Abstract
Idiopathic hypogonadotropic hypogonadism (IHH) has an incidence of 1–10 cases per 100,000 births. About 60% of patients with IHH present with associated anosmia, also known as Kallmann syndrome, characterized by total or partial loss of olfaction. Many of the gene mutations associated with Kallmann syndrome have been mapped to KAL1 or FGFR1. However, together, these mutations account for only about 15% of Kallmann syndrome cases. More recently, mutations in PROK2 and PROKR2 have been linked to the syndrome and may account for an additional 5–10% of cases. The remaining 40% of patients with IHH have a normal sense of smell. Prior to 2003, the only gene linked to normosmic IHH was the gonadotropin-releasing hormone receptor gene. However, mutations in this receptor are believed to account for only 10% of cases. Subsequently, mutations in KISS1R, TAC3 and TACR3 were identified as causes of normosmic IHH. Certain genes, including PROK2 and FGFR1, are associated with both anosmic and normosmic IHH. Despite recent advances in the field, the genetic causes of the majority of cases of IHH remain unknown. This Review discusses genes associated with hypogonadotropic disorders and the molecular mechanisms by which mutations in these genes may result in IHH.
Introduction
Idiopathic hypogonadotropic hypogonadism (IHH) is characterized by delayed or absent sexual development associated with inappropriately low gonadotropin and sex steroid levels in the absence of anatomical or functional abnormalities of the hypothalamic–pituitary–gonadal axis. The major underlying cause of IHH is failure to activate pulsatile secretion of gonadotropin-releasing hormone (GnRH) during puberty, a developmental stage characterized by a substantial increase in the frequency and amplitude of pulses of this hormone.
To date, the greatest insights into the molecular mechanisms that regulate activation of GnRH have been provided by the identification and study of genetic abnormalities in patients with pubertal disorders or infertility.1 Other abnormal phenotypes that are commonly associated with and segregate with hypogonadism in affected families have also helped to identify the underlying molecular defects. The most common associated phenotype is anosmia, an inability to perceive smells, which is explained by the common embryonic origins and developmental pathways of GnRH and olfactory neurons. The development of GnRH neurons is unusual in that they originate outside the brain. Similar to the development of olfactory fibers, GnRH neurons are formed in the nasal placode (olfactory epithelium), from which they migrate by use of the olfactory pathway to help guide them to their ultimate destination in the hypothalamus.2
Impaired migration of GnRH and olfactory neurons is the underlying cause of Kallmann syndrome (IHH associated with anosmia). Accordingly, gene mutations associated with Kallmann syndrome encode proteins that regulate GnRH and olfactory neuronal migration. Other associated neurological and somatic abnormalities, such as synkinesia, cerebellar ataxia, sensorineural deafness, mental retardation, unilateral renal agenesis and cleft palate, may segregate with the Kallmann syndrome phenotype, which suggests a common genetic origin of these abnormalities.3 The genetic mechanisms that underlie IHH in patients with a normal sense of smell are diverse and may involve genes that regulate development and/or GnRH secretion or action.
In some cases, the severity of hypogonadism and the presence of associated phenotypes in a pedigree present in a dimorphic distribution with incomplete penetrance. This pattern suggests that changes may occur in more than one gene and/or that sex-associated modifying factors may contribute to the phenotype.4–6 Evidence for the contribution of sex-associated factors in IHH is reinforced by the 5:1 male predominance of the disorder. In addition, male patients often present with a more severe phenotype than affected female individuals within a given family.
In this Review, we will discuss genes associated with IHH. The genes have been categorized according to whether the proteins they encode are involved in the development and migration of GnRH neurons, regulation of GnRH secretion or GnRH action (Table 1, Figure 1). We have focused primarily on genes that encode ligand–receptor pairs. Functional characterization of these mutations has shed light on the pathophysiology of IHH. Additional genes implicated in IHH include NELF (nasal embryonic LHRH factor) and genes that encode a host of transcription factors, most notably nuclear receptor DAX-1 and steroidogenic factor 1, but discussion of these is beyond the scope of this Review.
Table 1.
The genetic basis of idiopathic hypogonadotropic hypogonadism
| Genes related to IHH | Gene product | Mode of inheritance | Cases for this specific form of IHH (%) | Cases of IHH overall (%) |
|---|---|---|---|---|
| Kallmann syndrome ~60% of total | ||||
| KAL1 | Anosmin 1 | X-linked | 5–10 | 3–6 |
| FGFR1 | Fibroblast growth factor receptor 1 | Autosomal dominant | 10 | 6 |
| FGF8 | Fibroblast growth factor 8 | Autosomal dominant | <5 | <2 |
| PROK2, PROKR2 | Prokineticin 2, prokineticin receptor 2 | Autosomal recessive | 5–10 | 3–6 |
| CHD7 | Chromodomain-helicase-DNA-binding protein 7 | NR | 10 | 6 |
| Unidentified | NA | NA | 60–75 | NR |
| Normosmic IHH ~40% of total | ||||
| GNRHR | Gonadotropin-releasing hormone receptor | Autosomal recessive | 16–40 | 6.5–16 |
| KISS, KISS1R | Kisspeptin 1, KiSS-1 receptor | Autosomal recessive | 5 | 2 |
| TAC3, TACR3 | Neurokinin B, Neurokinin B receptor | Autosomal recessive | NR | NR |
| LEP, LEPR | Leptin, leptin receptor | Autosomal recessive | <5 | <2 |
| PCSK1 | Neuroendocrine convertase 1 | Autosomal recessive | <5 | <2 |
| Unidentified | NA | NA | ~50 | NR |
IHH incidence in the population is 1–10 cases in 100,000 births. Patients with Kallmann syndrome have partial or total loss of sense of smell, whereas in normosmic IHH, sense of smell is not affected. In the unidentified group, the genetic basis of the IHH phenotype has not been identified. Abbreviations: IHH, idiopathic hypogonadotropic hypogonadism; NA, not applicable; NR, not reported.
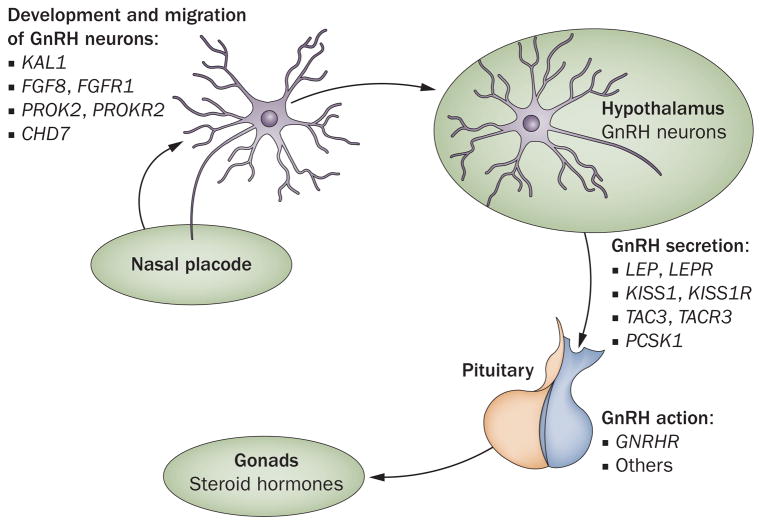
Figure 1.
The genetic basis of idiopathic hypogonadotropic hypogonadism. The mechanisms that underlie hypogonadotropic hypogonadism involve mutations in many genes. Some of these genes encode proteins that regulate GnRH neuronal migration, other genes encode proteins that regulate GnRH secretion or GnRH action. Abbreviation: GnRH, gonadotropin-releasing hormone.
Migration of GnRH neurons
KAL1
KAL1 is located on the X chromosome and encodes an extracellular cell adhesion protein, anosmin 1, essential for axonal guidance and migration of olfactory and GnRH neurons from the nasal placode to their final location in the brain.7 Anosmin 1 co-localizes with basic fibroblast growth factor receptor 1 (FGFR-1, discussed further below), in the olfactory bulb during development, which suggests that this protein is a component of FGFR-1 signaling.8 Mutations in KAL1 disrupt the migration of olfactory and GnRH neurons and are reported to be present in approximately 5–10% of patients with Kallmann syndrome (Table 1),7,9–11 with the highest prevalence seen in families with X-linked inheritance patterns.
The KAL1 gene comprises 14 exons and encodes a 680 amino acid protein with a complex structure that includes an N-terminal signal peptide followed by a cysteine-rich region, a whey acidic protein-like domain, four tandem repeats of fibronectin-like type III domains, and a C-terminal histidine-rich region.12 The KAL1 mutations identified to date predominantly consist of nucleotide deletions or insertions that result in sequence frameshifts or the introduction of premature stop codons. Less than 20% of the different mutations associated with Kallmann syndrome result in amino acid substitutions.13 Two amino acid changes in the whey acidic protein-like domain have been described: Cys163Tyr and Cys172Arg. This domain contains two conserved disulfide bond motifs (Cys151–Cys163 and Cys157–Cys172). The Cys163Tyr and Cys172Arg mutations are expected to prevent disulfide bond formation, which disrupts the tertiary structure of anosmin 1 and might result in misfolding and/or destabilization of the protein.
All the other amino acid substitutions in anosmin 1 that are associated with Kallmann syndrome are located in the fibronectin-like type III domains. All four of these domains have N-glycosylation sites as well as potential heparin binding sites. Two mutations (Asn267Lys and Glu514Lys) are expected to enhance heparin binding, which compromises the flexibility of anosmin 1.13 Reversal of hypogonadism has been described in a patient with a frameshift mutation in KAL1 that introduced a premature stop codon at amino acid 85. If expressed, this mutant anosmin 1 encodes a truncated protein that lacks 525 of its 610 amino acids. Reversibility of the phenotype in this case raises the possibility that this truncated mutant may somehow retain some biological activity, or that other pathways may compensate for the lack of activity of the defective anosmin 1.14
FGFR1
The protein encoded by FGFR1—FGFR-1—is a member of the tyrosine kinase superfamily of receptors. FGFR-1 contains an extracellular domain that has three immunoglobulin-like domains (D1, D2 and D3); these domains dictate the receptor’s affinity and specificity for its ligands. The protein also includes a single transmembrane helix and an intracellular domain with tyrosine kinase activity. In the presence of heparin sulfate, fibroblast growth factor binds FGFR-1 with high affinity, stimulating receptor dimerization and trans-autophosphorylation of tyrosine residues in the intracellular domain. These steps result in activation of downstream signaling. The major signaling pathway activated is the mitogen-activated protein kinase pathway. FGFR-1 signaling through this pathway regulates neuronal migration, differentiation, and survival, as well as cell proliferation during embryonic development.15–17
Kallmann syndrome caused by mutations in FGFR1 is typically transmitted in an autosomal dominant fashion and can be associated with failed morphogenesis of the olfactory bulbs, cleft palate and dental agenesis. The severity of the hypogonadism and the presence of associated phenotypes have variable expressivity with incomplete penetrance.18 Mutations in FGFR1 account for approximately 10% of Kallmann cases (Table 1). Unlike the situation with KAL1 mutations, mutations in FGFR1 have been reported in patients with IHH without anosmia, termed normosmic IHH.
The majority of FGFR1 mutations associated with Kallmann syndrome are single amino acid substitutions located in the immunoglobulin-like domains or tyrosine kinase domain. A Gly237Asp substitution in the second immunoglobulin-like domain removes a highly conserved glycine that is predicted to maintain structural integrity of the protein. In vitro studies indicate that a serine substitution at this position destabilizes the receptor, which results in misfolding and retention of the protein in the endoplasmic reticulum.4 An Asp224His substitution, also in the second immunoglobulin-like domain, is believed to prevent ligand-induced or heparin-sulfate-induced FGFR-1 dimerization and signaling.5 Amino acid substitutions that map to the third immunoglobulin-like domain of FGFR-1, such as Val273Met, Glu274Gly, Tyr339Cys, and Ser346Cys, are predicted to disrupt a characteristic Cys277–Cys341 disulfide bond in this domain.4,5 Mutations in the tyrosine kinase domain of FGFR-1 are predicted to decrease or inhibit kinase activity by disrupting receptor conformation (Ile538Val, Asn724Lys, and Gly703Arg) and/or altering the normal pattern of phosphorylation of the kinase domain (Ala520Thr, Gly703Ser, Pro722Ser, Pro745Ser, and Pro772Ser).5,13,17
Reversal of IHH after testosterone treatment has been reported in a patient harboring a heterozygous mutation in the tyrosine kinase domain of FGFR-1 (Arg622X). The creation of the stop codon in the tyrosine kinase domain by this mutation results in a truncated protein.19 Two family members carrying the same mutation were reported to have delayed puberty (mother) and anosmia (maternal grandfather). Reversal of IHH in this case suggests that FGFR-1 might regulate reproduction beyond the stage of embryonic GnRH neuronal migration.19 The reversal also raises the possibility that hormone replacement with testosterone may provide the assistance needed for compensation for GnRH deficiency in such cases.
Mutations in the gene FGF8, which encodes one of the many ligands of FGFR-1, have recently been linked to Kallmann syndrome. This finding suggests that FGF-8 is the FGFR-1 ligand responsible for GnRH neuronal development and/or migration.20
PROK2 and PROKR2
PROKR2 encodes a G-protein-coupled receptor, prokineticin receptor 2 (PK-R2), and PROK2 encodes one of its ligands, prokineticin-2 (PK2). When PK2 binds to PK-R2, signaling cascades that are important for development of the olfactory system and GnRH neuronal progenitors are initiated.21–24 Mutations in PK2 or PK-R2 are estimated to account for 5–10% of all Kallmann syndrome cases (Table 1). Similar to patients with PROKR2 mutations, Prokr2−/− mice exhibit olfactory bulb hypoplasia, hypogonadotropic hypogonadism, and a lack of GnRH neurons in the hypothalamus.25 Likewise, Prok2−/− mice fail to enter puberty, have low gonadotropin levels, and have a significant decrease in hypothalamic GnRH neurons.25,26 Interestingly, GnRH neurons in Prok2−/− mice were able to cross the cribriform plate (a horizontal plate of the ethmoid bone) to enter the central nervous system during embryonic development, but the neurons did not migrate into and populate the hypothalamus.24,26
Human mutations in PROK2 and PROKR2 have been found in the heterozygous, homozygous, or compound heterozygous state.26–30 Frameshift mutations that inactivate PK2 have been reported in sporadic cases of Kallmann syndrome28 and in families with a history of Kallmann syndrome or normosmic IHH.26,30 Three siblings (two brothers with Kallmann syndrome and a sister with normosmic IHH) have been identified who have a homozygous deletion in the PROK2 gene.26 The frameshift mutation introduces a premature stop codon, which results in the production of a truncated PK2 protein that lacks the cysteine-rich domains that are critical for the protein’s biological activity.31 A single amino acid substitution in PK2 (Arg73Cys) that greatly reduces activation of PK-R2-mediated stimulation of calcium signaling is predicted to disrupt a disulfide bond. This substitution has been identified in both the homozygous and the heterozygous state in patients with Kallmann syndrome.28
Many of the mutations identified in PROKR2 map to the highly conserved transmembrane domains of the receptor encoded by this gene. In a study of 192 patients with Kallmann syndrome, 10 mutations in PROKR2 were identified,27 five (50%) of which resulted in single amino acid substitutions in the transmembrane domains (Leu173Arg, Trp178Ser, Pro290Ser, Met323Ile, and Val331Met). Trp178 and Pro290 are the most conserved amino acids in transmembrane domains four and six, respectively. In vitro studies have shown that substitution of Leu173, Trp178, and Pro290 in PROKR2 impairs the cell surface targeting of the receptor, probably because the receptor protein is incorrectly folded. Calcium signaling by these mutated receptors is equally impaired.29,32 Other mutants with amino acid substitutions in the first, second and third intracellular loops, or in the carboxyl tail of PK-R2 have similarly impaired calcium signaling but normal expression levels and cell surface targeting. These mutants probably have impaired G-protein coupling.27,29,32
Interestingly, in mice, the IHH phenotype has been reported only in those homozygous for the Prok2 deletion, which suggests that one normal allele may compensate for the inactivity of the other. Similarly, homozygous mutations in PROK226,29 or PROKR230 have been found in patients with Kallmann syndrome who have asymptomatic parents or other family members with heterozygous mutations of this ligand or receptor. The apparent requirement of two mutant alleles for the phenotypic manifestation raises the possibility that an additional mutation may be present in patients with Kallmann syndrome in whom only a single heterozygous mutation in PROK2 or PROKR2 has been identified to date.
CHD7
The gene CHD7 encodes chromodomain-helicase-DNA-binding protein 7 (CHD-7). CHD7 is expressed in olfactory epithelium, the hypothalamus and the pituitary gland, which suggests that the encoded protein may have a role in olfactory bulb and GnRH neuronal development. Mutations in CHD7 are predicted to account for 6% of all IHH cases (Table 1).33 These mutations are hypothesized to be involved in the pathogenesis of IHH by disruption of GnRH neuronal development or migration. Mutations in CHD7 have been found in patients with CHARGE syndrome, a multisystem disorder that may include IHH. Furthermore, seven mutations in CHD7 were recently identified in patients with sporadic, normosmic IHH or Kallmann syndrome who were not diagnosed as having CHARGE syndrome.33 Three of these mutations affect domains critical for chromatin remodeling and transcription regulation by CHD-7. These mutations are believed to result in a milder form of CHARGE syndrome that manifests as normosmic IHH or Kallmann syndrome.
Regulation of GnRH secretion
KISS1 and KISS1R
The reproductive roles of the G-protein-coupled KiSS-1 receptor (also known as GPR54, encoded by KISS1R) and its endogenous ligand, kisspeptin 1 (encoded by KISS1) were revealed in 2003 when inactivating mutations in the KiSS-1 receptor were found in the homozygous state in members of two large, consanguineous families with a history of normosmic IHH.34,35 Subsequent studies have shown that both the ligand and receptor are expressed in areas of the hypothalamus that control reproduction.36 Furthermore, centrally or peripherally injected kisspeptin 1 results in a robust stimulation of GnRH and gonadotropin secretion in the rodent36–38 and primate39 and accelerates puberty in prepubertal rodents.40 In fact, kisspeptin 1 is recognized as the most potent regulator of GnRH secretion in humans and animal models. Kisspeptin 1 was identified by three independent groups in 2001 as a 54-amino acid peptide—kisspeptin 54—which corresponds to residues 68–121 of the KISS1 gene product; however, shorter peptides of kisspeptin also show biological activity.41–43
In rodents, the hypothalamic regions with the highest levels of kisspeptin-expressing neurons are the arcuate nucleus and the anteroventral periventricular nucleus. Kisspeptin 1 expression increases at the time of puberty in both the arcuate and the anteroventral periventricular nuclei. Levels in the latter are significantly higher in female rodents.44,45 Migration of GnRH neurons and GnRH content in the hypothalamus of mice with targeted disruption of KISS1R is normal. The hypothalamic–pituitary axis of these mice is functional, responding with robust luteinizing hormone secretion after GnRH treatment, which suggests that GnRH is the mediator of the stimulatory effects of kisspeptins on gonadotropin secretion.46 Indeed, Kiss1r−/− mice have underdeveloped gonads, low gonadotropins and steroid hormone levels, and incomplete gametogenesis.35,47 Kiss1−/− mice have a similar phenotype, which suggests that kisspeptins are the only physiological ligands of the KiSS-1 receptor.48
Inactivating mutations in KISS1R show an autosomal recessive pattern of transmission. Mutations in KISS1R represent about 5% of cases of normosmic IHH (Table 1). In 2003, de Roux et al. reported a mutation of KISS1R in a family with individuals with normosmic IHH.34 This deletion mutation eliminates most of the third cytoplasmic loop of the KiSS-1 receptor. Loss of this loop is predicted to impair the receptor’s G-protein coupling even if the receptor is correctly localized in the membrane. In a different family with individuals with normosmic IHH, a single amino acid substitution mutation (Leu148Ser) of the KiSS-1 receptor was identified.35 This substitution is located near the DRW(Y) motif, a motif that has an important role in regulating the activation and inactivation of many G-protein-coupled receptors.49,50 Another single amino acid substitution of the KiSS-1 receptor (Leu102Pro) identified in patients with IHH is located in the first extracellular loop of the receptor. The mutation decreases the expression of the receptor and impairs its signaling ability.51
A compound heterozygous mutation identified in a patient with IHH included an insertion of a stop codon at 331Arg in one allele and the replacement of the normal stop codon by an arginine in the other allele (X399Arg). The introduction of a stop codon at 331Arg is expected to generate a truncated KiSS-1 receptor. On the other hand, elimination of the normal stop codon might result in receptor misfolding. Accordingly, in vitro studies showed impaired function of both the Arg331X and X399Arg mutant KiSS-1 receptors.35 Another pair of compound heterozygous mutations (Arg297Leu and Cys223Arg) was identified in a boy with hypogonadism. Cys223 is a highly conserved amino acid located at the beginning of the third intracellular loop of the KiSS-1 receptor. Accordingly, receptor signaling by this mutated protein is profoundly impaired. Although the mutation in the other allele—Arg297Leu—only slightly decreased the activity of the KiSS-1 receptor, the combination of the two mutated alleles was enough to cause hypogonadism.52
The key role of the KiSS-1 receptor in the regulation of the onset of puberty was recently reinforced when the first identifiable genetic cause of central precocious puberty was recognized as a gain-of-function mutation in the receptor.53 The gain-of-function in this case was not achieved by constitutive activation of the KiSS-1 receptor. Instead, the apparent ‘activation’ of the receptor was the result of a reduced rate of receptor desensitization. Mutations in kisspeptin 1 might be expected to alter reproductive function in a similar fashion to those in the KiSS-1 receptor; however, no reports of kisspeptin mutations associated with IHH have been published to date.
LEP and LEPR
Leptin, encoded by LEP, is a fat-derived hormone that regulates food intake, energy expenditure and reproduction at the hypothalamic level. Exogenous administration of leptin accelerates puberty in mice and normalizes reproductive deficiencies in leptin-null (ob/ob) mice, which suggests that leptin may be a link between body fat and reproductive capability.54,55 Accordingly, loss of body fat owing to starvation or excessive exercise is known to suppress reproduction and results in amenorrhea and infertility.56,57 Likewise, inactivating mutations in LEP58 or the gene encoding its receptor, LEPR,59 have been found in patients with severe obesity and hypo-gonadotropic hypogonadism. These inactivating mutations account for less than 5% of normosmic IHH and have an autosomal recessive pattern of transmission (Table 1). The central role of leptin in these cases is highlighted by the recovery of gonadotropin secretion and menstrual cycles after treatment with recombinant leptin in females with amenorrhea due to congenital leptin deficiency60 or hypothalamic amenorrhea.61
In humans, leptin has a permissive role in the control of reproduction, being necessary but not sufficient for the onset of puberty and maintenance of fertility. Evidence from studies of animal models points to a role for kisspeptin 1 in the reproductive effects of leptin.62
TAC3 and TACR3
Neurokinin B (NKB), which is encoded by TAC3, is a member of the substance-P-related tachykinin family. NKB is expressed in the arcuate nucleus and its receptor is expressed in GnRH neurons of rodents, which suggests it has a possible role in the regulation of GnRH secretion.63 The NKB receptor, NKR (also known as NK-3R) is encoded by TACR3. This receptor is a member of the rhodopsin family of G-protein-coupled receptors.
A recent genome-wide single nucleotide polymorphism analysis identified homozygous mutations in TAC3 or TACR3 that segregated with the IHH phenotype in four (of nine) unrelated consanguineous families. In one family, a Met90Thr amino acid substitution of NKB affected individuals homozygous for the mutation but not those who were heterozygous. This mutation is located within the canonical tachykinin motif in the carboxyl terminus of the mature ligand. The tachykinin motif is predicted to undergo post-translational amidation for full activation of NKB.63 In vitro studies showed that the stimulation of NKR by NKB is markedly impaired by the Met90Thr substitution.63
Two mutations found in TACR3 replace highly conserved amino acids located in the first (Gly93Asn) or sixth (Pro353Ser) transmembrane domain of NKR. The Gly93Asn mutation was found in all affected members of one family, whereas the Pro353Ser mutation was found in all affected members of two other families.63 In vitro studies showed that these two mutations impair the ability of NKR to activate signal transduction.63
Taken together, these results strongly suggest that NKB and NKR have important roles as central regulators of reproductive capability. While the underlying mechanism has not been established, it probably involves the regulation of GnRH secretion, perhaps via interactions with the kisspeptin 1–KiSS-1 receptor system given the co-expression of NKB with kisspeptin in neurons in the arcuate nucleus of the hypothalamus.64
PCSK1
Neuroendocrine convertases (also known as prohormone convertases) are enzymes that process larger, usually inactive, precursor peptides to release bioactive fragments. For example, the pro-opiomelanocortin gene product is processed by neuroendocrine convertase 1 (NEC 1; encoded by PCSK1) in the corticotroph to produce adrenocorticotropic hormone and lipotropin and by neuroendocrine convertase 2 in the hypothalamus to produce melanocyte-stimulating hormone and endorphin. The first report of a patient with congenital NEC 1 deficiency associated with obesity and hypogonadotropic hypogonadism was published in 1995.65 A compound heterozygous mutation in PCSK1 was later identified in this patient in which a Gly483Arg substitution impairs the maturation of the inactive NEC 1 propeptide so that the propeptide is trapped in the endoplasmic reticulum. The second mutation causes a frameshift and the introduction of a premature stop codon in the catalytic domain of the enzyme.66
Only two other cases with a similar phenotype associated with PCSK1 mutations have been identified in humans. A compound heterozygous mutation (Glu250X and Ala213del) was identified in a female infant. Glu250X is predicted to truncate the protein within its catalytic domain, whereas the mutation in the other allele (Ala213del) eliminates a highly conserved alanine near His208, a residue known to be critical for catalytic activity.67 The second case involved a 6-year-old boy with a homozygous Ser307Leu substitution. This mutation is also within the catalytic domain and severely impairs the catalytic activity of the enzyme.68 On the basis of phenotypic similarities, infertility is expected in these two patients once they reach pubertal age. Accordingly, mice homozygous for an Asn222Asp NEC 1 substitution, which is also within the catalytic domain of the enzyme, are obese and have decreased fertility.69 Although no reports have been published on the molecular mechanisms by which mutations in PCSK1 cause hypogonadism, the impairment of GnRH prohormone precursor processing is probably involved in these cases.
GnRH action: GNRHR
The GnRH receptor (encoded by GNRHR) is a 328 amino acid protein that belongs to the rhodopsin family of G-protein-coupled receptors. Activation of the GnRH receptor increases calcium mobilization and stimulates influx of extracellular calcium. This increase in intracellular calcium induces pituitary luteinizing hormone and follicle-stimulating hormone secretion.70 Germ-line mutations in GNRHR were among the first genetic mutations identified in patients with IHH.71 During the past 10 years, at least 19 additional loss-of-function mutations in the GnRH receptor have been identified in patients with IHH.72 Large-scale screening indicates that GNRHR mutations account for 3.5–16% of sporadic cases of normosmic IHH and up to 40% of familial cases of normosmic IHH (Table 1).72 Inheritance is autosomal recessive and most patients have compound heterozygous GNRHR mutations.70
All but two known GNRHR mutations are missense, leading to single amino acid substitutions. These mutations impair GnRH signaling through loss of receptor expression, ligand binding, G-protein coupling, and/or abnormal intracellular trafficking of the receptor. About 82% of all transmembrane domain mutations cause complete impairment of ligand binding and signaling. Only two of 11 mutations identified in transmembrane domains encode a partially functional receptor; the other nine encode completely inactive receptors. On the other hand, only one of the mutations reported in other domains encodes a completely inactive receptor.
Some GNRHR mutations with partial effects on signaling were found in patients with compound heterozygous mutations, such as Gln106Arg–Arg262Gln. Gln106Arg maps to the first extracellular loop of the GnRHR and decreases GnRH binding.71 The Arg262Gln substitution is located in the third intracellular loop, the main site of G-protein interaction. This mutation impairs signal transduction, suggesting that activation of G proteins is affected. The parents of the affected siblings were phenotypically normal, despite each being heterozygous for one of these substitutions.73 Functional responses and structure–function relationships of GnRH receptor mutations have been described in detail.72,74 The reversal of IHH in a patient carrying a homozygous mutation in GNRHR has been reported by Pitteloud and colleagues.75 The reversal of the phenotype after treatment suggests that hormonal replacement may compensate for an underlying genetic abnormality in cases where the mutations result in only partial loss of function.
Conclusions
Mutations in a number of genes have been identified in patients as the primary genetic cause of IHH. These genes encode proteins that regulate GnRH neuronal development, migration from the nasal placode to the hypothalamus, GnRH secretion, or GnRH action. While some genetic mutations are sufficient to cause IHH, other cases are predicted to result from the combination of more than one genetic abnormality. In such cases, the partial loss of function caused by one genetic mutation may not manifest with a reproductive phenotype, whereas the presence of a second mutation overwhelms the ability to compensate for the effects of the first mutation, resulting in IHH. Many of the genetic causes of IHH have been identified only recently and their individual and/or combined roles in the regulation of reproduction are not yet completely understood. Despite these recent advances, the genetic basis of the vast majority of cases of IHH remains unknown, and much room remains for further discovery.
Key points
The genetic basis of the vast majority of cases of idiopathic hypogonadotropic hypogonadism (IHH) remains unknown
IHH with anosmia (Kallmann syndrome) is caused by the defective developmental migration of gonadotropin-releasing hormone (GnRH) and olfactory neurons; associated genes encode proteins involved in this migration
Molecular mechanisms that underlie IHH in patients with a normal sense of smell are diverse and may involve genes that regulate development and/or GnRH secretion or action
A considerable number of IHH cases probably involve mutations in more than one gene; that is, these cases are polygenic
Sex-associated factors also contribute to the IHH phenotype
Review criteria
Literature searches of Google Scholar and MEDLINE databases from 1990 to 2009 were based on the keywords: “IHH”, “hypogonadism”, “Kallmann syndrome”, “anosmia”, “FGFR1”, “KAL1”, “KAL2”, “GnRH”, “GPR54”, “kisspeptin”, “metastin”, “PROK2”, “PROKR2”, “leptin”, “leptin receptor”, and “prohormone convertase”.
Acknowledgments
This work was supported in part by grants from the NICHD/NIH through cooperative agreement U54 HD28138 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (U. B. Kaiser), NIH R01 HD19938 (U. B. Kaiser), NIH BIRCWH K12HD051959 (S. D. C. Bianco), NIH R21 HD059015-01 (S. D. C. Bianco) and by the Charles H. Hood Foundation Child Health Research Award (S. D. C. Bianco).
Footnotes
Competing interests
The authors declare no competing interests.
Contributor Information
Suzy D. C. Bianco, Department of Molecular and Cellular Pharmacology, Leonard M. Miller School of Medicine, University of Miami, Miami, FL, USA
Ursula B. Kaiser, Division of Endocrinology, Diabetes and Hypertension, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA
References
- 1.Cadman SM, Kim SH, Hu Y, González-Martínez D, Bouloux PM. Molecular pathogenesis of Kallmann’s syndrome. Horm Res. 2007;67:231–242. doi: 10.1159/000098156. [DOI] [PubMed] [Google Scholar]
- 2.Schwanzel-Fukuda M. Origin and migration of luteinizing hormone-releasing hormone neurons in mammals. Microsc Res Tech. 1999;44:2–10. doi: 10.1002/(SICI)1097-0029(19990101)44:1<2::AID-JEMT2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Cariboni A, Maggi R. Kallmann’s syndrome, a neuronal migration defect. Cell Mol Life Sci. 2006;63:2512–2526. doi: 10.1007/s00018-005-5604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitteloud N, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2006;103:6281–6286. doi: 10.1073/pnas.0600962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitteloud N, et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol. 2006;254–255:60–69. doi: 10.1016/j.mce.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 6.Pitteloud N, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Legouis R, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 8.Ayari B, Soussi-Yanicostas N. FGFR1 and anosmin-1 underlying genetically distinct forms of Kallmann syndrome are co-expressed and interact in olfactory bulbs. Dev Genes Evol. 2007;217:169–175. doi: 10.1007/s00427-006-0125-0. [DOI] [PubMed] [Google Scholar]
- 9.Franco B, et al. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 10.Hardelin JP, et al. Heterogeneity in the mutations responsible for X chromosome-linked Kallmann syndrome. Hum Mol Genet. 1993;2:373–377. doi: 10.1093/hmg/2.4.373. [DOI] [PubMed] [Google Scholar]
- 11.Albuisson J, et al. Kallmann syndrome: 14 novel mutations in KAL1 and FGFR1 (KAL2) Hum Mutat. 2005;25:98–99. doi: 10.1002/humu.9298. [DOI] [PubMed] [Google Scholar]
- 12.Soussi-Yanicostas N, et al. Initial characterization of anosmin-1, a putative extracellular matrix protein synthesized by definite neuronal cell populations in the central nervous system. J Cell Sci. 1996;109:1749–1757. doi: 10.1242/jcs.109.7.1749. [DOI] [PubMed] [Google Scholar]
- 13.Kim SH, Hu Y, Cadman S, Bouloux P. Diversity in fibroblast growth factor receptor 1 regulation: learning from the investigation of Kallmann syndrome. J Neuroendocrinol. 2008;20:141–163. doi: 10.1111/j.1365-2826.2007.01627.x. [DOI] [PubMed] [Google Scholar]
- 14.Ribeiro RS, Vieira TC, Abucham J. Reversible Kallmann syndrome: report of the first case with a KAL1 mutation and literature review. Eur J Endocrinol. 2007;156:285–290. doi: 10.1530/eje.1.02342. [DOI] [PubMed] [Google Scholar]
- 15.Ford-Perriss M, Abud H, Murphy M. Fibroblast growth factors in the developing central nervous system. Clin Exp Pharmacol Physiol. 2001;28:493–503. doi: 10.1046/j.1440-1681.2001.03477.x. [DOI] [PubMed] [Google Scholar]
- 16.Gill JC, Moenter SM, Tsai PS. Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology. 2004;145:3830–3839. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- 17.Böttcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 18.Salenave S, et al. Kallmann’s syndrome: a comparison of the reproductive phenotypes in men carrying KAL1 and FGFR1/KAL2 mutations. J Clin Endocrinol Metab. 2008;93:758–763. doi: 10.1210/jc.2007-1168. [DOI] [PubMed] [Google Scholar]
- 19.Pitteloud N, et al. Reversible Kallmann syndrome, delayed puberty, and isolated anosmia occurring in a single family with a mutation in the fibroblast growth factor receptor 1 gene. J Clin Endocrinol Metab. 2005;90:1317–1322. doi: 10.1210/jc.2004-1361. [DOI] [PubMed] [Google Scholar]
- 20.Falardeau J, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Bullock CM, Knauer DJ, Ehlert FJ, Zhou QY. Identification of two prokineticin cDNAs: recombinant proteins potently contract gastrointestinal smooth muscle. Mol Pharmacol. 2001;59:692–698. doi: 10.1124/mol.59.4.692. [DOI] [PubMed] [Google Scholar]
- 22.Lin DC, et al. Identification and molecular characterization of two closely related G protein-coupled receptors activated by prokineticins/endocrine gland vascular endothelial growth factor. J Biol Chem. 2002;277:19276–19280. doi: 10.1074/jbc.M202139200. [DOI] [PubMed] [Google Scholar]
- 23.Cheng MY, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 24.Prosser HM, Bradley A, Caldwell MA. Olfactory bulb hypoplasia in Prokr2 null mice stems from defective neuronal progenitor migration and differentiation. Eur J Neurosci. 2007;26:3339–3344. doi: 10.1111/j.1460-9568.2007.05958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto S, et al. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc Natl Acad Sci USA. 2006;103:4140–4145. doi: 10.1073/pnas.0508881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitteloud N, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dodé C, et al. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leroy C, et al. Biallelic mutations in the prokineticin-2 gene in two sporadic cases of Kallmann syndrome. Eur J Hum Genet. 2008;16:865–868. doi: 10.1038/ejhg.2008.15. [DOI] [PubMed] [Google Scholar]
- 29.Cole LW, et al. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93:3551–3559. doi: 10.1210/jc.2007-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abreu AP, et al. Loss-of-function mutations in the genes encoding prokineticin-2 or prokineticin receptor-2 cause autosomal recessive Kallmann syndrome. J Clin Endocrinol Metab. 2008;93:4113–4118. doi: 10.1210/jc.2008-0958. [DOI] [PubMed] [Google Scholar]
- 31.Bullock CM, Li JD, Zhou QY. Structural determinants required for the bioactivities of prokineticins and identification of prokineticin receptor antagonists. Mol Pharmacol. 2004;65:582–588. doi: 10.1124/mol.65.3.582. [DOI] [PubMed] [Google Scholar]
- 32.Monnier C, et al. PROKR2 missense mutations associated with Kallmann syndrome impair receptor signalling activity. Hum Mol Genet. 2009;18:75–81. doi: 10.1093/hmg/ddn318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HG, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Roux N, et al. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seminara SB, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 36.Gottsch ML, et al. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- 37.Irwig MS, et al. Kisspeptin activation of gonadotropin releasing hormone neurons and regulation of KiSS-1 mRNA in the male rat. Neuroendocrinology. 2004;80:264–272. doi: 10.1159/000083140. [DOI] [PubMed] [Google Scholar]
- 38.Matsui H, Takatsu Y, Kumano S, Matsumoto H, Ohtaki T. Peripheral administration of metastin induces marked gonadotropin release and ovulation in the rat. Biochem Biophys Res Commun. 2004;320:383–388. doi: 10.1016/j.bbrc.2004.05.185. [DOI] [PubMed] [Google Scholar]
- 39.Shahab M, et al. Increased hypothalamic GPR54 signaling: a potential mechanism for initiation of puberty in primates. Proc Natl Acad Sci USA. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro VM, et al. Advanced vaginal opening and precocious activation of the reproductive axis by KiSS-1 peptide, the endogenous ligand of GPR54. J Physiol. 2004;561:379–386. doi: 10.1113/jphysiol.2004.072298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotani M, et al. The metastasis suppressor gene KiSS1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 42.Ohtaki T, et al. Metastasis suppressor gene KiSS1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 43.Clements MK, et al. FMRF amide-related neuropeptides are agonists of the orphan G-protein-coupled receptor GPR54. Biochem Biophys Res Commun. 2001;284:1189–1193. doi: 10.1006/bbrc.2001.5098. [DOI] [PubMed] [Google Scholar]
- 44.Clarkson J, Herbison AE. Postnatal development of kisspeptin neurons in mouse hypothalamus; sexual dimorphism and projections to gonadotropin-releasing hormone neurons. Endocrinology. 2006;147:5817–5825. doi: 10.1210/en.2006-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kauffman AS, et al. Sexual differentiation of Kiss1 gene expression in the brain of the rat. Endocrinology. 2007;148:1774–1783. doi: 10.1210/en.2006-1540. [DOI] [PubMed] [Google Scholar]
- 46.Roa J, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. New frontiers in kisspeptin/GPR54 physiology as fundamental gatekeepers of reproductive function. Front Neuroendocrinol. 2008;29:48–69. doi: 10.1016/j.yfrne.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Funes S, et al. The KiSS-1 receptor GPR54 is essential for the development of the murine reproductive system. Biochem Biophys Res Commun. 2003;312:1357–1363. doi: 10.1016/j.bbrc.2003.11.066. [DOI] [PubMed] [Google Scholar]
- 48.d’Anglemont de Tassigny X, et al. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci USA. 2007;104:10714–10719. doi: 10.1073/pnas.0704114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol. 2007;71:959–964. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 50.Wacker JL, et al. Disease-causing mutation in GPR54 reveals the importance of the second intracellular loop for class A G-protein-coupled receptor function. J Biol Chem. 2008;283:31068–31078. doi: 10.1074/jbc.M805251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tenenbaum-Rakover Y, et al. Neuroendocrine phenotype analysis in five patients with isolated hypogonadotropic hypogonadism due to a L102P inactivating mutation of GPR54. J Clin Endocrinol Metab. 2007;92:1137–1144. doi: 10.1210/jc.2006-2147. [DOI] [PubMed] [Google Scholar]
- 52.Semple RK, et al. Two novel missense mutations in G protein-coupled receptor 54 in a patient with hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2005;90:1849–1855. doi: 10.1210/jc.2004-1418. [DOI] [PubMed] [Google Scholar]
- 53.Teles MG, et al. A GPR54-activating mutation in a patient with central precocious puberty. N Engl J Med. 2008;358:709–715. doi: 10.1056/NEJMoa073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chehab FF, Lim ME, Lu R. Correction of the sterility defect in homozygous obese female mice by treatment with the human recombinant leptin. Nat Genet. 1996;12:318–320. doi: 10.1038/ng0396-318. [DOI] [PubMed] [Google Scholar]
- 55.Caprio M, Fabbrini E, Isidori AM, Aversa A, Fabbri A. Leptin in reproduction. Trends Endocrinol Metab. 2001;12:65–72. doi: 10.1016/s1043-2760(00)00352-0. [DOI] [PubMed] [Google Scholar]
- 56.Licinio J. Leptin in anorexia nervosa and amenorrhea. Mol Psychiatry. 1997;2:267–269. doi: 10.1038/sj.mp.4000298. [DOI] [PubMed] [Google Scholar]
- 57.Hill JW, Elmquist JK, Elias CF. Hypothalamic pathways linking energy balance and reproduction. Am J Physiol Endocrinol Metab. 2008;294:e827–e832. doi: 10.1152/ajpendo.00670.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clément K, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 59.Strobel A, Issad T, Camoin L, Ozata M, Strosberg AD. A leptin missense mutation associated with hypogonadism and morbid obesity. Nat Genet. 1998;18:213–215. doi: 10.1038/ng0398-213. [DOI] [PubMed] [Google Scholar]
- 60.Licinio J, et al. Phenotypic effects of leptin replacement on morbid obesity, diabetes mellitus, hypogonadism, and behavior in leptin-deficient adults. Proc Natl Acad Sci USA. 2004;101:4531–4536. doi: 10.1073/pnas.0308767101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welt CK, et al. Recombinant human leptin in women with hypothalamic amenorrhea. N Engl J Med. 2004;351:987–997. doi: 10.1056/NEJMoa040388. [DOI] [PubMed] [Google Scholar]
- 62.Tena-Sempere M. KiSS-1 and reproduction: focus on its role in the metabolic regulation of fertility. Neuroendocrinology. 2006;83:275–281. doi: 10.1159/000095549. [DOI] [PubMed] [Google Scholar]
- 63.Topaloglu AK, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rance NE. Menopause and the human hypothalamus: evidence for the role of kisspeptin/neurokinin B neurons in the regulation of estrogen negative feedback. Peptides. 2009;30:111–122. doi: 10.1016/j.peptides.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Rahilly S, et al. Brief report: impaired processing of prohormones associated with abnormalities of glucose homeostasis and adrenal function. N Engl J Med. 1995;333:1386–1390. doi: 10.1056/NEJM199511233332104. [DOI] [PubMed] [Google Scholar]
- 66.Jackson RS, et al. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 67.Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocr Rev. 2006;27:710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 68.Farooqi IS, et al. Hyperphagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. J Clin Endocrinol Metab. 2007;92:3369–3373. doi: 10.1210/jc.2007-0687. [DOI] [PubMed] [Google Scholar]
- 69.Lloyd DJ, Bohan S, Gekakis N. Obesity, hyperphagia and increased metabolic efficiency in Pc1 mutant mice. Hum Mol Genet. 2006;15:1884–1893. doi: 10.1093/hmg/ddl111. [DOI] [PubMed] [Google Scholar]
- 70.de Roux N. GnRH receptor and GPR54 inactivation in isolated gonadotropic deficiency. Best Pract Res Clin Endocrinol Metab. 2006;20:515–528. doi: 10.1016/j.beem.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 71.de Roux N, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 72.Bédécarrats GY, Kaiser UB. Mutations in the human gonadotropin-releasing hormone receptor: insights into receptor biology and function. Semin Reprod Med. 2007;25:368–378. doi: 10.1055/s-2007-984743. [DOI] [PubMed] [Google Scholar]
- 73.Beranova M, et al. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2001;86:1580–1588. doi: 10.1210/jcem.86.4.7395. [DOI] [PubMed] [Google Scholar]
- 74.Conn PM, Ulloa-Aguirre A, Ito J, Janovick J. A G protein-coupled receptor trafficking in health and disease: lessons learned to prepare for therapeutic mutant rescue in vivo. Pharmacol Rev. 2007;59:225–250. doi: 10.1124/pr.59.3.2. [DOI] [PubMed] [Google Scholar]
- 75.Pitteloud N, et al. The fertile eunuch variant of idiopathic hypogonadotropic hypogonadism: spontaneous reversal associated with a homozygous mutation in the gonadotropin-releasing hormone receptor. J Clin Endocrinol Metab. 2001;86:2470–2475. doi: 10.1210/jcem.86.6.7542. [DOI] [PubMed] [Google Scholar]