated.
Results
Functional characterization of RyR2S437-YFP/S2367-CFP
Based on the structural information from 3D cryo-EM, we constructed a cDNA with dual fluorescent protein insertions in the RyR2 primary sequence – a yellow fluorescent protein (YFP) after Ser437 (in the N-terminal mutation region) and a cyan fluorescent protein (CFP) after Ser2367 (in the central region) – to form a new RyR2 chimera, RyR2S437-YFP/S2367-CFP (Fig. 1B). The cDNAs was then expressed in HEK293 cells.
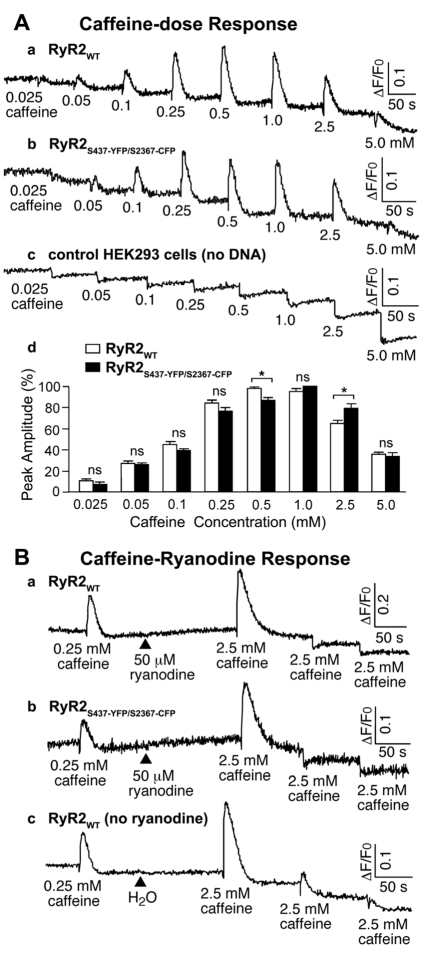
We have previously shown that a single insertion of GFP after Ser437 or Ser2367 has little effect on the structure and function of the heterologously expressed RyR2 channel (Liu et al., 2005; Wang et al., 2007). To determine whether a dual insertion of YFP after residue Ser437 and CFP after residue Ser2367 affects RyR2 function, we measured Ca2+ release induced by sequential additions of increasing concentrations of caffeine in HEK293 cells transfected with wild-type RyR2 (RyR2WT) or RyR2S437-YFP/S2367-CFP, using the fluorescent Ca2+ indicator dye fluo-3 AM. As shown in Fig. 2A,a, the level of intracellular Ca2+ release in HEK293 cells transfected with RyR2WT cDNA was hardly detected after the first addition of caffeine (0.025 mM). The second addition of caffeine (0.05 mM) induced a clearly detectable level of Ca2+ release, suggesting that the threshold for the activation of the wt RyR2 channel by caffeine is ~0.05 mM. The level of Ca2+ release increased progressively with each consecutive addition of caffeine from 0.05 mM up to 1.0 mM, and then decreased with further additions of caffeine (2.5 and 5 mM). The reduced level of Ca2+ release seen after additions of 2.5 and 5.0 mM caffeine, as compared with that seen after the addition of 1.0 mM caffeine, are probably due to the depletion of the intracellular Ca2+ stores by the previous additions of caffeine (0.025 to 1.0 mM). As seen in Fig. 2A,b, HEK293 cells expressing RyR2S437-YFP/S2367-CFP exhibited a pattern of response to repeated caffeine stimulations (from 0.025 to 5 mM) nearly identical to that seen in HEK293 cells expressing RyR2WT. The peak amplitudes, normalized to the maximal caffeine response in each experiment, of Ca2+ release events induced by various concentrations of caffeine were similar in RyR2WT- and RyR2S437-YFP/S2367-CFP-expressing cells (Fig. 2A,d). No caffeine-induced Ca2+ release was detected in non-transfected HEK293 cells (Fig. 2A,c). The immediate drops in fluorescence after the additions of caffeine were caused by fluorescence quenching of fluo-3 AM by caffeine (Chen et al., 2002).
Fig. 2.
Functional characterization of RyR2S437-YFP/S2367-CFP. (A,B) The response of HEK293 cells transfected with wild type RyR2 (RyRWT; A,a, B,a, and B,c), RyR2S437-YFP/S2367-CFP (A,b and B,b), or no DNA (A,c) to different concentrations of caffeine (A), or caffeine and ryanodine (B). Fluorescence intensity of the fluo-3-loaded transfected cells was monitored continuously before and after sequential additions of increasing concentrations of caffeine (0.025-5.0 mM) or before and after the sequential additions of 0.25 mM caffeine, 50 μM ryanodine, and 2.5 mM caffeine. (A,d) The peak amplitudes (%) of caffeine responses, normalized to the maximal peak value (100%) for each experiment, at different concentrations of caffeine in HEK293 cells expressing RyR2WT (white bars) and RyR2S437-YFP/S2367-CFP (black bars). Data are mean ± s.e.m. (n=5; *P<0.05; ns, not significant).
The responses of RyR2WT and RyR2S437-YFP/S2367-CFP to ryanodine are shown in Fig. 2B. The addition of 0.25 mM caffeine to HEK293 cells transfected with RyR2WT elicited a small increase in fluo-3 fluorescence, corresponding to a release of Ca2+ from intracellular stores. A subsequent addition of ryanodine caused a slow release of Ca2+ (Fig. 2B,a). This is probably due to the binding of ryanodine to a small population of RyR2 channels that are open under these conditions and consequently an increase in open probability of these channels. Importantly, the ryanodine-pretreated cells responded only to the first, but not to the second or third caffeine stimulation. This pattern of response is most probably due to ryanodine binding to caffeine-induced open channels and holding the channels in the open state such that the ryanodine modified ‘open’ channels cannot be further stimulated by caffeine (Kong et al., 2008; Zhang et al., 2003). Consistent with these explanations, RyR2WT-expressing HEK293 cells without ryanodine treatment showed no slow release of Ca2+ and responded to multiple caffeine stimulations (Fig. 2B,c). As seen in Fig. 2B,b, HEK293 cells expressing RyR2S437-YFP/S2367-CFP showed a response to ryanodine identical to that seen in HEK293 cells expressing RyR2WT. Taken together, these observations demonstrate that RyR2S437-YFP/S2367-CFP forms a functional Ca2+ release channel with responses to caffeine and ryanodine nearly identical to those of the RyR2WT.
FRET measurement in live HEK293 cells that express RyR2S437-YFP/S2367-CFP
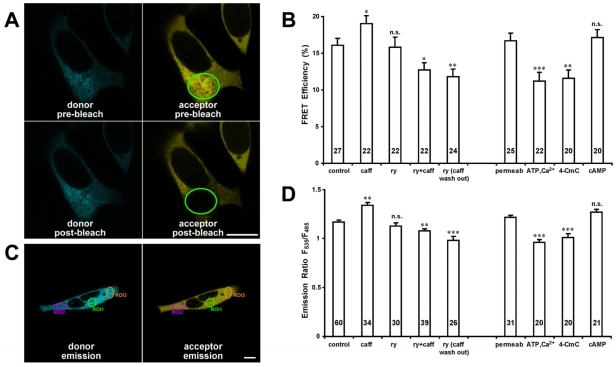
Fig. 3A illustrates the acceptor photobleaching method to measure FRET efficiency in the HEK293 cells that expressed RyR2S437-YFP/S2367-CFP. Cyan (donor) and yellow (acceptor) fluorescence images were recorded before and after photobleaching of the acceptor and used to calculate the FRET efficiency. There is one potential concern in measuring FRET in live cells using the photobleaching approach: the recovery of fluorescence through diffusion of RyR2s into the bleached field may lower the actual donor emission intensity after photobleaching, which would result in an underestimated FRET efficiency. To rule out the possibility that diffusion of RyR2s may contribute to changes in FRET, we performed a FRAP (fluorescence recovery after photobleaching) study. We continually monitored the recovery of YFP fluorescence after applying the same photobleaching that was used for FRET study. Our data showed that the fluorescence intensity of photobleached YFP recovered less than 3% after 50 seconds (the same time frame of the photobleaching FRET experiments), and less than 5% after 3 minutes. The results from FRAP demonstrate that the diffusion, both laterally and axially, of RyR2s in the live cells would not significantly interfere with FRET efficiency determined by the photobleaching approach.
Fig. 3.
FRET analysis of HEK293 cells expressing RyR2S437-YFP/S2367-CFP. (A) Confocal images showing cyan and yellow fluorescence before and after photobleaching from a HEK293 cell expressing RyR2S437-YFP/S2367-CFP cDNA. The green ellipse demarcates the area selected for photobleaching. Scale bar: 10 μm. (B) Changes of averaged FRET efficiency of the RyR2S437-YFP/S2367-CFP upon treatment with various RyR2 channel modulators. *P<0.05, **P<0.01 and ***P<0.001, respectively, compared with control. (C) Confocal images of CFP and YFP fluorescence were acquired under CFP excitation only (458 nm). The emitted fluorescence intensities were measured at 485±15 nm (CFP) and 535±15 nm (YFP). At least three regions of interest (ROI) were selected within each HEK293 cell, and the fluorescence intensities of each ROI were analyzed by LAS AF, and mean intensities were used to calculate the emission ratio F535/F485. Scale bar: 10 μm. (D) Changes in averaged emission ratio upon treatment with RyR2 modulators. Data are mean ± s.e.m., with the number of cells indicated on the bars.
As shown in Fig. 3B, the average FRET efficiency in the cells that expressed RyR2S437-YFP/S2367-CFP in the absence of any treatment was 16.1±0.9% (n=27 cells, mean ± s.e.m.). The addition of 5 mM caffeine, a pharmacological activator of channel activity, significantly increased the FRET efficiency to 19.0±1.1% (n=22, P<0.05). There was no significant change in the FRET efficiency in the cells that were treated with 100 μM ryanodine alone (15.8±1.4%, n=22, P=0.83). However, in cells that were pre-treated with ryanodine, the addition of caffeine decreased FRET efficiency to 12.7±1.0% (n=22, P<0.05), and the FRET efficiency further decreased to 11.8±1.0% (n=24, P<0.01) after the removal of caffeine. For cells that were permeabilized with saponin (100 μg/ml, 4-5 minutes), the FRET efficiency was 16.7±1.0% (n=25), and it decreased to 11.2±1.2% (n=22, P<0.001) or 11.6±1.1% (n=20, P<0.01) after addition of the channel activators 5 mM ATP and 50 μM Ca2+ or 0.4 mM 4-chloro-m-cresol (4-CmC). There was no significant change in the FRET efficiency after addition of 10 μM cAMP (17.1±1.1%, n=20, P=0.79), which is not a direct activator of RyR2. These observations indicate that caffeine induces conformational changes in RyR2 different from those induced by ryanodine, ATP and Ca2+, and 4-CmC (see Discussion).
Fig. 3C illustrates another method of detecting FRET in the HEK293 cells that involves calculating the donor and acceptor emission ratio. The changes in the F535/F485 ratio reflect dynamic changes in FRET (Bossuyt et al., 2006). As shown in Fig. 3D, the average F535/F485 in the cells without any treatment was 1.17±0.02 (n=60 cells). After treating the cells with 5 mM caffeine, the F535/F485 ratio increased to 1.34±0.03 (n=34 cells, P<0.01). Changes in F535/F485 were negligible in cells treated with 100 μM ryanodine alone (1.13±0.03, n=30, P=0.21). However, upon pre-treatment with ryanodine, caffeine decreased F535/F485 to 1.08±0.02 (n=39, P<0.01), and the F535/F485 further decreased to 0.98±0.04 (n=26, P<0.001) after the removal of caffeine. For cells that were permeabilized with saponin, the F535/F485 ratio was 1.22±0.02 (n=31), and decreased to 0.96±0.03 (n=20, P<0.001) or 1.01±0.04 (n=20, P<0.001) after addition of 5 mM ATP and 50 μM Ca2+ or 0.4 mM 4-CmC. There was a slight (but not statistically significant) increase in the F535/F485 ratio after the addition of 10 μM cAMP (1.27±0.03, n=21, P=0.09). Therefore, similar results were obtained using both the acceptor photobleaching and the donor and acceptor emission ratio methods of FRET detection.
Effect of domain peptides on the FRET efficiency of RyR2S437-YFP/S2367-CFP
Next, we tested whether domain peptides that mimic CPVT-causing mutations in the central region of the primary structure of RyR2 affect the FRET efficiency of RyR2S437-YFP/S2367-CFP. Domain peptides are short peptides that match a region of the sequence of one ‘mutation’ domain; they competitively interfere with the interaction between the native domains (Ikemoto and Yamamoto, 2002). Domain peptides are thought to cause structural changes (unzipping) by disrupting domain-domain interactions. DPc10, a synthetic peptide corresponding to the sequence Gly-2460 to Pro-2495 in RyR2, was found to enhance the ryanodine binding activity, and to increase the sensitivity of RyR2 to activating Ca2+ (Yamamoto and Ikemoto, 2002). A single point mutation, Arg to Ser in DPc10 (DPc10-mut), which is analogous to the reported Arg2474Ser CPVT mutation, abolished both of these effects (Yamamoto and Ikemoto, 2002). This mutant domain peptide was used as a negative control. The primary sequence of the mutation regions in RyR1 and RyR2 is highly conserved. DP4 (corresponding to RyR1 residues 2442-2477) is a well-characterized domain peptide that disrupts the normal domain-domain interactions in RyR1 (Lamb et al., 2001; Shtifman et al., 2001; Yamamoto et al., 2000). Interestingly, DP4 activates RyR2 as well as RyR1. This observation suggests that essentially the same regulatory domain-domain interaction is operating in both RyR2 and RyR1 channels. Therefore, we also tested the impact of DP4 on FRET in our experiments.
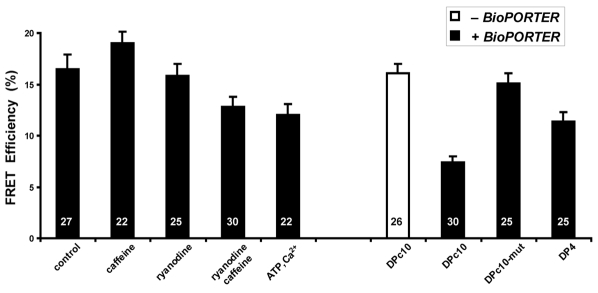
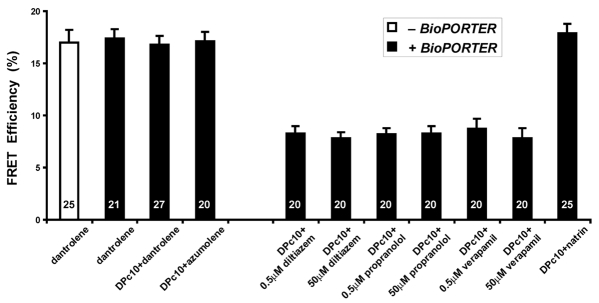
If domain unzipping occurs when domain peptides are added to RyR2S437-YFP/S2367-CFP, we would anticipate an increased domain-domain distance, and this would result in decreased FRET. To ensure the delivery of domain peptides into HEK293 cells, we used BioPORTER, a reagent for intracellular delivery of bioactive molecules (Hamada et al., 2009). Fig. 4 shows the observed changes in the FRET efficiency in the presence of BioPORTER or domain peptides. As a control (left-hand side of Fig. 4), we found that BioPORTER itself did not affect the FRET changes found for the experiments documented in Fig. 3B, which were done in the absence of BioPORTER. DPc10 alone also showed no statistically significant effects on FRET. Notably, in the presence of BioPORTER, domain peptide DPc10 significantly reduced the FRET efficiency (P<0.001), consistent with an increasing distance between Ser437-YFP and Ser2367-CFP, and suggesting that DPc10 causes inter-domain unzipping. DPc10-mut, as a negative control, only slightly reduced the FRET in the HEK293 cells (P=0.44). DP4, the skeletal homolog of DPc10, reduced the FRET signals (P<0.01); however, the effect of DP4 on RyR2 was weaker than that of DPc10.
Fig. 4.
Sensitivity of FRET efficiency of the RyR2S437-YFP/S2367-CFP to domain peptides reveals dynamic interaction between RyR2 structural domains. The FRET efficiency was determined by the photobleaching method. HEK293 cells were treated with BioPORTER, a protein-delivery reagent for intracellular delivery of domain peptides. The concentrations of domain peptides DPc10, DPc10-mut, and DP4 were each 50 μM.
Effect of drugs that correct defective domain-domain interactions
After exposure to conditions that induce domain-unzipping, the addition of a drug (e.g. dantrolene or azumolene) to the system is predicted to restore improperly unzipped domains back to a normal zipped mode. Dantrolene, a drug that suppresses RyR1 Ca2+ release from SR in skeletal muscle, is used as a therapeutic agent in individuals susceptible to malignant hyperthermia. Dantrolene and its more water-soluble analog, azumolene, have been shown to strongly inhibit DP4-induced unzipping of the domain-domain interactions, and stabilize the domain switch in the zipped configuration of RyR1 (Kobayashi et al., 2005). Dantrolene has little or no effect on normal RyR2 (Paul-Pletzer et al., 2001; Paul-Pletzer et al., 2005); however, it has an effect on diseased RyR2 (Paul-Pletzer et al., 2005; Tian et al., 1991). The dantrolene-binding site on RyR1 has been identified in an N-terminal region of the RyR1 sequence encompassing amino acids 590-609 (Paul-Pletzer et al., 2002). Interestingly, this sequence is within the N-terminal mutation region. Since the dantrolene binding sequence is exactly the same in the two RyR isoforms (Kobayashi et al., 2009; Paul-Pletzer et al., 2005), a reasonable assumption is that the potentially active dantrolene binding site in RyR2 becomes accessible for drug binding only when the inter-domain interaction becomes defective (Paul-Pletzer et al., 2005). Consistent with this idea, we observed that both dantrolene and azumolene completely inhibited the FRET decrease in RyR2S437-YFP/S2367-CFP that is caused by DPc10 (Fig. 5).
Fig. 5.
Impact of drugs on inter-domain dynamics. The domain unzipping was induced by the addition of domain peptide DPc10. HEK293 cells were incubated in medium containing DPc10 and BioPORTER for 3 hours, and various agents were then added to the cells and incubated for another hour before examination of the FRET signals. The concentrations used in the experiments were: DPc10, 50 μM; dantrolene, 10 μM; azumolene, 10 μM; propranolol, verapamil, and diltiazem, 0.5 μM and 50 μM each; and natrin, 10 μM.
We have also tested the effects on FRET of several drugs that are thought to inhibit intracellular Ca2+ leakage (Wakimoto et al., 2007), namely, propranolol, verapamil and diltiazem. Propranolol blocks β-adrenergic receptors, and it is an anti-hypertensive agent that is used in the treatment of cardiac arrhythmias. Propranolol has been reported to inhibit [3H]ryanodine binding to RyR1 (Zchut et al., 1996), restore RyR conformational changes and prevent Ca2+ leakage from RyR2 (Doi et al., 2002). Verapamil blocks the L-type Ca2+ channel in smooth muscle and cardiac muscle. Verapamil has also been shown to directly interact with RyR1 and to block the activity of RyR1as a Ca2+ channel (Valdivia et al., 1990), and it suppresses arrhythmias characteristic of CPVT that arise from RyR2 mutations (Swan et al., 2005). Diltiazem acts as an L-type Ca2+ channel antagonist, similar in function to verapamil, and has been shown to inhibit Ca2+ release from intracellular stores in neutrophils. Diltiazem has been shown to partially inhibit both the FK506-induced Ca2+ leakage in normal SR vesicles and the spontaneous Ca2+ leakage in failing SR vesicles (Yano et al., 2003).
By monitoring the FRET signals in RyR2S437-YFP/S2367-CFP, we directly tested the impact of various drugs on the domain interactions between the N-terminal and the central regions of RyR2. Our results are shown in Fig. 5. The unzipped RyR2s (induced by DPc10, revealed as reduced FRET efficiency) could be converted back to the zipped state, after addition of 10 μM dantrolene or azumolene (a typical clinical concentration). Addition of propranolol, verapamil, or diltiazem did not produce such an effect. The concentrations of propranolol, verapamil and diltiazem used in our tests were 0.5 μM (approximately the typical clinical concentration) and 50 μM. The latter concentration is close to the toxic level in plasma, and is about 100 times higher than the normal effective plasma concentrations found associated with typical clinical doses (Jefferson et al., 1979; Ochs and Knuchel, 1984; Vogelgesang et al., 1984). It has been reported that high concentrations of verapamil (intraperitoneal 8 μg/g body mass in mouse) can reduce spontaneous Ca2+ release (Wakimoto et al., 2007), and high concentrations of propranolol (2 mM) can inhibit [3H]ryanodine binding to RyR1 (Zchut et al., 1996). However, both of these cited concentrations are higher than the maximal toxic level in the plasma, and the results may, therefore, not be physiologically significant. We found that natrin also reversed the DPc10-induced reduction in FRET, similar to the effects of dantrolene and azumolene (Fig. 5). Natrin is a toxin from snake venom that strongly inhibits both the binding of ryanodine to RyR1 and the Ca2+ channel activity of RyR1. As revealed by cryo-EM, natrin binds to a region in RyR1 that is close to the N-terminal and central mutation domains (Zhou et al., 2008). Our results indicate that propranolol, verapamil and diltiazem have no effect on restoring the defective domain-domain interaction, but dantrolene, azumolene, and natrin do stabilize the interaction, and could conceivably prevent CPVT and ARVD2 through their correction of the defective domain-domain interaction in RyR2. It will be interesting to test whether dantrolene and azumolene, and perhaps natrin, are effective in suppressing cardiac arrhythmias in animal models of CPVT and ARVD2. Taken together, our data indicate that FRET of RyR2S437-YFP/S2367-CFP is sensitive to modifications by RyR activators, domain peptides and various drugs.
The domain switch formed by the N-terminal and central domains is an inter-subunit interaction
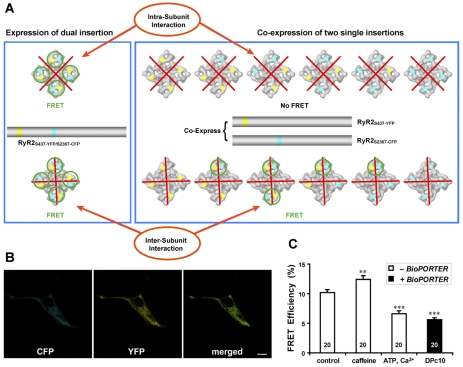
Functional RyR2 is a homo-tetramer that is composed of four subunits. However, it is not known, whether the domain switch is formed within one subunit, or between two neighboring subunits In other words, do the interacting N-terminal region and central region both belong to one subunit (an intra-subunit interaction), or do they belong to two different subunits (an inter-subunit interaction)? In Fig. 6A, red crosses are used to highlight the two possibilities, that an inter-subunit boundary exists or does not exist between the N-terminal and central regions.
Fig. 6.
The interaction between N-terminal and central mutation regions is an inter-subunit interaction. (A) Proposed model of domain-domain interaction. The site of N-terminal YFP insertion after Ser437 (yellow dots) and the central domain CFP insertion after Ser2367 (cyan dots) are adjacent in the clamp region of RyR2, and form a domain switch that is important for channel gating. What was unclear was whether this domain switch is formed by two domains located in one subunit (top row), or from two domains belonging to two neighboring subunits (bottom row). For the cDNA construct that contains both YFP inserted after Ser437 and CFP after Ser2367, FRET is predicted when the cDNA is expressed, regardless of whether the interaction is within a subunit or between two subunits (left panel; FRET pairs indicated with green circles). We constructed two RyR2 cDNAs, one with CFP inserted after Ser437, the other with YFP inserted after Ser2367. When these two cDNAs are coexpressed in the HEK293 cells, six possible hybrid RyR2 molecules can result (right panel). The top row shows the six possible structures if the domain switch is formed by two domains contained within one subunit (i.e. an intra-subunit interaction); in this case, no FRET signal will be detected, because the distance between CFP and YFP in two separate clamp corners is over 200 Å. The bottom row shows the six possible structures if the domain switch is formed at the interface of two neighboring subunits (i.e. an inter-domain interaction); in this case, four out of six have at least one CFP-YFP pair within the same clamp region (highlighted by light green circles). FRET is predicted in these four cases, similar to what is expected (left panel), and to what we indeed observed (data shown in Fig. 3) for RyR2S437-YFP/S2367-CFP. (B) Images of live HEK293 cells co-transfected with cDNAs of RyR2S437-YFP and RyR2S2367-CFP show the colocalization of CFP and YFP. Scale bar: 10 μm. (C) The FRET efficiency as determined by the photobleaching method. FRET signals were detected in HEK293 cells, demonstrating that the domain switch formed between the N-terminal and central mutation domains is an inter-subunit interaction. The average FRET efficiency for the control cells was 10.2±0.5 (n=20 cells); for caffeine-treated cells, 12.4±0.6 (n=20), for ATP and Ca2+, 6.6±0.5 (n=20), and for cells treated with DPc10 (with BioPORTER), 5.6±0.3 (n=20). **P<0.01, and ***P<0.001, compared with control.
When the cDNA for RyR2S437-YFP/S2367-CFP is expressed in the HEK293 cells, a FRET signal occurs regardless of whether the inter-domain interaction is between subunits or within a subunit (left panel in Fig. 6A). To investigate whether the domain-domain interaction is an inter-subunit or intra-subunit interaction, we coexpressed two cDNAs in HEK293 cells, with one cDNA having Ser437 labeled with YFP only, and the other one having Ser2367 labeled with CFP only. When these two cDNAs are coexpressed in the HEK293 cell, there are six possible hybrid RyR2 molecules (right panel in Fig. 6A). We expect no FRET in the case of an intra-subunit interaction, but we expect FRET to occur in the case of an inter-subunit interaction (compare the top and bottom panels in Fig. 6A).
The results are shown in Fig. 6B,C. We detected FRET signals in the HEK cells that coexpressed cDNAs of RyR2S437-YFP and RyR2S2367-CFP. Furthermore, the FRET signals were altered by caffeine, ATP and Ca2+, and domain peptide DPc10. These results indicate that the N-terminal domain of one subunit interacts with the central domain of the neighboring subunit, rather than the same subunit. This inter-subunit nature of domain-domain interactions lends support to a model of subunit organization that we have predicted previously (see Discussion below).
Similar experiments using other site-specific CFP-YFP pairs could be used to further clarify the subunit boundaries of RyR2. For example, GFPs inserted at Ser437 and at Tyr846 were mapped close to one another in the 3D structure (Liu et al., 2006; Wang et al., 2007). FRET signals have been detected in cells expressing a dual insertion of RyR2S437-YFP/Y846-CFP (FRET efficiency 19.5±1.5%, n=25 cells). However, there were no FRET signals in cells that co-expressed the two single insertions, RyR2S437-YFP and RyR2Y846-CFP, although both CFP and YFP fluorescence as detected, and colocalized. This indicates that these two sites are located at an intra-subunit boundary.
Discussion
RyR2S437-YFP/S2367-CFP: a conformational probe of RyR2
In the present study, we generated a RyR2 fusion protein (RyR2S437-YFP/S2367-CFP) containing CFP and YFP that are suitably located for FRET studies on the conformational dynamics of RyR2. We have previously mapped the N-terminal and central mutation hotspots of RyR2 using GFP, inserted either after Ser437 or Ser2367, as a structural marker in the 3D cryo-EM maps. Here, we replaced the GFP with CFP and YFP to form a FRET probe. The distance between the CFP and YFP was predicted to be 50 Å (center to center) based on the 3D locations of single insertions of GFP (Wang et al., 2007); thus, the two juxtaposed probes should be suitable for FRET, which is effective for a range between 30 and 100 Å in 3D space. One potential problem with our FRET experiments on RyR2S437-YFP/S2367-CFP is that the insertion of fluorescent proteins into the interacting domains itself could induce domain-unzipping, since the CFP and YFP are inserted close to the interface, and both are bulky proteins (the molecular masses of CFP and YFP are ~28 kDa). If this occurred, then introducing exogenous domain-unzipping probes would not produce any further effect. On the contrary, as shown in the Result section, FRET does occur in the HEK293 cells expressing RyR2S437-YFP/S2367-CFP and it is sensitive to RyR2 modulators, to domain-unzipping probes, and to drugs. Thus, S437-YFP/S2367-CFP can be used as an internal probe for monitoring conformational changes in functional RyR2 channels.
Effects of RyR channel activators on FRET of RyR2S437-YFP/S2367-CFP: caffeine induces unique conformational changes in RyR2
RyR undergoes global conformational changes when the Ca2+ channel switches between the open and closed states that involve allosteric coupling of the trans-membrane pore region and the cytoplasmic region (Samso et al., 2005; Samso et al., 2009; Serysheva et al., 1999; Sharma et al., 2006). Cryo-EM studies have shown that ATP and Ca2+ together produce almost maximum activation of RyR, and drive the channel population toward a predominately open state (Serysheva et al., 1999; Sharma et al., 2006). Consistent with this observation, we found that ATP and Ca2+ decreased the FRET efficiency and the emission ratio of F535/F485 in RyR2S437-YFP/S2367-CFP, indicating that these two structural domains move apart upon activation by ATP and Ca2+ (Fig. 3). We also found that 4-CmC, another potent RyR activator, had a similar effect on FRET. By contrast, we observed that caffeine alone increased the FRET signal. These data suggest that as RyR channel activators, the action of caffeine on the domain switch conformation is different from that of 4-CmC, or ATP and Ca2+. Previous biochemical studies have demonstrated that caffeine has a different activation site from those of 4-CmC (Du et al., 2000; Fessenden et al., 2006; Herrmann-Frank et al., 1996), and ATP and Ca2+ (Chen et al., 1998; Sitsapesan and Williams, 1990). Caffeine is able to activate all three RyR isoforms, whereas 4-CmC activates only RyR1 and RyR2, but not RyR3 (Choisy et al., 2000; Fessenden et al., 2000). Caffeine also has a different binding site from ryanodine (Wang et al., 2003). Our FRET studies show that ryanodine alone does not affect FRET, which is consistent with ryanodine's known inability to bind to RyRs in the closed state (Du et al., 1998). However, in the presence of caffeine, ryanodine caused a decreased in the FRET (Fig. 3B,D). This probably resulted from binding of ryanodine to the caffeine-activated channels, thereby producing a marked increase in the channel open probability and inducing similar conformational changes in the domain switch region to those induced by 4-CmC, and ATP and Ca2+. Interestingly, removal of caffeine with a continuous wash of ryanodine buffer resulted in a further decrease in FRET (instead of a return to the baseline), suggesting that, once bound, ryanodine keeps RyR2s in their higher open probability state. Together with the finding that caffeine alone increased the FRET, the results suggest that the caffeine and ryanodine cause two distinct effects on the conformational changes in the domain switch region. Consistent with these different conformational changes induced by caffeine and ryanodine, we have previously shown that caffeine preferentially sensitizes the RyR2 channel to activation by luminal, but not cytosolic, Ca2+ (Kong et al., 2008), whereas ryanodine dramatically sensitizes the RyR2 channel to activation by cytosolic Ca2+ (Masumiya et al., 2001). These FRET and functional studies suggest that activation of RyR2 by cytosolic and luminal Ca2+ are different and may involve different conformational changes in the receptors.
Ikemoto's ‘domain switch’ hypothesis
The decreased FRET that we observed for RyR2S437-YFP/S2367-CFP in the presence of the domain peptides DPc10 and DP4 provides direct support for the domain switch hypothesis (Ikemoto and Yamamoto, 2002). The decrease in FRET is consistent with the notion that the amino and central ‘disease’ domains, which bear the inserted YFP and CFP, move apart (‘unzip’) when the interaction between the domains is weakened by the peptides, which competitively interfere with the interaction. Further supporting this interpretation are our findings that both dantrolene (or azumolene), a drug that putatively strengthens the interdomain interaction, and DPc10-mut, a peptide containing an amino acid substitution that is analogous to a mutation occurring in people with the disease CPVT, inhibit the decrease in FRET. A new finding is that the amino and central disease domains interact across an inter-subunit boundary. Like the potassium channel KcsA, the functional RyR is a homo-tetramer that is composed of four identical subunits that are arranged symmetrically around the pore. In KcsA, there is evidence to suggest that direct interaction between the four subunits leads to a cooperative opening and closing of the ion-conducting pore (Blunck et al., 2008), which is structurally similar to the pore region of the RyR (Samso et al., 2005; Samso et al., 2009; Welch et al., 2004). Here we have identified a region of inter-subunit interaction that is of critical importance for normal channel function in RyR. Evidence has been presented that interactions between the N-terminal domain and the central domain involve two adjacent subunits, and we suggest that mutations in either domain may weaken the normal subunit-subunit interactions, thus altering the stability of the channel.
The subunit organization within tetrameric RyR
RyRs are homo-tetrameric structures, but even the highest resolution attained to date in 3D reconstructions of RyRs (Serysheva et al., 2008) is insufficient to unambiguously resolve the boundaries between the subunits (Hamilton and Serysheva, 2009). Thus, the precise shape of the subunit is unknown. Alternatively, we may be able to gain some clues to the assembly of RyR subunits, by mapping specific residues or sequences within the linear sequence of RyR2 onto the 3D structure. Based on the 3D information obtained through these mappings, we have proposed three models of subunit organization (Jones et al., 2008). The N-terminal and central mutation segments are predicted to involve an inter-subunit interaction in one model, but to involve an intra-subunit interaction in the other two. The inter-subunit interaction model was favored by Serysheva et al. based on their analysis of the density distribution in a 3D reconstruction (Serysheva et al., 2008). Nevertheless, direct evidence for support this model was lacking.
In the present study, we have assessed whether the N-terminal and central mutation regions are involved in an inter-subunit interaction or an intra-subunit interaction using a FRET-based approach. The concept is quite simple: if two structural domains are close to one another, FRET signals will be detected when the two domains are labeled with a fluorophore donor and a fluorophore acceptor respectively (i.e. a dual insertion). The distance between the two domains must be less than about 100 Å for an acceptor to take energy from a donor. Moreover, the distances between this acceptor and the other three donors that are present in a tetramer must be great than 100 Å to avoid any crosstalk FRET. If we construct two individually labeled RyRs (i.e. two single insertions), the expressed hybrid RyR molecules will display FRET signals only when the two structural domains belong to two different subunits, i.e. when the domain-domain interface involves an inter-subunit interaction. There will be no FRET signal when the domain-domain interface involves an intra-subunit interaction (see model in Fig. 6A). Using this approach, we have demonstrated that the interaction between Ser437 and Ser2367 is an inter-subunit interaction, whereas the interaction between Ser437 and Tyr-846 is an intra-subunit interaction. By this method, we could determine whether there is a subunit boundary between any two closely apposed structural domains which can be labeled with FRET probes.
Materials and Methods
cDNAs construction
The cloning and construction of the 15 kb full-length cDNA encoding mouse cardiac RyR2 sequence (NCBI reference sequence: NP_076357.2, GI:124430578) has been described previously (Zhao et al., 1999). cDNAs encoding RyR2S437-YFP and RyR2S2367-CFP were constructed according to the previously described procedure (Liu et al., 2005; Wang et al., 2007). To generate the cDNA construct of RyR2S437-YFP/S2367-CFP, we removed the AflII(915)-ClaI(2350) fragment with YFP in RyR2S437-YFP by digesting with AflII and ClaI. The AflII(915)-ClaI(2350) fragment with YFP was used to replace the AflII(915)-ClaI(2350) fragment in RyR2S2367-CFP.
Cell culture and cDNA transfection
HEK293 cells were maintained in Dulbecco's modified Eagle's medium as described previously (Chen et al., 1997). HEK293 cells grown in 35-mm glass bottom culture dishes for 20-24 hours, and after subculture were transfected with 1.8 μg cDNA of RyR2S437-YFP/S2367-CFP or a mixture of RyR2S437-YFP and RyR2S2367-CFP (molar ratio 1:1), using Ca2+ phosphate precipitation.
Intracellular Ca2+ release measurements in transfected HEK293 cells
Free cytosolic Ca2+ concentration in transfected HEK293 cells was measured using the fluorescent Ca2+ indicator dye fluo-3 AM as described previously, with some modifications (Chen et al., 1997). Cells grown for ~18 hours after transfection were washed five times with PBS (137 mM NaCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 2.7 mM KCl) and incubated in Krebs-Ringer-Hepes (KRH) buffer without MgCl2 or CaCl2 (KRH buffer: 125 mM NaCl, 5 mM KCl, 1.2 mM KH2PO4, 6 mM glucose, 1.2 mM MgCl2, 2 mM CaCl2, and 25 mM Hepes, pH 7.4) at room temperature for 45 minutes with gentle shaking and then at 37°C for 45 minutes. After being detached from culture dishes by gentle pipetting, cells were collected by centrifugation at 1000 r.p.m. for 5 minutes in a Thermo/EC Centra CL2 centrifuge. Cell pellets were resuspended in supplemented Dulbecco's modified Eagle's medium and loaded with 10 μM fluo-3 at room temperature for 1 hour. The fluo-3-loaded cells were washed with KRH buffer three times and resuspended in KRH buffer plus 0.1 mg/ml BSA and 250 μM sulfinpyrazone. An aliquot of fluo-3 loaded cells was then added to 2 ml (final volume) KRH buffer in a cuvette, and the fluorescence intensity of fluo-3 before and after sequential additions of various concentrations of caffeine or ryanodine was monitored at 530 nm in an SLM-Aminco series 2 luminescence spectrometer using 480 nm excitation at 25°C.
Domain peptides
The amino acid sequences of domain peptides, DPc10, DPc10-mut, and DP4, were previously described (Yamamoto et al., 2000; Yamamoto and Ikemoto, 2002). Domain peptides were synthesized on an Applied Biosystems model 431A synthesizer employing Fmoc as the α-amino protecting group. The peptide was cleaved and de-protected with 95% trifluoroacetic acid and purified by reversed-phase high-pressure liquid chromatography.
FRET measurements
HEK293 cells grown for 24-48 hours after transfection were washed three times with KRH buffer without MgCl2 or CaCl2 and examined on a Leica TCS SP5 confocal laser scanning microscope with a 63×/1.4 NA oil-immersion objective lens. Cells were kept at 37°C using a water-heated stage incubator. To test the effect of RyR2 modulators, domain peptides, and various drugs, buffer in the cultured dishes was exchanged by peristaltic pumps. We utilized two experimental approaches, acceptor photobleaching and donor-acceptor emission ratio, to detect and measure FRET signals in the live cells. For the acceptor photobleaching method, CFP and YFP were excited with separate laser channels of 458 nm and 514 nm, respectively. Emission fluorescence intensity data were obtained at 465-495 nm (CFP) and 520-550 nm (YFP). We used a 700 Hz line frequency scan speed with bidirectional scan mode in combination with an image format of 1024×1024 pixels, which can record one image every 754 mseconds. Repeated scans (30-60) with maximum laser intensity at 514 nm were used to photobleach YFP which lasted about 23-45 seconds, and the FRET efficiency was calculated according to the equation:
where ICFPpre and ICFPpost are the respective background-corrected CFP fluorescence intensities before and after photobleaching YFP (Papadopoulos et al., 2004). The photobleaching, fluorescence intensity measurements, and FRET efficiency calculation were controlled automatically by the software Leica Application Suite Advanced Fluorescence (LAS AF).
For the donor-acceptor emission ratio method, confocal fluorescent images of CFP and YFP were acquired under CFP excitation only (458 nm). The emitted fluorescence intensities were measured at 485±15 nm (CFP) and 535±15 nm (YFP). At least three regions of interest (ROI) were selected within each HEK293 cell, the fluorescent intensities of each ROI were analyzed by LAS AF, and mean intensities were used to calculate the emission ratio F535/F485.
FRAP experiments were performed to investigate the possibility that diffusion of non-bleached RyRs into bleached areas interfere with the FRET analysis in live HEK293 cells. Photobleaching was performed in the same way as in FRET experiments, and fluorescent intensity of YFP was continuously monitored at 520-550 nm. The FRAP protocol was also controlled by LAS SF. FRAP was performed in 10 separate cells and then averaged for analysis.
Data are presented as mean ± s.e.m. Data analysis was performed using the unpaired Student's t-test. A P-value below 0.05 was considered statistically significant.
Acknowledgments
The project described was supported by the American Heart Association grant 0430076N to Z.L.; by National Institutes of Health grants R01HL095541 to Z.L., R01AR040615 to T.W. and R01HL072841 to N.I.; and by research grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Alberta to S.R.W.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute, National Institute of Arthritis and Musculoskeletal and Skin Diseases, or the National Institutes of Health. We thank Wadsworth Center's Advanced Light Microscopy and Image Analysis Core Facility, Electron Microscopy Core Facility, and the Resource for Visualization of Biological Complexity (NIH Biotechnological Resource Grant P41RR01219). Deposited in PMC for release after 12 months.
References
- Bers D. M. (2001). Excitation-Contraction Coupling and Cardiac Contractile Force Dordrecht, Netherlands: Kluwer Academic; [Google Scholar]
- Blunck R., McGuire H., Hyde H. C., Bezanilla F. (2008). Fluorescence detection of the movement of single KcsA subunits reveals cooperativity. Proc. Natl. Acad. Sci. USA 105, 20263-20268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt J., Despa S., Martin J. L., Bers D. M. (2006). Phospholemman phosphorylation alters its fluorescence resonance energy transfer with the Na/K-ATPase pump. J. Biol. Chem. 281, 32765-32773 [DOI] [PubMed] [Google Scholar]
- Chen S. R., Li X., Ebisawa K., Zhang L. (1997). Functional characterization of the recombinant type 3 Ca2+ release channel (ryanodine receptor) expressed in HEK293 cells. J. Biol. Chem. 272, 24234-24246 [DOI] [PubMed] [Google Scholar]
- Chen S. R. W., Ebisawa K., Li X., Zhang L. (1998). Molecular identification of the ryanodine receptor Ca2+ sensor. J. Biol. Chem. 273, 14675-14678 [DOI] [PubMed] [Google Scholar]
- Chen S. R. W., Li P., Zhao M., Li X., Zhang L. (2002). Role of the proposed pore-forming segment of the Ca2+ release channel (ryanodine receptor) in ryanodine interaction. Biophys. J. 82, 2436-2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi G., Kopplin L. J., Tester D. J., Will M. L., Haglund C. M., Ackerman M. J. (2004). Spectrum and frequency of cardiac channel defects in swimming-triggered arrhythmia syndromes. Circulation 110, 2119-2124 [DOI] [PubMed] [Google Scholar]
- Choisy S., Huchet-Cadiou C., Leoty C. (2000). Differential effects of 4-chloro-m-cresol and caffeine on skinned fibers from rat fast and slow skeletal muscles. J. Pharmacol. Exp. Ther. 294, 884-893 [PubMed] [Google Scholar]
- Dirksen R. T., Avila G. (2005). Pathophysiology of muscle disorders linked to mutations in the skeletal muscle ryanodine receptor. In Ryanodine Receptors-Structure, Function and Dysfunction in Clinical Disease New York: Springer Science and Business Media; [Google Scholar]
- Doi M., Yano M., Kobayashi S., Kohno M., Tokuhisa T., Okuda S., Suetsugu M., Hisamatsu Y., Ohkusa T., Kohno M., et al. (2002). Propranolol prevents the development of heart failure by restoring FKBP12.6-mediated stabilization of ryanodine receptor. Circulation 105, 1374-1379 [DOI] [PubMed] [Google Scholar]
- Du G. G., Imredy J. P., MacLennan D. H. (1998). Characterization of recombinant rabbit cardiac and skeletal muscle Ca2+ release channels (ryanodine receptors) with a novel [3H]ryanodine binding assay. J. Biol. Chem. 273, 33259-33266 [DOI] [PubMed] [Google Scholar]
- Du G. G., Khanna V. K., MacLennan D. H. (2000). Mutation of divergent region 1 alters caffeine and Ca2+ sensitivity of the skeletal muscle Ca2+ release channel (ryanodine receptor). J. Biol. Chem. 275, 11778-11783 [DOI] [PubMed] [Google Scholar]
- Fessenden J. D., Wang Y., Moore R. A., Chen S. R. W., Allen P. D., Pessah I. N. (2000). Divergent functional properties of ryanodine receptor types 1 and 3 expressed in a myogenic cell line. Biophys. J. 79, 2509-2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessenden J. D., Feng W., Pessah I. N., Allen P. D. (2006). Amino acid residues Gln4020 and Lys4021 of the ryanodine receptor type 1 are required for activation by 4-chloro-m-cresol. J. Biol. Chem. 281, 21022-21031 [DOI] [PubMed] [Google Scholar]
- Hamada T., Gangopadhyay J. P., Mandl A., Erhardt P., Ikemoto N. (2009). Defective regulation of the ryanodine receptor induces hypertrophy in cardiomyocytes. Biochem. Biophys. Res. Comm. 380, 493-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton S. L., Serysheva I. I. (2009). Ryanodine receptor structure: progress and challenges. J. Biol. Chem. 284, 4047-4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann-Frank A., Richter M., Sarközi S., Mohr U., Lehmann-Horn F. (1996). 4-chloro-m-cresol, a potent and specific activator of the skeletal muscle ryanodine receptor. Biochim. Biophys. Acta 1289, 31-40 [DOI] [PubMed] [Google Scholar]
- Ikemoto N., Yamamoto T. (2002). Regulation of calcium release by interdomain interaction within ryanodine receptors. Front. Biosci. 7, D659-D670 [DOI] [PubMed] [Google Scholar]
- Jefferson D., Jenner P., Marsden C. D. (1979). Relationship between plasma propranolol concentration and relief of essential tremor. J. Neurol. Neurosurg. Psychiatry 42, 831-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. P., Meng X., Xiao B., Cai S., Bolstad J., Wagenknecht T., Liu Z., Chen S. R. W. (2008). Localization of PKA phosphorylation site, Ser2030, in the three-dimensional structure of cardiac ryanodine receptor. Biochem. J. 410, 261-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Bannister M. L., Gangopadhyay J. P., Hamada T., Parness J., Ikemoto N. (2005). Dantrolene stabilizes domain interactions within the ryanodine receptor. J. Biol. Chem. 280, 6580-6587 [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Yano M., Suetomi T., Ono M., Tateishi H., Mochizuki M., Xu X., Uchinoumi H., Okuda S., Yamamoto T., et al. (2009). Dantrolene, a therapeutic agent for malignant hyperthermia, markedly improves the function of failing cardiomyocytes by stabilizing interdomain interactions within the ryanodine receptor. J. Am. Coll. Cardiol. 53, 1993-2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H., Jones P. P., Koop A., Zhang L., Duff H. J., Chen S. R. W. (2008). Caffeine induces Ca2+ release by reducing the threshold for luminal Ca2+ activation of the ryanodine receptor. Biochem. J. 414, 441-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen P. J., Brown K. M., Piippo K., Swan H., Devaney J. M., Brahmbhatt B., Donarum E. A., Marino M., Tiso N., Viitasalo M., et al. (2001). Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation 103, 485-490 [DOI] [PubMed] [Google Scholar]
- Lamb G. D., Posterino G. S., Yamamoto T., Ikemoto N. (2001). Effects of a domain peptide of the ryanodine receptor on Ca2+ release in skinned skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 281, C207-C214 [DOI] [PubMed] [Google Scholar]
- Liu Z., Wang R., Zhang J., Chen S. R. W., Wagenknecht T. (2005). Localization of a disease-associated mutation site in the three-dimensional structure of the cardiac muscle ryanodine receptor. J. Biol. Chem. 280, 37941-37947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang R., Chen S. R. W., Wagenknecht T. (2006). The amino-terminal and central mutation hotspots are adjacent to each other in the three-dimensional structure of RyR2 as revealed by cryo-electron microscopy. Biophys. J. 90, 389 [Google Scholar]
- Masumiya H., Li P., Zhang L., Chen S. R. W. (2001). Ryanodine sensitizes the Ca2+ release channel (ryanodine receptor) to Ca2+ activation. J. Biol. Chem. 276, 39727-39735 [DOI] [PubMed] [Google Scholar]
- Ochs H. R., Knuchel M. (1984). Pharmacokinetics and absolute bioavailability of diltiazem in humans. J. Mol. Med. 62, 303-306 [DOI] [PubMed] [Google Scholar]
- Papadopoulos S., Leuranguer V., Bannister R. A., Beam K. G. (2004). Mapping sites of potential proximity between the dihydropyridine receptor and RyR1 in muscle using a cyan fluorescent protein-yellow fluorescent protein tandem as a fluorescence resonance energy transfer probe. J. Biol. Chem. 279, 44046-44056 [DOI] [PubMed] [Google Scholar]
- Paul-Pletzer K., Palnitkar S. S., Jimenez L. S., Morimoto H., Parness J. (2001). The skeletal muscle ryanodine receptor identified as a molecular target of [3H]azidodantrolene by photoaffinity labeling. Biochemistry 40, 531-542 [DOI] [PubMed] [Google Scholar]
- Paul-Pletzer K., Yamamoto T., Bhat M. B., Ma J., Ikemoto N., Jimenez L. S., Morimoto H., Williams P. G., Parness J. (2002). Identification of a dantrolene-binding sequence on the skeletal muscle ryanodine receptor. J. Biol. Chem. 277, 34918-34923 [DOI] [PubMed] [Google Scholar]
- Paul-Pletzer K., Yamamoto T., Ikemoto N., Jimenez L. S., Morimoto H., Williams P. G., Ma J., Parness J. (2005). Probing a putative dantrolene-binding site on the cardiac ryanodine receptor. Biochem. J. 387, 905-909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priori S. G., Napolitano C., Tiso N., Memmi M., Vignati G., Bloise R., Sorrentino V., Danieli G. A. (2001). Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103, 196-200 [DOI] [PubMed] [Google Scholar]
- Priori S. G., Napolitano C., Memmi M., Colombi B., Drago F., Gasparini M., DeSimone L., Coltorti F., Bloise R., Keegan R., et al. (2002). Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation 106, 69-74 [DOI] [PubMed] [Google Scholar]
- Rosamond W., Flegal K., Furie K., Go A., Greenlund K., Haase N., Hailpern S. M., Ho M., Howard V., Kissela B., et al. (2008). Heart disease and stroke statistics-2008 update: A report from the American Heart Association statistics committee and stroke statistics subcommittee. Circulation 117, e25-e146 [DOI] [PubMed] [Google Scholar]
- Samso M., Wagenknecht T., Allen P. D. (2005). Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat. Struct. Mol. Biol. 12, 539-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samso M., Feng W., Pessah I. N., Allen P. D. (2009). Coordinated movement of cytoplasmic and transmembrane domains of RyR1 upon gating. PLoS Biol. 7, e1000085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva I. I., Schatz M., van Heel M., Chiu W., Hamilton S. L. (1999). Structure of the skeletal muscle calcium release channel activated with Ca2+ and AMP-PCP. Biophys. J. 77, 1936-1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva I. I., Ludtke S. J., Baker M. L., Cong Y., Topf M., Eramian D., Sali A., Hamilton S. L., Chiu W. (2008). Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl. Acad. Sci. USA 105, 9610-9615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M. R., Jeyakumar L. H., Fleischer S., Wagenknecht T. (2006). Three-dimensional visualization of FKBP12.6 binding to an open conformation of cardiac ryanodine receptor. Biophys. J. 90, 164-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtifman A., Ward C. W., Yamamoto T., Wang J., Olbinski B., Valdivia H. H., Ikemoto N., Schneider M. F. (2001). Interdomain interactions within ryanodine receptors regulate Ca2+ spark frequency in skeletal muscle. J. Gen. Physiol. 119, 15-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitsapesan R., Williams A. J. (1990). Mechanisms of caffeine activation of single calcium-release channels of sheep cardiac sarcoplasmic reticulum. J. Physiol. 423, 425-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan H., Laitinen P., Kontula K., Toivonen L. (2005). Calcium channel antagonism reduces exercise-induced ventricular arrhythmias in catecholaminergic polymorphic ventricular tachycardia patients with RyR2 mutations. J. Cardiovasc. Electrophys. 16, 162-166 [DOI] [PubMed] [Google Scholar]
- Tester D. J., Spoon D. B., Valdivia H. H., Makielski J. C., Ackerman M. J. (2004). Targeted mutational analysis of the RyR2-encoded cardiac ryanodine receptor in sudden unexplained death: A molecular autopsy of 49 medical examiner/coroner's cases. Mayo Clin. Proc. 79, 1380-1384 [DOI] [PubMed] [Google Scholar]
- Tian Q., Katz A. M., Kim D. H. (1991). Effects of azumolene on doxorubicin-induced Ca2+ release from skeletal and cardiac muscle sarcoplasmic reticulum. BBA – Mol. Cell Res. 1094, 27-34 [DOI] [PubMed] [Google Scholar]
- Valdivia H. H., Valdivia C., Ma J., Coronado R. (1990). Direct binding of verapamil to the ryanodine receptor channel of sarcoplasmic reticulum. Biophys. J. 58, 471-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelgesang B., Echizen H., Schmidt E., Eichelbaum M. (1984). Stereoselective first-pass metabolism of highly cleared drugs: studies of the bioavailability of L- and D-verapamil examined with a stable isotope technique. Br. J. Clin. Pharmacol. 18, 733-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto H., Alcalai R., Song L., Arad M., Seidman C. E., Seidman J. G., Berul C. I. (2007). Prevention of fatal arrhythmia in a catecholaminergic polymorphic ventricular tachycardia mouse model carrying calsequestrin-2 mutation. Circulation 116, 216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang L., Bolstad J., Diao N., Brown C., Ruest L., Welch W., Williams A. J., Chen S. R. W. (2003). Residue Gln4863 within a predicted transmembrane sequence of the Ca2+ release channel (ryanodine receptor) is critical for ryanodine interaction. J. Biol. Chem. 278, 51557-51565 [DOI] [PubMed] [Google Scholar]
- Wang R., Chen W., Cai S., Zhang J., Bolstad J., Wagenknecht T., Liu Z., Chen S. R. W. (2007). Localization of an NH2-terminal disease-causing mutation hot spot to the “clamp” region in the three-dimensional structure of the cardiac ryanodine receptor. J. Biol. Chem. 282, 17785-17793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch W., Rheault S., West D. J., Williams A. J. (2004). A model of the putative pore region of the cardiac ryanodine receptor channel. Biophys. J. 87, 2335-2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Ikemoto N. (2002). Peptide probe study of the critical regulatory domain of the cardiac ryanodine receptor. Biochem. Biophys. Res. Comm. 291, 1102-1108 [DOI] [PubMed] [Google Scholar]
- Yamamoto T., El-Hayek R., Ikemoto N. (2000). Postulated role of interdomain interaction within the ryanodine receptor in Ca2+ channel regulation. J. Biol. Chem. 275, 11618-11625 [DOI] [PubMed] [Google Scholar]
- Yano M., Kobayashi S., Kohno M., Doi M., Tokuhisa T., Okuda S., Suetsugu M., Hisaoka T., Obayashi M., Ohkusa T., et al. (2003). FKBP12.6-mediated stabilization of calcium-release channel (ryanodine receptor) as a novel therapeutic strategy against heart failure. Circulation 107, 477-484 [DOI] [PubMed] [Google Scholar]
- Yano M., Yamamoto T., Ikemoto N., Matsuzaki M. (2005). Abnormal ryanodine receptor function in heart failure. Pharmacol. Ther. 107, 377-391 [DOI] [PubMed] [Google Scholar]
- Zchut S., Feng W., Shoshan-Barmatz V. (1996). Ryanodine receptor/calcium release channel conformations as reflected in the different effects of propranolol on its ryanodine binding and channel activity. Biochem. J. 315, 377-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu Z., Masumiya H., Wang R., Jiang D., Li F., Wagenknecht T., Chen S. R. W. (2003). Three-dimensional localization of divergent region 3 of the ryanodine receptor to the clamp-shaped structures adjacent to the FKBP binding sites. J. Biol. Chem. 278, 14211-14218 [DOI] [PubMed] [Google Scholar]
- Zhao M., Li P., Li X., Zhang L., Winkfein R. J., Chen S. R. W. (1999). Molecular identification of the ryanodine receptor pore-forming segment. J. Biol. Chem. 274, 25971-25974 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Wang Q. L., Meng X., Shu Y., Jiang T., Wagenknecht T., Yin C. C., Sui S. F., Liu Z. (2008). Structural and functional characterization of ryanodine receptor-natrin toxin interaction. Biophys. J. 95, 4289-4299 [DOI] [PMC free article] [PubMed] [Google Scholar]