Abstract
Many RNAs undergo large conformational changes in response to the binding of proteins and small molecules. However, when RNA functional dynamics occur in the ns-μs time scale they become invisible to traditional solution NMR relaxation methods. Residual dipolar couplings methods have revealed the presence of extensive ns-μs domain motions in HIV-1 TAR RNA, but this technique lacks information on the rates of motions. We have used solid-state deuterium NMR to quantitatively describe trajectories of key residues in TAR by exploiting the sensitivity of this technique to motions that occur in the ns-μs regime. Deuterium lineshape and relaxation data were used to model motions of residues within the TAR binding interface. The resulting motional models indicate that two functionally essential bases within the single stranded bulge sample both the free and Tat-bound conformations on the microsecond timescale in the complete absence of the protein. Thus, our results strongly support a conformational capture mechanism for recognition: the protein does not induce a new RNA structure, but instead captures an already-populated conformation.
Introduction
Functional dynamics in HIV-1 TAR occurs on a timescale slower than ~10 ns1, 2 but faster than ~100 μs3, 4. These motions allow the RNA to adapt its structure to generate the conformation recognized by Tat and cyclin T1 proteins5-7 (Figure 1a), a key regulatory checkpoint in the viral lifecycle. Dynamics occurring on the ns-μs time scale are invisible to conventional solution-state NMR relaxation measurements because they are masked by global rotational diffusion1. Additionally, such motions are too fast to be detected by relaxation dispersion methods that are powerful in studies of slower protein dynamics8-13. Residual dipolar couplings (RDCs) provide rich information on conformational dynamics in this timescale14, and their application to RNA has recently demonstrated that the helical domains of TAR experience extensive motions that connect the conformations of TAR observed in the absence and presence of protein and small molecule ligands1, 2. However, RDCs do not provide information on rates; therefore, the time scale of motion can only be broadly and indirectly inferred by a process of elimination when faster (sub-ns) and slower (ms) motions are not observed1, 2, 14.
Figure 1.
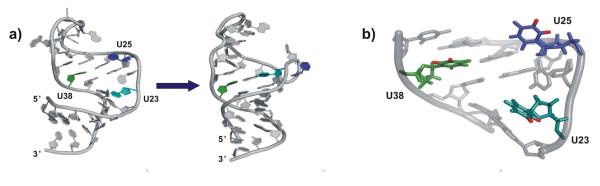
(a) HIV-1 TAR changes conformation when Tat protein or small molecules bind to it, forming a structure conducive to binding of the protein and activation of transcription. (b) Location of the three nucleotides whose motions were probed using solid state NMR. U23 (teal) is critical for binding of Tat; U25 (blue) is a single stranded residue whose conformation changes considerably when tat binds; U38 (green) belongs to the upper helix and is base paired with A27. Sites of deuterium labeling (the 5 and 6 base positions) are shown in red.
Unlike these solution-state methods, solid-state deuterium NMR is very sensitive to motions within the intermediate (ns-μs) timescale15. By studying samples in a highly hydrated solid-state gel that preserves the conformational dynamics, but avoids lineshape sharpening by molecular tumbling, information on anisotropic motions is retained. In addition, solid-state relaxation measurements provide information on the rates or timescales of motions faster than those that affect the lineshapes. Thus, the combined measurement of quadrupole echo lineshape, Zeeman spin-lattice relaxation (T1Z) and quadrupolar order relaxation (T1Q) allows the quantitative characterization of atomic motions occurring at rates between 104 and 1010 s−1 16, 17.
In this work, we apply solid state NMR measurements to obtain motional rates and amplitudes in the intermediate ns-μs regime typically inaccessible to solution NMR. Using this approach we find that the extrahelical residues, U23 and U25, experience significant conformational exchange over the ns-μs timescale. Additionally, by probing the helical site U38, we determine that TAR experiences significant interhelical motions even in the solid state on an intermediate timescale, coincident with the results of RDC measurements2.
Results
We probed motions of 5,6-2H labeled uridine bases at three positions in TAR RNA (U23, U25 and U38, Figure 1b) at high levels of hydration to reproduce conditions observed in solution18. The lineshapes for all three sites clearly reveal modulations by intermediate time scale (ns-μs) motions15 (Figure 2a). In order to identify the underlying atomic motions and define their rates and amplitudes, we modeled dynamics at each site by generating model trajectories; the procedure by which model trajectories were obtained is described in detail in the methods section. Briefly, for each site, modeling was performed by hypothesizing that the motions that modulate the lineshape correspond to transitions between the two conformations observed for these single stranded nucleotides in free TAR, and in the complex of TAR with peptide mimics of Tat protein.19, 20 For U38, we assumed that this residue might experience helical domain motions similar to those observed using RDCs1, 2. Once initial trajectories were generated under these assumptions, motional rates and amplitudes were defined by systematically fitting the experimental lineshapes and relaxation rates.
Figure 2.
(a) Experimental deuterium solid-state NMR spectra of selectively 5,6 deuterated U23 (left), U25 (center) and U38 (right) in TAR RNA. The curves through the experimental data represent best fits using the motional models and parameters described in the text; the line in the center of each spectrum corresponds to the residual deuterium signal from the deuterium-depleted solvent water. (b) Experimental relaxation times (left side of each column) are compared with those calculated using the motional models described in the text (right side of each column) for both T1z and T1Q.
For all three nucleotides, we were able to obtain high-quality fits to the lineshapes (Figure 2a) and to the two independent relaxation rates (Figure 2b) simultaneously. As described in the methods section and presented graphically in supplementary Figures S1-S3, the experimental data constrain the parameters of the fit within very narrow boundaries because multiple observables are fit simultaneously. Furthermore, because two deuterated sites per residue were monitored simultaneously, a model capable of producing a satisfactory fit to the data must define a single trajectory for the base plane which simultaneously reproduces superposition lineshape data, superposition T1Z data, and superposition T1Q data arising from two labeled sites. Therefore, while we cannot formally exclude the possibility that other models may also describe the dynamic processes presented here, the model trajectories we report robustly satisfy multiple independent experimental observations.
Amplitudes and Rates of Motions for the Non-Helical Bases
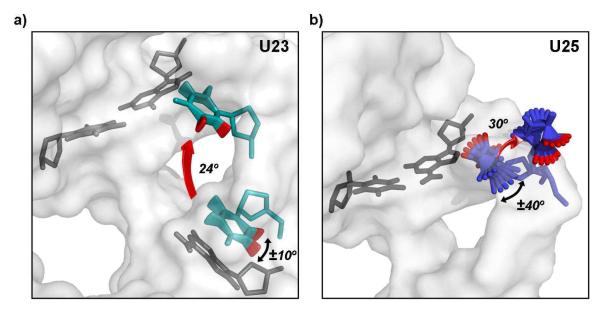
The spectroscopic data for U23 are well-modeled by a large-amplitude (24°) intermediate-time scale (6.7×107 s−1) hop of the base, in addition to a rapid (1010 s−1) small-amplitude (±11°) twisting of the base plane about the glycosidic bond occurring at both ends of the hop (Figure 3a). This trajectory would correspond to transient unstacking of U23 from A22, as observed in ligand-free TAR21; it is also consistent with sampling of positions near A27 and U38, the base-triple partners U23 must find in order to form the tat- and argininimide-bound structures19,22. The NMR data for U25 are well-modeled by a 30 degree jump at a rate of 6×107 s−1; such a motion is consistent with an outward and downward rotation of the backbone away from the intrahelical conformation observed in free TAR21 as it is flipped out of the helix, as observed in bound TAR19 (Figure 3b). In addition, U25 experiences much slower, large-amplitude twisting of the base (6×105 s−1; ± 40°). Extensive ps-ns motions were previously observed for this same residue3 and for C24 immediately adjacent to it4. However, the large amplitude motions described here for both U23 and U25 occur at rates that make them undetectable to traditional solution-state NMR relaxation experiments, as well as T1ρ power-dependence4 experiments.
Figure 3.
(a) The motions of U23 (in teal with sites monitored by selective deuteration in red) can be accurately modeled by a 24° jump linking the conformation observed for this residue in the free structure of TAR21 (at the bottom) to its position observed in the complex of TAR with peptides derived from the tat protein (at the top) 19; in addition to these large scale motions we observe much faster ±11° rotations of the base with respect to the sugar. (b) The motions of U25 (green with sites monitored by selective deuteration in red) can be accurately modeled as a slow 30° outward jump to generate the extrahelical conformation observed in many TAR complexes, together with a slower ± 40° rotation about the glycosidic bond.
Amplitudes and Rates of Motions for the Helical U38 Base
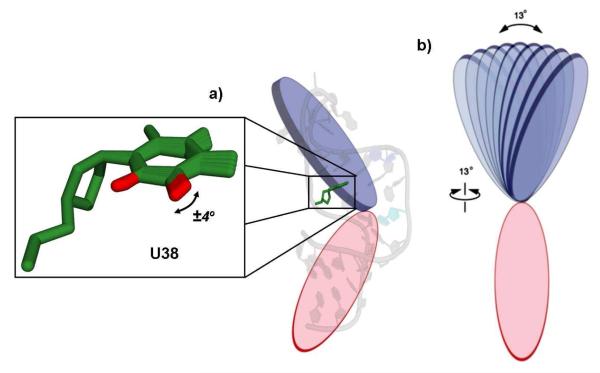
The spectroscopic data for U38 are well described by a combination of 13° twisting and 13° bending, corresponding to a movement of the entire upper helix which carries the U38:A27 base pair, occurring at a rate of 1.4×106 s−1. In addition, we observe small-amplitude local motions (± 4°) at a much faster rate of 2.2×108 s−1 (Figure 4). There was no evidence of substantial fast (ns–ps) motions in this residue when solution relaxation data were analyzed4, but these motions are similar to the global motions of the upper helix observed using RDCs1, 2. A model for the overall inter-helical bending and twisting motions of the TAR construct in the solid state cannot be constructed based on the motions of the upper helix alone. However, due to the absence of an apical loop, the more compact lower helix likely undergoes motions of amplitude similar to or even larger than those we observe for the upper helix. These combined helical motions in the solid samples will correspond to significantly larger inter-helical excursions, perhaps comparable in amplitude to those obtained using RDCs under solution conditions. Thus, while the amplitude we find for the helical motion carrying the A27:U38 base pair is relatively small, the motions we see are likely more representative of those in solution than this would suggest.
Figure 4.
(a) Residue U38 (green) is represented inside a pseudo-cylindrical plane (blue) representative of the upper helical domain of TAR, while the lower pseudo-cylindrical plane (red) represents the lower helix. (b) The motions of U38 correspond to a 13° twist and 13° bend of the entire upper helix, in addition to much faster smaller librational motions of the base.
Consistent with the global nature of the motions responsible for the U38 lineshape, the spectral signature of these slow collective motions only becomes visible as hydration increases to the point where sample conditions approach those observed in solution18; the more localized motions of U23 and U25 are instead observed even at low hydration levels18.
Discussion
The essential functional dynamics of TAR RNA occur on a timescale (ns-μs) that is invisible to current solution relaxation NMR techniques3, 4 and that is also too fast to be studied by relaxation dispersion10. Thus, the motions we have reported for U23 and U25 were never described before. The helical domain motions we observe for U38 are analogous to those previously described using RDCs1, 2, although possibly of reduced amplitude. However, the RDC technique cannot provide information on the rates of these motions; instead they are only broadly and indirectly inferred by a process of elimination when motions on other time scales are not observed. In contrast, our approach yields both amplitudes and rates of motion.
The sensitivity of the deuterium solid state lineshape to anisotropic motions allows us to extract time-dependent trajectories for atomic motions in TAR RNA and to quantitatively characterize conformational exchange processes occurring on a μs time scale. Conformational exchange has been extensively characterized in proteins on the much slower time window (ms) that is accessible via relaxation dispersion experiments9-11, 13, but not at μs rates. The widespread observation of extensive conformational rearrangement in RNA recognition and function makes it highly probable that ns-μs conformational exchange will be found in other RNAs. Similarly, there is emerging evidence14 that functional motion in proteins is also rich in the ns-μs time window. Solid state NMR approaches provide a bridge in covering RNA and protein dynamics23 by providing atomic-level information on motions occurring in this intermediate time window.
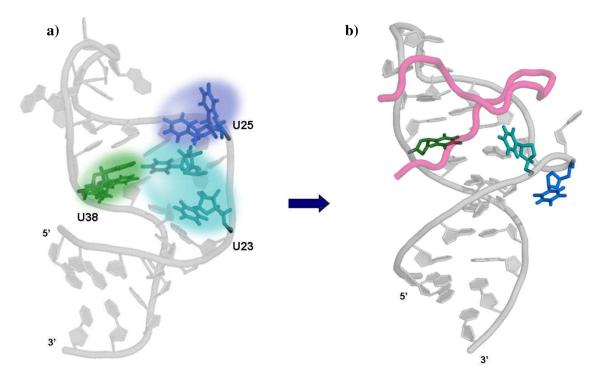
Our data strongly suggests the recruitment of Tat occurs by a conformational capture mechanism (Figure 5), as was previously suggested based on data on the motions of the TAR helices2, 24. In this picture, the protein does not need to alter the conformation of TAR, but instead selects a conformation in the RNA that is already substantially populated in its absence. The motions we have identified in U23 and U25 are consistent with the very motions which would connect the structure of TAR in its ligand-free form21 to that observed in the presence of peptide mimics of Tat proteins7, 20, 22. This second structure is essential to provide a binding site to recruit the Tat5 and cyclin T1 regulatory proteins to TAR, thus these motions likely underlie the functional rearrangements critical for transcription and thus for HIV replication8. The motions of U38 are thus quite likely to be essential as well, since they correspond to the helical domain motions allowing TAR to sample the manifold of conformational states observed in its complexes with small molecules and in particular, with Tat2. Global reorientation of the upper helix, as monitored at U38, occurs on a very similar time scale to the hopping of U23, suggesting that these motions may be correlated and, together, may create the unique RNA structure required to recruit Tat protein to TAR.
Figure 5.
(a) In the absence of ligands, the three residues in TAR experience a wide range of conformations (indicated by the clouds colored according to Figure 1), which include the conformation observed in the complex with peptides derived from Tat protein. The two conformations of TAR are represented as the two alternative structures of U23 (in teal) and U25 (green); the complex with a 37-mer peptide derived from Tat protein is shown in (b), emphasizing the conformational capture mechanism of the Tat protein.
Materials and Methods
Preparation of RNA samples for solid state investigation
[5,6- 2H]-labeled uridine nucleotides for deuterium dynamics experiments were prepared in house using well-established methods25. Conversion of the deuterated uridine nucleotide to phosphoramidites for RNA synthesis was done by Dharmacon (Lafayette, Colorado, USA). RNA oligonucleotides were deprotected and desalted by Dharmacon. Each sample was checked for homogeneity using analytical denaturing polyacrylamide gel electrophoresis, and used without further purification. The following three TAR RNA 29-mer constructs were used: 5′-GGCCAGA-2’FU-CUG(pS)AGCGAAAGC*UCUCUGGCC-3′ (“U38”), 5′-GGCCAGA*U-2’FC-UG(pS)AGCGAAAGCUCUCUGGCC-3′ (“U23”), and 5′-GGCCAGA-2’FU-C*UG(pS)AGCGAAAGCUCUCUGGCC-3′ (“U25”).
Here pS indicates a phosphorothioate label, -2’FU- or -2’FC- indicates a 2′ fluorine substitution, and *U indicates a [5,6- 2H] base-labeled Uridine). Previous work has demonstrated that phosphorothioate and 2′-fluorine substitutions at those positions do not perturb the structure of TAR26-28. These labels were added to allow us to verify the conformation of TAR RNA using REDOR methods28. Although perturbations to the dynamics arising from these modifications cannot be ruled out completely, the combination of the high degree of structural similarity between wild-type and substituted TAR, as well as our focus on base motions, suggest that perturbations to the base dynamics by these substitutions are not substantial.
NMR sample preparation
5.70 μmol of the U38 oligonucleotide was dissolved in buffer (50 mM NaCl, 10 mM sodium cacodylate, pH 5.5), then frozen using liquid nitrogen and lyophilized (final composition 10% NaCl, and 4.7% cacodylate respectively, by weight upon lyophilization). To remove residual HDO and D2O, samples were re-dissolved in deuterium-depleted water then lyophilized 3-4 times. Each sample was then re-dissolved a final time in deuterium-depleted water, annealed in a 90°C water bath for five minutes, allowed to cool to room temperature for 30 minutes, and then frozen using liquid nitrogen. All samples were again lyophilized extensively to remove residual water (~80 mtorr, 7-15 days) before transfer to the NMR sample rotor. The dry sample was packed into a 5 mm Kel-F sample chamber, and re-lyophilized to remove water absorbed during sample packing.
Samples were hydrated to 16 waters per nucleotide in sealed chambers over saturated salt solutions containing deuterium-depleted water29. Water addition to the samples was monitored gravimetrically. Samples were then sealed and allowed to equilibrate an additional week before use in NMR experiments. The hydration levels were chosen after carefully conducting a thorough investigation of the line shape as a function of water content18. Water introduced to salted nucleic acid samples progressively populates the grooves, bases, and backbone in a well-characterized fashion, beginning with the phosphates and eventually reaching limiting local hydration so that subsequent addition of water contributes primarily to the surrounding water shells30. At W=16, local hydration of the nucleic acid backbone and bases is substantially complete18, 30-32. Following that hydration level, the solid state NMR spectra display much smaller changes indicative of relatively small changes in dynamics between W=16 and the highest hydration we have investigated, W=3018,.
Samples for analysis of U23 (4.3 μmol) and U25 (6.6 μmol) were prepared using the procedure described above, with different sample sizes as indicated.
Solid-state NMR Experiments
All experiments were performed at 11.7 Tesla field (76.76 MHz deuterium resonance frequency), using a homebuilt single-channel static deuterium probe. Pulses were generated using an AMT (American Microwave Technologies) M3446 (1 kW and 10-130 MHz) amplifier. Typical 90-degree pulse times were 2.0-2.4 μs. All measurements were made at room temperature (26°C), with continuous application of 20°C cooling air. 5mm kel-F sample rotors were made in-house. Dwell time for all experiments was 0.2 μs. Recycle delay for all experiments was one second.
Deuterium line shapes were recorded using a quadrupole echo pulse sequence33 (90°×- τ -90°y- τ -echo), where the second half of the echo signal was acquired and Fourier-transformed following left-shift of all time-domain datasets to their respective echo maxima. Quadrupole echo experiments used eight-step phase cycling, with an echo delay of 40 μs. Typical spectra resulted from acquisition of 300,000-500,000 scans, collected as 80,000-scan datasets then co-added.
Spin-lattice relaxation times were determined using saturation-recovery experiments. An initial train of five 90x pulses at 2 ms intervals was used, followed by a variable relaxation delay and quadrupole echo detection ((90°×)n-delay-90°×- τ -90°y- τ -echo; n = 5). Typical delay times for hydrated samples were 1, 100, 500, 1,000, and 2,000 ms.
T1Q relaxation times were determined using the broadband Jeener-Broekaert pulse sequence34, 35, with eight-step phase cycling. The pulse sequence used is as follows: 90 – tau1 – 67 – tau1 – 45 – tau2 – 45 – [variable relaxation delay] – 45 – echo delay – 90 – echo delay – acquisition. Typical delays were 5 s (tau1), 2.5 usec (tau2), 1, 100, 500, 1,000, and 2,000 ms (variable relaxation delay), with 40 s echo delays.
Data Processing
Data processing was performed using in-house written NMR processing software. Data acquisition was begun prior to the echo maximum for all experiments, and all time-domain datasets were left-shifted prior to Fourier Transformation. Lineshape spectra were multiplied with an exponential corresponding to 500 kHz line broadening. Relaxation data were analyzed using a non-linear least squares fit to a single exponential decay to obtain powder-averaged spin-lattice relaxation times.
Modeling RNA Dynamics
To construct the models, we make the assumption that the motions are composed of independent components which can be represented using multi-axis models. This permits a representation of the complex motions in which local motions are represented by jumps of the principal axis system with respect to an intermediate frame, while conformational exchange jumps of the bulge residues or collective motions of the helix (which would carry the U38:A27 pair) are treated by jumps of this intermediate frame with respect to the crystal frame. Motions of the C-D bond vector are then described by two successive independent transformations, where one set of angles describes the local motion and a second set describes the motions of the intermediate frame, representing a conformational exchange motion in each model.
Motional models for each site were constructed based upon known structural constraints. Each base is confined to move within a steric ‘box’ defined by surrounding residues which limit its available motional space: U38 local motion, for example, is restricted to positions between base pairs above and below the A27:U38 pair; motions of this local jumping frame itself thus represent motions of the upper helix. For bulge residues U23 and U25, the picture is more complicated. These two residues reside in the single-stranded bulge linking two helices, thus both should be directly dependent – both structurally and dynamically – on the motions of the adjacent helices. This dependence should be transmitted by fluctuations of the sugar-phosphate backbone, involving numerous torsion angles. Motions for U23, however, must be sterically bounded from below by A22 and from above by the upper helix, thus we can treat motions of this site as delimited by its stacking interactions atop A22 and base-pairing interactions with A27:U38 of the upper helix, while restricting lateral motion of the residue to rotations of the bulge backbone. Therefore, to construct a model for the conformational exchange motions of U23, we first extracted coordinates for the C1’-N1 bond vector in bound and unbound TAR (1ANR, 1ARJ), to define likely endpoints for conformational exchange. Vectors representing the C1′-N1 bond axis were defined with respect to a frame attached to the A27:U38 base pair for each TAR conformation; these were then used to construct initial-guess jump sites representing the conformational exchange motion. Remarkably, refinement of this initial model to fit the observed solid state NMR data required a reduction in jump amplitude of only 3 degrees. The motions of residue U25 are not nearly so well-defined, and this is reflected in the models we report here. It has been shown that U25 can be substituted by a short linker without significant impairment of TAR function, suggesting that it functions largely as a spacer residue. Solution NMR structures suggest that, while motions of this residue must be bounded from above by adjacent residue G26 and laterally by the conformation of the adjacent bulge backbone, it is not restrained by either stacking or base-pairing interactions. Because the bulge is known to be extruded from between the two helices during tat recognition, it would be expected that motions of U25 would be relatively unrestricted and determined largely by those of the local backbone alone. This is consistent with the large-amplitude unrestrained motions we have used to model the U25 data.
Angular excursions corresponding to structural transitions bounded by those constraints were represented as site geometries and used to construct multi-axis jump models. Line shape and T1Z simulations were then performed using the programs MXET137, EXPRESS38, and Turbopowder39. Fitting of the model trajectories against the experimental results was performed iteratively, by systematically varying jump rates and trajectories (amplitudes of motions) in the simulations until a satisfactory fit was obtained (examples are shown in supplementary materials).
To generate the corresponding T1Q values, model parameters were input to correlation functions derived from the formalism of Torchia and Szabo40, specific to the motional model derived for each site. Relaxation times corresponding to the resulting spectral densities for each model were then obtained using the following relationships41, 42:
| (1) |
| (2) |
where is the static quadrupolar coupling constant.
The requirement that candidate motional models fit the combined relaxation and lineshape data restricts possible motional models drastically. Separately, relaxation or lineshape data for each site could typically be fit by more than one model; however, systematic grid searches of the parameter space revealed that, for U38, concatenated two-site jump models simultaneously consistent with both the observed lineshape and relaxation data could be restricted to a single motional model. For U23, we are able to identify two models consistent with the data. In addition to the model described above, a second model in which the motional rates are approximately reversed can also reproduce the data. In such a scenario, however, the rate of the conformational exchange motion must be increased to 2.9e10 s−1(and the local twisting about the glycosidic bond is slowed to 5.4e7 s−1). Given the large amplitude of the deformations of the backbone required for conformational exchange in TAR, such a rapid rate for this component of the motion is physically implausible. Moreover, it is also unlikely that the local motions within the respective potential minima at each end of the exchange jump should be so slow.
While the fit for U25 is less satisfactory, as seen in figure S3, this model too is tightly constrained by the data. Given the lack of restraints on the dynamics of this residue, and the resulting absence of a well-defined potential landscape, definition of more finely-detailed motional trajectories than that which we provide here would be difficult to support. Obtaining a full representation of the more complicated motions undergone by U25 would likely require incorporation of additional motional axes. Thus far, our attempts to add a third dynamic axis have not produced improvement in the fits to date.
The goodness of fit is shown by χ2 plots for fits to the combined T1Z and T1Q relaxation data for each of the three sites, as shown in supplementary Figures S1, S2 and S3 for each of the three residues. Figure S4 shows mean quadrupole echo amplitudes for the three sites at 16 waters per nucleotide.
Supplementary Material
Acknowledgements
This work was supported by grants from the NSF (MCB-0642253) and NIH (RO1-EB03152) to GD and GV. We are grateful to Dr. Zahra Shajani for many stimulating discussions and to members of the Varani research group for reading the manuscript.
Footnotes
Supporting Information Available: Illustration of the fitting procedure used to obtain the models presented in the main text. χ2 plots for fits to the combined T1Z and T1Q relaxation data for each of the three sites, as a function of model parameters (rates or amplitudes), and quadrupole echo amplitudes for the three sites at 16 waters per nucleotide. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhang Q, Sun X, Watt E, Al-Hashimi HM. Resolving Complex Motional Modes that code for RNA Adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1268. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 3.Hansen AL, Al-Hashimi HM. Dynamics of Larger Elongated RNA by NMR Carbon Relaxation. J. Am. Chem. Soc. 2007:1–11. doi: 10.1021/ja0757982. [DOI] [PubMed] [Google Scholar]
- 4.Bardaro MF, Jr, Shajani Z, Patora-Komisarska K, Robinson JA, G. V. How binding of small molecule and peptide ligands to HIV-1 TAR alters the RNA motional landscape. Nucleic Acids Res. 2009;37:1529–1540. doi: 10.1093/nar/gkn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puglisi JD, Chen L, Frankel AD, Williamson JR. Role of RNA Structure in Arginine recognition of TAR RNA. Proceedings of the National Academy of Science USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24(20):3974–81. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol. 1995;253(2):313–32. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 8.Karn J. Tackling Tat. J. Mol. Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 9.Korzhnev DM, Salvatella X, Vendruscolo M, Nardo AAD, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. NATURE. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 10.Palmer AGI, Kroenke CD, Loria JP. Nuclear Magnetic Resonance Methods for Quantifying Microsecond-to-Millisecond Motions in Biological Macromolecules. Meth. Enzym. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 11.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. NATURE. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 12.Boehr DD, McElheny D, Dyson HJ, Wright PE. The Dynamic Energy Landscape of Dihydrofolate Reductase Catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 13.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. NATURE. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 14.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KFA, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320(5882):1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 15.Olsen G, Echodu D, Shajani Z, Bardaro M, Varani G, Drobny GP. Solid-state deuterium NMR studies reveal micros-ns motions in the HIV-1 transactivation response RNA recognition site. J. Am. Chem. Soc. 2008;130:2896–2897. doi: 10.1021/ja0778803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiess HW. In: NMR Basic Principles and Progress, D. N. s., Rotation of Molecules and Nuclear Spin Relaxation. Diehl P, Fluck E, Kosfeld R, editors. Vol. 15. Springer Verlag; Berlin: 1978. pp. 59–214. [Google Scholar]
- 17.Vold RR, Vold RL. Deuterium Relaxation in Molecular Solids, in Advances in Magnetic and Optical Resonance. Vol. 16. Academic Press; San Diego: 1991. pp. 85–171. [Google Scholar]
- 18.Olsen GL, Jr, B. M, Echodu CD, Drobny GP, Varani G. Hydration dependent dynamics in RNA. J Biomol NMR. 2009;45(1-2):133–142. doi: 10.1007/s10858-009-9355-6. [DOI] [PubMed] [Google Scholar]
- 19.Aboul-ela F, Karn J, Varani G. The Structure of the Human Immunodeficiency Virus Type-1 TAR RNA Reveals Principles of RNA Recognition by Tat Protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 20.Long KS, Crothers DM. Evidence for Base Triple Formation in the HIV-1 Tat-TAR Complex. Journal of Molecular Biology. 1998 [Google Scholar]
- 21.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the Absence of Ligands Reveals a Novel Conformation of the Trinucleotide Bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 23.Lorieau JL, McDermott AE. Conformational flexibility of a microcrystalline globular protein: order parameters by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2006;128:11505–11512. doi: 10.1021/ja062443u. [DOI] [PubMed] [Google Scholar]
- 24.Leulliot N, Varani G. Current Topics in RNA-Protein Recognition: Control of Specificity and Biological Function through Induced Fit and Conformational Capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Yu P, LeProust E, Gao X. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruse WBT, Salisbury SA, Brown T, Cosstick R, Eckstein F, Kennard O. Chiral phosphorothioate analogues of B-DNA: the crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C] Journal of Molecular Biology. 1986;192:891–905. doi: 10.1016/0022-2836(86)90035-5. [DOI] [PubMed] [Google Scholar]
- 27.Merritt ME, Sigurdsson ST, Drobny GP. Long-range distance measurements to the phosphodiester backbone of solid nucleic acids using P-31-F-19 REDOR NMR. J Am Chem Soc. 1999;121:6070–6071. [Google Scholar]
- 28.Olsen GL, Edwards TE, Deka P, Varani G, Sigurdsson ST, Drobny GP. Monitoring tat peptide binding to TAR RNA by solid-state 31P-19F REDOR NMR. Nucleic Acids Res. 2005;33(11):3447–54. doi: 10.1093/nar/gki626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weast RC. In: CRC Handbook of Chemistry and Physics. 60 ed CRC, editor. CRC Press; Boca Raton, FL: 1979. p. E-46. [Google Scholar]
- 30.Falk M, Hartman KA, Lord RC. Hydration of Deoxyribonucleic Acid .1. A Gravimetric Study. J Am Chem Soc. 1962;84:3843–3846. [Google Scholar]
- 31.Wang AC, Kennedy MA, Reid DR, P. DG. A Solid-State 2H NMR Investigation of Purine Motion in a 12-Base-Pair RNA Duplex. Journal of Magnetic Resonance B. 1994;105:1–10. doi: 10.1006/jmrb.1994.1092. [DOI] [PubMed] [Google Scholar]
- 32.Schurr JM, Fujimoto BS, Diaz R, Robinson BH. Manifestations of Slow Site Exchange Processes in Solution NMR. A Continuous Gaussian Exchange Model. Journal of Magnetic Resonance. 1999;140:404–431. doi: 10.1006/jmre.1999.1858. [DOI] [PubMed] [Google Scholar]
- 33.Davis JH. Deuterium magnetic resonance study of the gel and liquid crystalline phases of dipalmitoyl phosphatidylcholine. Biophys J. 1979;27:339–358. doi: 10.1016/S0006-3495(79)85222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wimperis S, Bodenhausen G. Broadband excitation of quadrupolar order by modified jeener-broekaert sequences. Chemical Physics Letters. 1986;132:194–199. [Google Scholar]
- 35.Hoatson GL. Broadband composite excitation sequences for creating quadrupolar order in 2H NMR. Journal of Magnetic Resonance. 1990;94:152–159. [Google Scholar]
- 36.Aboul-ela F, Varani G. Recognition of HIV-1 TAR RNA by Tat protein and Tat-derived peptides. J. Mol. Struct. 1998;423:29–39. [Google Scholar]
- 37.Greenfield MS, Ronemus AD, Vold RL, Vold RR, Ellis PD, Raidy TE. Deuterium quadrupole-echo NMR spectroscopy. III. Practical aspects of lineshape calculations for multiaxis rotational processes. Journal of Magnetic Resonance. 1987;72:89–107. [Google Scholar]
- 38.Vold RL, Hoatson GL. Effects of jump dynamics on solid state nuclear magnetic resonance line shapes and spin relaxation times. Journal of Magnetic Resonance. 2009;198:57–72. doi: 10.1016/j.jmr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Wittebort RJ, Olejniczak ET, Griffin RG. Analysis of 2H Nuclear Magnetic Resonance Lineshapes in Anisotropic Media. Journal of Chemical Physics. 1987;86:5411–5420. [Google Scholar]
- 40.Torchia DA, Szabo A. Spin-lattice relaxation in solids. Journal of Magnetic Resonance. 1982;49:107–121. [Google Scholar]
- 41.Palmer AG, III, Williams J, McDermott A. Nuclear Magnetic Resonance Studies of Biopolymer Dynamics. J. Phys. Chem. 1996;100:13293–13310. [Google Scholar]
- 42.Alam TM, Drobny GP. Solid-State NMR Studies of DNA Structure and Dynamics. Chem Rev. 1991;91:1545–1590. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.