Abstract
5-Aminolevulinate synthase is a homodimeric pyridoxal 5’-phosphate-dependent enzyme that catalyzes the first step of the heme biosynthetic pathway in animals, fungi, and the α-subclass of the photosynthetic purple bacteria. The reaction cycle involves condensation of glycine with succinyl-coenzyme A to yield 5-aminolevulinate, carbon dioxide, and CoA. Mutations in the human erythroid-specific aminolevulinate synthase gene are associated with the erythropoietic disorder X-linked sideroblastic anemia. Recent kinetic and crystallographic data have facilitated an unprecedented understanding of how this important enzyme produces 5-aminolevulinate, and suggest possible directions for future research that may lead to treatments not only for X-linked sideroblastic anemia, but also other diseases.
Keywords: 5-Aminolevulinate synthase, pyridoxal 5’ phosphate, heme, X-linked sideroblastic anemia, transient kinetics
INTRODUCTION
5-Aminolevulinate (ALA) is the universal precursor for all biologically synthesized tetrapyrroles, including hemes, chlorophylls, cobalamin (vitamin B12), siroheme, and coenzyme F430 (5, 18). In plants, archaea, and the vast majority of bacteria, ALA is produced in three enzymatic steps from glutamate, while in animals, fungi, and the α-subclass of the photosynthetic purple bacteria, ALA is produced in a single step by 5-aminolevulinate synthase (ALAS; E.C. 2.3.1.37) (16). This PLP-dependent enzyme functions as a homodimer and converts glycine and succinyl-CoA into CoA, carbon dioxide, and ALA. The reaction mechanism is highly unusual for a PLP-dependent enzyme; it involves both proton transfers as well as a decarboxylation step at the amino acid α-carbon. ALAS catalyzes the rate-determining step of porphyrin biosynthesis, and its overexpression leads to accumulation of protoporphyrin IX in vivo (55).
In eucaryotes, the ALAS mRNA is translated into a precursor protein containing an N-terminal signal sequence that directs import across the mitochondrial membranes and into the inner mitochondrial matrix, where the signal sequence is removed and the citric acid cycle intermediate succinyl-CoA is available as a substrate. This requirement for succinyl-CoA metabolically associates heme biosynthesis with oxidative respiration, and represents a key evolutionary branchpoint between plants and other eucaryotes (44). Vertebrate genomes encode two highly conserved but differentially expressed ALAS genes, one for the ubiquitously expressed housekeeping isoform (ALAS1) and the other for the erythroid-specific (ALAS2) isoform (6). Mutations in the human ALAS2 gene can result in X-linked sideroblastic anemia, which is phenotypically characterized by iron accumulation within erythroblast mitochondria (7, 8, 19). Approximately one-third of X-linked sideroblastic anemia patients are pyridoxine responsive, and in these patients ALAS mutations are commonly observed in or near the PLP-binding site (4). Recently it has been shown that genetic alterations can lead to overexpression of ALAS2 activity and cause a completely different disease, protoporphyria (55). The disease states caused by under- or over-expression of ALAS2 emphasize the requirement for tight regulatory control of enzyme activity.
STRUCTURAL FEATURES OF ALAS
ALAS is classified as a member of the α-family of PLP-dependent enzymes. These enzymes require PLP as an obligatory cofactor and catalyze transformations of amino acids in which the bonding rearrangements occur at the carbon atom adjacent to the amine that forms an aldimine linkage to the cofactor (3). On the basis of amino acid sequence data, ALAS was initially categorized as part of the aminotransferase fold-type I subfamily branch of the α-family tree (9), but recent analyses based on crystallographic data indicate that ALAS is more appropriately considered to be part of the fold-type II subfamily branch (44). This subtle, but important, distinction greatly increases the likelihood that ALAS evolved from glutamate-1-semialdehyde aminotransferase, the PLP-dependent enzyme responsible for ALA synthesis in all organisms not containing an ALAS gene, and thus identifies the evolutionary link between the heme and chlorophyll biosynthetic pathways.
ALAS is most closely related to the other three members of the CoA-dependent acetyltransferase, or α-oxoamine synthase, subfamily. These enzymes all catalyze reactions involving small amino acids, CoA esters, and aminoketones, and although only four members are known, the α-oxoamine synthases are collectively of tremendous biological significance, because in addition to the important role of ALAS in heme biosynthesis, serine palmitoyltransferase and 8-amino-7-oxononanoate synthase catalyze the key committed steps of sphingolipid (24) and biotin synthesis (43), respectively.
Crystal structures of Rhodobacter capsulatus ALAS as the holoenzyme, and with glycine and succinyl-CoA bound, have been solved (4). The solution of these structures has helped shed unprecedented light on the enzymological properties of the enzyme, including the relative positions of the substrates, cofactor, active site residues, and a conformationally mobile loop of amino acids that closes over the active site in the substrate bound structures, all of which are illustrated in Figure 1. The cofactor resides nearly 20Ǻ down an active site cleft formed at the subunit interface, almost precisely in the center of the structure in a nearly non-aqueous environment, which is presumably essential for preventing side reactions. Importantly, the 49% sequence identity between the R. capsulatus enzyme and the catalytic core of human ALAS2 makes the R. capsulatus structures excellent models for detailed studies of mammalian ALAS, as well as for predicting the relationships between human ALAS2 mutations and sideroblastic anemia.
Figure 1. Structural features of the ALAS catalytic core.
A and B: side and top views, respectively, illustrating the dimeric structure, the position of the active site at the subunit interface (50), and the substrates. Note that the cofactor is almost exactly at the center of the structure. The conformationally mobile active site loop is colored light brown. C: Monomeric surface structure from the perspective of the adjacent subunit with PLP-glycine external aldimine (with yellow carbons) and succinyl-CoA (with white carbons) bound. Open and closed conformations of the active site loop are in red. D: Structural model for hydrogen bonding interactions with the PLP-bound α-amino-β-ketoadipate intermediate.
ALAS: AN ENZYME WITH A HISTORY
Our understanding of the broad features of ALAS reaction chemistry is founded upon the inspired and even heroic efforts of a number of eminent scientists. Early work revolved around identifying the biological precursors to heme, which were eventually found to be the substrates and product of ALAS. The bulk of this work was carried out in the laboratories of Shemin and Neuberger, dating back to 1945, when Shemin, in the laboratory of Rittenberg, personally synthesized and ingested 66 grams of 32.4% 15N-labeled glycine in order to follow the fate of glycine in the body (34, 35, 45, 47). This led to the discovery that glycine is utilized to synthesize the four pyrrole rings of heme. Subsequent experiments using 14C-labeled acetate, which Shemin did not ingest, led him to the remarkably prescient postulate that succinyl-CoA, which was not even known to exist at the time, was the second substrate for heme biosynthesis (42, 46). This was followed a few years later by the discoveries that ALA was the key precursor to heme and that enzymatic formation of ALA was observable in bacterial and avian extracts (21, 22, 33, 48). Work in Neuberger’s laboratory independently validated and extended these discoveries (36, 39, 40).
Initially three reasonable reaction mechanisms were envisioned whereby ALAS might convert glycine and succinyl-CoA into ALA, based upon the presumption that PLP was utilized as a cofactor, and the recognition that the conversion essentially requires replacement of the carboxyl group of glycine with a succinyl moiety (33) (Figure 2). Initial formation of an aldimine linkage between glycine and PLP is followed by generation of a stabilized carbanion that condenses with succinyl-CoA. The carbanion intermediate might arise either from loss of carbon dioxide (Fig. 2A) or loss of a proton (Fig. 2B). Loss of carbon dioxide would lead to ALA directly (path 1), whereas loss of a proton would lead to an α-amino-β-ketoadipic acid intermediate, a molecule that could either be decarboxylated enzymatically (path 2) or spontaneously in solution to yield ALA (path 3).
Figure 2. Early hypothetical ALAS reaction mechanisms.
See text for details.
Resolution of these uncertainties was accomplished in a series of brilliant radiolabeling experiments led by Jordan and Akhtar (2). The stereospecifically tritiated pro-R and pro-S enantiomers of glycine were synthesized enzymatically and utilized to conclusively demonstrate that ALAS catalyzes removal of the pro-R proton of glycine, supporting the existence of an enzyme bound α-amino-β-ketoadipic acid intermediate in the reaction sequence (31, 57). Determination of whether or not this intermediate is enzymatically decarboxylated to yield ALA proved even more challenging, however, due to the high degree of difficulty in isolating and determining the absolute stereochemistry of the protons at carbon-5 in the product. In an extraordinary technical achievement it was discovered that the pro-S proton of glycine is found in the pro-S position of ALA, indicating that decarboxylation of the intermediate must occur enzymatically, since if it had occurred free in solution this position would have been found to be racemic (1).
These findings were solidified and expanded upon by purification of the enzyme from multiple sources (13, 54, 56), ultimately confirming PLP as an enzyme bound cofactor (13, 37) and allowing utilization of steady-state kinetics to demonstrate that the reaction mechanism is ordered, with glycine binding before succinyl-CoA, and ALA released last (12, 38). The recently solved crystal structures, wherein the active site is at the bottom of a deeply recessed cleft that is occluded upon binding of succinyl-CoA, provide a beautiful structural verification of these kinetic findings (4). Nandi confirmed the reversibility of the ALAS reaction by utilizing the dangerously volatile compound [14C]-sodium bicarbonate along with ALA and CoA to generate radiolabelled glycine, and he was also the first person to observe a quinonoid intermediate absorption band in spectrum of the purified enzyme with substrates (38).
With the advent of molecular cloning technologies, the stage was set for isolation of large quantities of recombinant ALAS for even more detailed characterization (14). In the absence of a crystal structure, identification of the active site lysine residue, that covalently binds the cofactor in the absence of substrates, was used as a starting point to construct a rudimentary, but surprisingly, accurate model for the ALAS active site, based on the known three dimensional structure of aspartate aminotransferase (15, 23). Several key active site residues involved in cofactor and substrate binding, and catalysis, were correctly identified and characterized using site-directed mutagenesis (23, 27, 49, 51). These studies were facilitated by development of a simple continuous activity assay and observation of a spectroscopically stable quinonoid intermediate in the presence of ALA (26, 27). Initial pre-steady state kinetic studies revealed that ALA release is rate-determining for the reaction cycle and led to the postulate that conformational changes were associated with substrate binding and product release, possibly even determining the rates of chemical events at the active site (28). The existence of catalytically important conformational changes was initially suggested primarily by the observation that the rate at which glycine, the first substrate, forms an external aldimine with PLP is accelerated by at least two orders of magnitude in the presence of the second substrate, succinyl-CoA, and on the difficulty in reasonably assigning the very slow release of ALA from the enzyme to a chemical event.
A VERY UNUSUAL PLP-DEPENDENT ENZYME
The reaction mechanism of ALAS is highly unusual for a PLP-dependent enzyme. Nearly all α-family PLP enzymes catalyze cleavage of a single α-carbon substrate bond during the reaction cycle, but ALAS cleaves two. Of the more than 140 distinct PLP-dependent enzymes known (41), we are aware of only three other PLP-enzymes that catalyze cleavage of two α-carbon bonds as part of a normal reaction cycle: 8-amino-7-oxononanoate synthase, serine palmitoyltransferase, and dialkylglycine decarboxylase. The first two of these enzymes are α-oxoamine synthases and catalyze reactions analogous to ALAS, while dialkylglycine decarboxylase is a related enzyme that catalyzes a ping-pong reaction mechanism involving transamination and decarboxylation half-reactions (44, 52). An explanation of why cleavage of two amino acid substrate α-carbon bonds is so rare requires a brief discussion of PLP biochemistry.
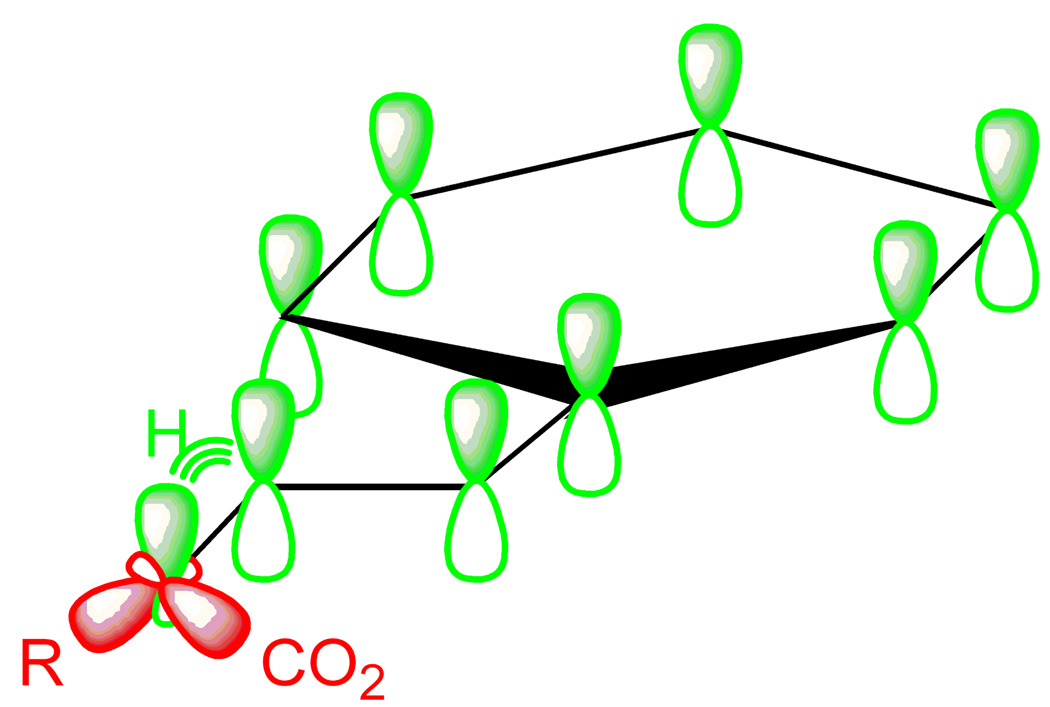
PLP-dependent reactions involving amino acids are initiated by formation of a covalent bond between the cofactor and the substrate amine to form a common intermediate known as an external aldimine (11, 25, 30). From the external aldimine several reaction pathways are possible for α-family enzymes, including transamination, decarboxylation, racemization, and elimination and replacement of electrophilic amino acid R-groups. Interestingly, a single unifying principle underlies all of these reactions: the cofactor stabilizes negative charge at the amino acid substrate α-carbon. The bonds to this carbon atom are labile in the external aldimine because bonding electrons can delocalize into the pi system of the cofactor, forming what is referred to as a quinonoid intermediate. This brings up the question as to how nature can accurately discriminate amongst a variety of different reactions which are all based on a common chemical property. For example, how does a transaminase prevent decarboxylation or racemization from occurring? The answer to this vital biological question was provided by Dunathan, and is simply that the bonds about the substrate α-carbon in the external aldimine are not all equally labile (Figure 3) (10). The lability of the bonds depends upon their stereoelectronic relationship to the planar cofactor pi system. Effective cleavage only occurs for a bond that is aligned perpendicular to the plane of the PLP-aldimine conjugated system. This stereoelectronic relationship most closely aligns the sigma orbitals of the target bond with the pi orbitals of the cofactor, and since these orbitals must overlap and rehybridize during the bond cleavage step, this orientation favors cleavage over the other α-carbon bonds, by a factor estimated to be about a million-fold (17, 53). Thus, binding interactions between the enzyme and the external aldimine complex orient the target bond perpendicular to the plane of the cofactor. Cleavage of more than one bond during a reaction cycle implies some intermediate torsion about the α-carbon bond, an alternate reaction pathway, or that Dunathan’s hypothesis is not entirely correct. Without direct evidence for any of these possibilities there was obviously a gap in our understanding of the ALAS reaction mechanism.
Figure 3. Stereoelectronic control of PLP-dependent reaction specificity.
The carbon-hydrogen bond most closely aligned with the π-orbitals of the cofactor, which is colored green, is approximately a million-fold more labilie than the bonds, which are colored in red.
The R. capsulatus ALAS crystal structure with glycine bound clearly indicates that the pro-R proton of glycine is oriented perpendicular to the plane of the cofactor ring, and points towards the active site lysine residue that removes this proton prior to condensation with succinyl-CoA, in perfect agreement with Dunathan’s hypothesis (4). Condensation with succinyl-CoA at the opposite face of the cofactor and expulsion of CoA leaves the bound α-amino-β-keto adipate intermediate, as modeled in Figure 1D (29). This structure predicts that the original carboxyl group of glycine, which is subsequently lost as carbon dioxide, producing an ALA-bound quinonoid intermediate (27), is only 30° out of the cofactor plane rather than the 90° theoretically required to use the cofactor as an electron sink. This discrepancy stimulated a detailed re-evaluation of the ALAS reaction mechanism involving extensive pre-steady state kinetic experimentation, the results of which benefitted from interpretation within the context of the crystal structures (29).
The pH-dependence of the individual steps of quinonoid intermediate formation and decay provide a deeper understanding of the subtleties of the ALAS reaction mechanism. Reaction of the substrates with ALAS results in a spectroscopically observable quinonoid intermediate on a time frame coincident with decarboxylation; the earlier reaction steps occurring too quickly to be observed using traditional stopped-flow spectroscopy (16, 59). Formation of the quinonoid intermediate is acid-catalyzed with an apparent pKa of 7.7, from which it has been concluded that histidine-207, located directly above the PLP ring in Figure 1D, acts as a general acid to promote decarboxylation through the substrate carbonyl, as depicted in Figure 4 step V-VI. ALAS therefore does not violate the stereoelectronic control hypothesis because the decarboxylation step does not directly utilize the PLP cofactor as an electron sink. Instead it is the enol (VI) that is in equilibrium with the observed quinonoid intermediate (VIb; electron movements 2), as well as the ALA-bound external aldimine (VII; electron movements 1), the latter of which releases ALA to complete the reaction sequence. The involvement of the enol (VI) in forming the quinonoid in the reverse reaction direction also explains why no quinonoid intermediate is observed upon reaction of the enzyme with ALA analogs devoid of the carbonyl functional group (59). The hydrogen bonding network depicted in Figure 1D spans four conserved amino acid residues bridging the carbonyl group and cofactor ring nitrogen electron sinks, and provides a structural rationale for suggesting an enol-quinonoid intermediate equilibrium may be a central feature in the evolution of ALAS and other α-oxoamine synthases.
Figure 4. Current understanding of the ALAS reaction mechanism.
Ironically, the spectroscopically visible quinonoid intermediate so central to elucidating the details of the reaction mechanism (VIb) is not directly on the reaction pathway. (See text for details.)
Quinonoid intermediate decay in single-turnover pH experiments occurs in two kinetic steps, the first of which is acid-catalyzed with an apparent pKa of 8.1, a value identical to that obtained by lysine-313-dependent pH titration of the ALAS-ALA complex (27). This step was therefore assigned to protonation of the enol (VI) by lysine-313, which would quench the quinonoid absorbance to form the cofactor-bound product (VII). Another revealing observation supports the assignments for quinonoid intermediate formation and decay. When glycine is replaced by deuteroglycine in single-turnover experiments there is no kinetic isotope effect observed on quinonoid intermediate formation, again consistent with this step reflecting the decarboxylation, but there is a small yet measurable effect (kH/kD = 1.26 ± 0.02) on the first step of quinonoid intermediate decay. This rather remarkable result may be interpreted as not only indicating that the first step of quinonoid intermediate decay represents a protonation step, but also that in a significant proportion of turnovers the proton removed from the pro-R position of glycine at the beginning of the reaction cycle is returned to the corresponding pro-R position of ALA. The active site lysine, observable at the bottom of Figure 1D, is deeply recessed within the active site at the center of the structure and below the cofactor, in a position suggesting a high degree of solvent exclusion. This and the small magnitude of the kinetic isotope effect support the possibility of partial proton conservation, and provide yet another illustration of the high degree of orchestration involved in ALAS reaction chemistry. The second step of quinonoid decay, which is pH-independent and represents the slowest step of the reaction cycle, must therefore reflect ALA release, and has been assigned to opening of the active site loop, which allows ALA to dissociate from the active site. Recently, this assignment has been supported by using protein fluorescence stopped-flow spectroscopy to resolve the on and off rates for ALA binding to murine ALAS2. These experiments verify that the changes in tryptophan fluorescence associated with ALA release are equivalent to kcat determined in separate steady-state kinetic experiments (data not shown).
Beyond the mechanistic details revealed by these studies, two important points emerge. First, the quinonoid intermediate that has proven central to our expanded understanding of the reaction mechanism is not itself directly on the reaction pathway! In fact, we have recently isolated a poorly active murine ALAS2 point mutant (R85L) involved in hydrogen bonding the carboxylate tail of succinyl-CoA that does not form any discernible quinonoid intermediate (T. Lendrihas and G. C. Ferreira, unpublished observations). Second and more importantly, the reaction cycle is believed to be kinetically limited by a conformational change associated with product release. This assignment is extremely noteworthy because ALAS activity limits the rate of protoporphyrin IX production, meaning that this conformational change would not only control the rate of ALA production, but also control flux through the entire porphyrin biosynthetic pathway. It also suggests that allosteric effectors may exist that bind ALAS and modulate activity by perturbing the conformational equilibrium associated with product release. It has already been observed that a conservative mutation in the active site loop (R433K in murine ALAS2) increases the catalytic rate two-fold (51), and linking the two subunits to construct a monomeric enzyme increases the catalytic rate five-fold (58). These findings support the possibility that ALAS activity could be up-regulated in vivo by one or more as yet undiscovered allosteric effectors. Good candidates include succinyl-CoA synthetase, iron, and porphyrins.
SUMMARY AND FUTURE DIRECTIONS
Several important findings now bring our understanding of ALAS to an unprecedented level. Combined kinetic and structural data strongly suggest that while the reaction chemistry is unusually complex, the kinetics of ALA formation are dominated by conformational flexibility of an active site loop located near the carboxy-terminus. This hypothesis should be verified in a rigorous fashion due its impact on heme biosynthesis, sideroblastic anemia, and protoporphyria. The possibility that the active site loop is a hot spot for mutations conferring hyperactivity could lead to enzyme variants with applications in photodynamic therapy of cancers and other diseases (20, 32).
ACKNOWLEDGEMENT
This work was supported by grants from the National Institutes of Health (DK63191 and GM080270).
Abbreviations
- ALA
5-aminolevulinate
- ALAS
5-aminolevulinate synthase
- PLP
pyridoxal 5’-phosphate
REFERENCES
- 1.Abboud MM, Jordan P, Akhtar M. Biosynthesis of 5-aminolevulinic acid: involvement of a retention–inversion mechanism. J. Chem. Soc. Chem. Comm. 1973;16:643–644. [Google Scholar]
- 2.Akhtar M, Abboud MM, Barnard G, Jordan P, Zaman Z. Mechanism and stereochemistry of enzymic reactions involved in porphyrin biosynthesis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1976;273:117–136. doi: 10.1098/rstb.1976.0005. [DOI] [PubMed] [Google Scholar]
- 3.Alexander FW, Sandmeier E, Mehta PK, Christen P. Evolutionary relationships among pyridoxal-5'-phosphate-dependent enzymes. Regio-specific alpha, beta and gamma families. Eur. J. Biochem. 1994;219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- 4.Astner I, Schulze JO, van den Heuvel J, Jahn D, Schubert WD, Heinz DW. Crystal structure of 5-aminolevulinate synthase, the first enzyme of heme biosynthesis, and its link to XLSA in humans. EMBO J. 2005;24:3166–3177. doi: 10.1038/sj.emboj.7600792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battersby AR. Tetrapyrroles: the pigments of life. Nat. Prod. Rep. 2000;17:507–526. doi: 10.1039/b002635m. [DOI] [PubMed] [Google Scholar]
- 6.Bishop DF, Henderson AS, Astrin KH. Human delta-aminolevulinate synthase: assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics. 1990;7:207–214. doi: 10.1016/0888-7543(90)90542-3. [DOI] [PubMed] [Google Scholar]
- 7.Bottomley SS, May BK, Cox TC, Cotter PD, Bishop DF. Molecular defects of erythroid 5-aminolevulinate synthase in X-linked sideroblastic anemia. J. Bioenerg. Biomembr. 1995;27:161–168. doi: 10.1007/BF02110031. [DOI] [PubMed] [Google Scholar]
- 8.Camaschella C. Recent advances in the understanding of inherited sideroblastic anaemia. Br. J. Haematol. 2008;143:27–38. doi: 10.1111/j.1365-2141.2008.07290.x. [DOI] [PubMed] [Google Scholar]
- 9.Christen P, Mehta PK. From cofactor to enzymes. The molecular evolution of pyridoxal-5'-phosphate-dependent enzymes. Chem. Rec. 2001;1:436–447. doi: 10.1002/tcr.10005. [DOI] [PubMed] [Google Scholar]
- 10.Dunathan HC. Conformation and reaction specificity in pyridoxal phosphate enzymes. Proc. Natl. Acad. Sci. U. S. A. 1966;55:712–716. doi: 10.1073/pnas.55.4.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 12.Fanica-Gaignier M, Clement-Metral J. 5-Aminolevulinic-acid synthetase of Rhodopseudomonas spheroides Y. Kinetic mechanism and inhibition by ATP. Eur. J. Biochem. 1973;40:19–24. doi: 10.1111/j.1432-1033.1973.tb03164.x. [DOI] [PubMed] [Google Scholar]
- 13.Fanica-Gaignier M, Clement-Metral J. 5-Aminolevulinic-acid synthetase of Rhodopseudomonas spheroides Y. Purification and some properties. Eur. J. Biochem. 1973;40:13–18. doi: 10.1111/j.1432-1033.1973.tb03163.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira GC, Dailey HA. Expression of mammalian 5-aminolevulinate synthase in Escherichia coli. Overproduction, purification, and characterization. J. Biol. Chem. 1993;268:584–590. [PubMed] [Google Scholar]
- 15.Ferreira GC, Neame PJ, Dailey HA. Heme biosynthesis in mammalian systems: evidence of a Schiff base linkage between the pyridoxal 5'-phosphate cofactor and a lysine residue in 5-aminolevulinate synthase. Protein Sci. 1993;2:1959–1965. doi: 10.1002/pro.5560021117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira GC, Zhang JS. Mechanism of 5-aminolevulinate synthase and the role of the protein environment in controlling the cofactor chemistry. Cell. Mol. Biol. (Noisy-le-grand) 2002;48:827–833. [PubMed] [Google Scholar]
- 17.Fogle EJ, Liu W, Woon ST, Keller JW, Toney MD. Role of Q52 in catalysis of decarboxylation and transamination in dialkylglycine decarboxylase. Biochemistry. 2005;44:16392–16404. doi: 10.1021/bi051475b. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda H, Casas A, Batlle A. Aminolevulinic acid: from its unique biological function to its star role in photodynamic therapy. Int. J. Biochem. Cell. Biol. 2005;37:272–276. doi: 10.1016/j.biocel.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Furuyama K, Sassa S. Multiple mechanisms for hereditary sideroblastic anemia. Cell. Mol. Biol. (Noisy-le-grand) 2002;48:5–10. [PubMed] [Google Scholar]
- 20.Gagnebin J, Brunori M, Otter M, Juillerat-Jeanneret L, Monnier P, Iggo R. A photosensitising adenovirus for photodynamic therapy. Gene Ther. 1999;6:1742–1750. doi: 10.1038/sj.gt.3300992. [DOI] [PubMed] [Google Scholar]
- 21.Gibson KD. Biosynthesis of delta-aminolaevulic acid by extracts of Rhodopseudomonas spheroides. Biochim. Biophys. Acta. 1958;28:451. doi: 10.1016/0006-3002(58)90502-x. [DOI] [PubMed] [Google Scholar]
- 22.Gibson KD, Laver WG, Neuberger A. Initial stages in the biosynthesis of porphyrins. 2. The formation of delta-aminolaevulic acid from glycine and succinyl-coenzyme A by particles from chicken erythrocytes. Biochem. J. 1958;70:71–81. doi: 10.1042/bj0700071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong J, Hunter GA, Ferreira GC. Aspartate-279 in aminolevulinate synthase affects enzyme catalysis through enhancing the function of the pyridoxal 5'-phosphate cofactor. Biochemistry. 1998;37:3509–3517. doi: 10.1021/bi9719298. [DOI] [PubMed] [Google Scholar]
- 24.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi H. Pyridoxal enzymes: mechanistic diversity and uniformity. J. Biochem. 1995;118:463–473. doi: 10.1093/oxfordjournals.jbchem.a124931. [DOI] [PubMed] [Google Scholar]
- 26.Hunter GA, Ferreira GC. A continuous spectrophotometric assay for 5-aminolevulinate synthase that utilizes substrate cycling. Anal. Biochem. 1995;226:221–224. doi: 10.1006/abio.1995.1217. [DOI] [PubMed] [Google Scholar]
- 27.Hunter GA, Ferreira GC. Lysine-313 of 5-aminolevulinate synthase acts as a general base during formation of the quinonoid reaction intermediates. Biochemistry. 1999;38:3711–3718. doi: 10.1021/bi982390w. [DOI] [PubMed] [Google Scholar]
- 28.Hunter GA, Ferreira GC. Pre-steady-state reaction of 5-aminolevulinate synthase. Evidence for a rate-determining product release. J. Biol. Chem. 1999;274:12222–12228. doi: 10.1074/jbc.274.18.12222. [DOI] [PubMed] [Google Scholar]
- 29.Hunter GA, Zhang J, Ferreira GC. Transient kinetic studies support refinements to the chemical and kinetic mechanisms of aminolevulinate synthase. J. Biol. Chem. 2007;282:23025–23035. doi: 10.1074/jbc.M609330200. [DOI] [PubMed] [Google Scholar]
- 30.John RA. Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta. 1995;1248:81–96. doi: 10.1016/0167-4838(95)00025-p. [DOI] [PubMed] [Google Scholar]
- 31.Jordan PM, Akhtar M. The mechanism of action of serine transhydroxymethylase. Biochem. J. 1970;116:277–286. doi: 10.1042/bj1160277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juarranz A, Jaen P, Sanz-Rodriguez F, Cuevas J, Gonzalez S. Photodynamic therapy of cancer. Basic principles and applications. Clin. Transl. Oncol. 2008;10:148–154. doi: 10.1007/s12094-008-0172-2. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi G, Kumar A, Talmage P, Shemin D. The enzymatic synthesis of delta-aminolevulinic acid. J. Biol. Chem. 1958;233:1214–1219. [PubMed] [Google Scholar]
- 34.Kresge N, Simoni RD, Hill RL. David Rittenberg: Exploring Porphyrin Synthesis with Duck Blood and Isotope Tracers. J. Biol. Chem. 2005;280:e12–e13. [Google Scholar]
- 35.Kresge N, Simoni RD, Hill RL. A Pathway for Heme Biosynthesis: the Work of David Shemin. J. Biol. Chem. 2006;281:e28–e29. [Google Scholar]
- 36.Muir HM, Neuberger A. The biogenesis of porphyrins. The distribution of N in the ring system. Biochem. J. 1949;45:163–170. [PMC free article] [PubMed] [Google Scholar]
- 37.Nandi DL. Delta-aminolevulinic acid synthase of rhodopseudomonas spheroides. Binding of pyridoxal phosphate to the enzyme. Arch. Biochem. Biophys. 1978;188:266–271. doi: 10.1016/s0003-9861(78)80008-3. [DOI] [PubMed] [Google Scholar]
- 38.Nandi DL. Studies on delta-aminolevulinic acid synthase of Rhodopseudomonas spheroides. Reversibility of the reaction, kinetic, spectral, and other studies related to the mechanism of action. J. Biol. Chem. 1978;253:8872–8877. [PubMed] [Google Scholar]
- 39.Neuberger A, Muir HM, Gray CH. Biosynthesis of porphyrins and congenital porphyria. Nature. 1950;165:948–950. doi: 10.1038/165948a0. [DOI] [PubMed] [Google Scholar]
- 40.Neuberger A, Scott JJ. Aminolaevulinic acid and porphyrin biosynthesis. Nature. 1953;172:1093–1094. doi: 10.1038/1721093a0. [DOI] [PubMed] [Google Scholar]
- 41.Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003;4:850–854. doi: 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radin NS, Rittenberg D, Shemin D. The role of acetic acid in the biosynthesis of heme. J. Biol. Chem. 1950;184:755–768. [PubMed] [Google Scholar]
- 43.Schneider G, Lindqvist Y. Structural enzymology of biotin biosynthesis. FEBS Lett. 2001;495:7–11. doi: 10.1016/s0014-5793(01)02325-0. [DOI] [PubMed] [Google Scholar]
- 44.Schulze JO, Schubert WD, Moser J, Jahn D, Heinz DW. Evolutionary relationship between initial enzymes of tetrapyrrole biosynthesis. J. Mol. Bio.l. 2006;358:1212–1220. doi: 10.1016/j.jmb.2006.02.064. [DOI] [PubMed] [Google Scholar]
- 45.Shemin D. An illustration of the use of isotopes: the biosynthesis of porphyrins. Bioessays. 1989;10:30–35. doi: 10.1002/bies.950100108. [DOI] [PubMed] [Google Scholar]
- 46.Shemin D, Kumin S. The mechanism of porphyrin formation; the formation of a succinyl intermediate from succinate. J. Biol. Chem. 1952;198:827–837. [PubMed] [Google Scholar]
- 47.Shemin D, Rittenberg D. The utilization of glycine for the synthesis of a porphyrin. J. Biol. Chem. 1945;159:567–568. [Google Scholar]
- 48.Shemin D, Russell CS. Aminolevulinic acid, its role in the biosynthesis of porphyrins and purines. J. Am. Chem. Soc. 1953;75:4873–4874. [Google Scholar]
- 49.Tan D, Barber MJ, Ferreira GC. The role of tyrosine 121 in cofactor binding of 5-aminolevulinate synthase. Protein Sci. 1998;7:1208–1213. doi: 10.1002/pro.5560070516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan D, Ferreira GC. Active site of 5-aminolevulinate synthase resides at the subunit interface. Evidence from in vivo heterodimer formation. Biochemistry. 1996;35:8934–8941. doi: 10.1021/bi952918m. [DOI] [PubMed] [Google Scholar]
- 51.Tan D, Harrison T, Hunter GA, Ferreira GC. Role of arginine 439 in substrate binding of 5-aminolevulinate synthase. Biochemistry. 1998;37:1478–1484. doi: 10.1021/bi971928f. [DOI] [PubMed] [Google Scholar]
- 52.Toney MD. Reaction specificity in pyridoxal phosphate enzymes. Arch. Biochem. Biophys. 2005;433:279–287. doi: 10.1016/j.abb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 53.Tsai MD, Weintraub HJ, Byrn SR, Chang C, Floss HG. Conformation-reactivity relationship for pyridoxal Schiff's bases. Rates of racemization and alpha-hydrogen exchange of the pyridoxal Schiff's bases of amino acids. Biochemistry. 1978;17:3183–3188. doi: 10.1021/bi00609a002. [DOI] [PubMed] [Google Scholar]
- 54.Warnick GR, Burnham BF. Regulation of prophyrin biosynthesis. Purification and characterization of -aminolevulinic acid synthase. J. Biol. Chem. 1971;246:6880–6885. [PubMed] [Google Scholar]
- 55.Whatley SD, Ducamp S, Gouya L, Grandchamp B, Beaumont C, Badminton MN, Elder GH, Holme SA, Anstey AV, Parker M, Corrigall AV, Meissner PN, Hift RJ, Marsden JT, Ma Y, Mieli-Vergani G, Deybach JC, Puy H. C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload. Am. J. Hum. Genet. 2008;83:408–414. doi: 10.1016/j.ajhg.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whiting MJ, Elliott WH. Purification and properties of solubilized mitochondrial -aminolevulinic acid synthetase and comparison with the cytosol enzyme. J. Biol. Chem. 1972;247:6818–6826. [PubMed] [Google Scholar]
- 57.Zaman Z, Jordan PM, Akhtar M. Mechanism and stereochemistry of the 5-aminolaevulinate synthetase reaction. Biochem. J. 1973;135:257–263. doi: 10.1042/bj1350257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Cheltsov AV, Ferreira GC. Conversion of 5-aminolevulinate synthase into a more active enzyme by linking the two subunits: spectroscopic and kinetic properties. Protein Sci. 2005;14:1190–1200. doi: 10.1110/ps.041258305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Ferreira GC. Transient state kinetic investigation of 5-aminolevulinate synthase reaction mechanism. J. Biol. Chem. 2002;277:44660–44669. doi: 10.1074/jbc.M203584200. [DOI] [PMC free article] [PubMed] [Google Scholar]