SUMMARY
Fragile X Syndrome (FXS), the most common genetic form of mental retardation and autism, is caused by loss of function mutations in an RNA binding protein, Fragile X Mental Retardation Protein (FMRP). Patients’ neurons, as well as those of the mouse model, Fmr1 knockout (KO), are characterized by an excess of dendritic spines, suggesting a deficit in excitatory synapse elimination. In response to neuronal activity, myocyte enhancing factor 2 (MEF2) transcription factors induce robust synapse elimination. Here, we demonstrate that MEF2 activation fails to eliminate functional or structural excitatory synapses in hippocampal neurons from Fmr1 KO mice. Similarly, inhibition of endogenous MEF2 increases synapse number in wildtype, but not Fmr1 KO neurons. MEF2-dependent synapse elimination is rescued in Fmr1 KO neurons by acute postsynaptic expression of FMRP, but not RNA binding mutants of FMRP. Our results reveal that active MEF2 and FMRP function together in an acute, cell autonomous mechanism to eliminate excitatory synapses.
Keywords: Fragile X Syndrome, FMRP, MEF2, synapse elimination, CA1
INTRODUCTION
The proper synaptic connectivity of neural circuits relies upon a dynamic process of synapse formation and elimination (Hua and Smith, 2004). During early postnatal brain development, synapse formation exceeds that of elimination which results in an excess of excitatory synapses. Excess synapses are subsequently eliminated or pruned in the adolescent brain that leads to fewer synapses in the adult (Rakic et al., 1986). Activity-dependent synapse elimination contributes to experience-dependent refinement of neural circuits during brain development (Hua and Smith, 2004; Sanes and Lichtman, 1999; Wiesel, 1982; Zuo et al., 2005) and may also function as a homeostatic mechanism, where neurons adapt to changes in global patterns of synaptic activity by adjusting the strength and number of synapses (Chandrasekaran et al., 2007; Turrigiano, 2008; Wierenga et al., 2006). Recent evidence indicates that neuronal activity decreases synapse number in part through activation of the Myocyte enhancing factor 2 (MEF2) family of transcription factors (Barbosa et al., 2008; Flavell et al., 2006; Pulipparacharuvil et al., 2008). MEF2 factors are activated upon neuronal depolarization, postsynaptic Ca2+ increases and a signaling process that involves the Ca2+/Calmodulin effectors, calcineurin and CaM kinase (Flavell et al., 2006; McKinsey et al., 2002; Pulipparacharuvil et al., 2008; Shalizi et al., 2006). Selective activation of MEF2 dependent transcription results in a rapid and robust synapse elimination and knockdown or gene deletion of MEF2 isoforms in either hippocampal or striatal neurons results in increased synapse number, an effect that requires neuronal activity (Barbosa et al., 2008; Flavell et al., 2006; Pulipparacharuvil et al., 2008). Importantly, recent studies suggest that the MEF2-dependent synapse elimination process is critical for normal learning and memory and behaviors associated with drug abuse (Barbosa et al., 2008; Li et al., 2008; Pulipparacharuvil et al., 2008). However, the cellular and molecular mechanisms by which MEF2 controls synapse number are currently unknown.
Fragile X Syndrome (FXS), a major genetic cause of mental retardation and autism, results from loss-of-function mutations in the Fmr1 gene (Abrahams and Geschwind, 2008; Bassell and Warren, 2008). Pyramidal neurons of FXS patients, as well as those of the mouse model of FXS, Fmr1 KO mice, display increased dendritic spine number, suggesting that aspects of FXS may be due in part to deficits in excitatory synapse elimination (Irwin et al., 2002; Irwin et al., 2001). Fmr1 encodes an RNA binding protein, Fragile X Mental Retardation Protein (FMRP) that associates with small messenger ribonucleoprotein complexes, as well as polyribosomes in neurons and dendrites. Accumulating evidence supports a role for FMRP in regulation of dendritic protein synthesis and plasticity of mature synaptic function (Bassell and Warren, 2008). Although recent work demonstrates a direct and cell autonomous role for FMRP in synapse elimination, it is unknown if or how FMRP interacts with other known pathways that regulate synapse number or development (Pfeiffer and Huber, 2007). Here, we report that MEF2 is incapable of regulating synapse number in Fmr1 KO neurons and provide evidence that FMRP functions downstream of MEF2 mediated transcription in a common or parallel, convergent pathway that is required for synapse elimination.
RESULTS
MEF2-mediated, bidirectional regulation of hippocampal synapse number requires FMRP
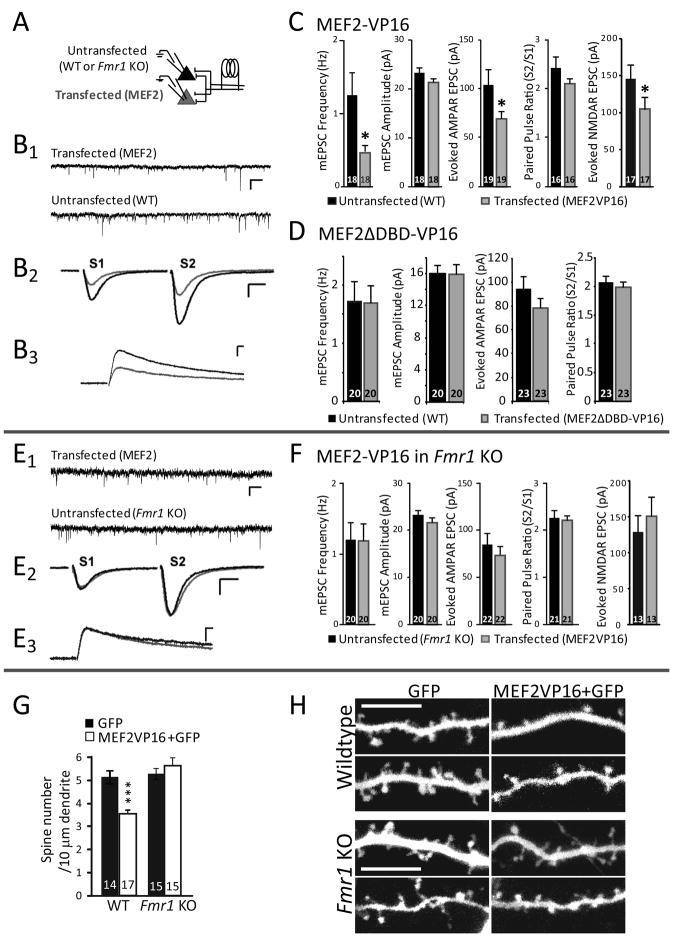
To test whether MEF2-dependent synapse elimination requires FMRP, we generated mouse organotypic hippocampal slice cultures from wild-type or Fmr1 KO littermates. Slice cultures were then cotransfected with plasmids that express a tamoxifen-inducible, constitutively-active form of MEF2 (MEF2-VP16-ER™) together with enhanced green fluorescent protein (GFP) (Flavell et al., 2006). To activate MEF2-dependent transcription in transfected neurons, slice cultures were treated with 4-hydroxytamoxifen (4OHT) or vehicle (0.1% EtOH) for 24–48 hours prior to simultaneous whole-cell patch-clamp recordings of transfected and untransfected pyramidal neurons in the CA1 region to measure synapse function (Fig. 1A). Wildtype neurons expressing activated MEF2-VP16-ER™ displayed a ~40% reduction in the amplitude of evoked EPSCs and mEPSC frequency (Fig. 1B-C), consistent with a decrease in the number of functional synapses. In contrast, mEPSC amplitudes were unchanged indicating that MEF2 activity did not alter the strength of individual synapses. Similar results were obtained with brief (16–30 hr) transfection of a non-inducible, constitutively active form of MEF2 (MEF2-VP16; without the ER™; Fig. 4A). Synaptic transmission in MEF2-VP16-ER™ transfected neurons treated with vehicle was unchanged (Fig. S1A, B). Similarly, transfection of a mutant form of MEF2 that lacks DNA-binding activity (MEF2ΔDBD-VP16-ER™) had no effect on synaptic function implicating MEF2-DNA interactions and transcription in the observed phenotype (Flavell et al., 2006) (Fig. 1D). Furthermore, these results indicate that expression of the VP16 transcriptional activator has no effect on synapse function. MEF2-induced decreases in synaptic function may result from either a decrease in presynaptic release probability, synapse silencing (Liao et al., 1995), or a reduction in synapse number. MEF2 activation did not alter presynaptic release probability as measured by paired-pulse facilitation of evoked EPSCs (Fig. 1B2, C). MEF2 was effective in reducing isolated NMDA receptor-mediated EPSCs to a similar extent as AMPAR EPSCs indicating that synapse silencing is unlikely (Fig. 1B3, C). MEF2-VP16 also did not alter the decay kinetics of NMDAR EPSCs (untransfected τ = 118 ± 8 msec; MEF2-VP16 transfected τ = 129 ± 9 msec; p = 0.28). These latter two results suggest that synapse maturation is not altered by MEF2 activation. Therefore, the decrease in synapse function by MEF2 is most consistent with a decrease in synapse number.
Figure 1. MEF2 activation induces functional and structural synapse elimination in wildtype, but not Fmr1 KO neurons.
A Experimental recording configuration. Organotypic hippocampal slice cultures from p6–7 wild-type (WT) or Fmr1 KO mice were biolistically transfected with MEF2-VP16-ER™ and treated with 4-hydroxytamoxifen (4-OHT) for 24–48 hours to induce MEF2-VP16-ER™ activation. Synaptic function was measured using dual simultaneous whole-cell patch clamp recordings of transfected and neighboring untransfected CA1 neurons. B1,B2, Representative traces of mEPSCs (B1; scale bar = 10 pA/500 ms.) and evoked AMPAR mediated EPSCs (B2; scale bar = 50 pA/10 ms) from a simultaneous recording from an untransfected and a neighboring MEF2-VP16-ER™ transfected WT neuron. B3 Pharmacologically isolated NMDAR mediated EPSCs from untransfected and neighboring MEF2-VP16 transfected WT neurons (scale bar = 50 pA/50 ms). C. Average mEPSC frequency, mEPSC amplitude, evoked AMPAR-mediated EPSC amplitude, and paired-pulse ratio (amplitude EPSC2/EPSC1) from untransfected WT and MEF2-VP16-ER™ transfected cells. Group data of pharmacologically isolated, evoked, NMDAR mediated EPSC amplitudes from untransfected and neighboring MEF2-VP16 transfected WT neurons. In this and all figures, averages are plotted + SEM and n (# of cell pairs) is indicated on each bar. * p< 0.05. D. Transfection of WT slice cultures with MEF2ΔDBD-VP16-ER™, a DNA binding-deficient mutant of MEF2, and treatment with 4OHT does not alter excitatory synaptic transmission. Plotted are average mEPSC frequency, mEPSC amplitude, evoked EPSC amplitude, and paired-pulse ratio from untransfected and MEF2ΔDBD-VP16-ER™ transfected Fmr1 KO neurons. E, F. MEF2-VP16-ER™ transfection into Fmr1 KO slice cultures. E1, E2. Representative traces of mEPSCs (E1; scale bar = 10 pA/500 ms.) and evoked EPSCs (E2; scale bar = 50 pA/10 ms) from a simultaneous recording of an untransfected and neighboring MEF2-VP16-ER™ transfected Fmr1 KO neuron. E3 Pharmacologically isolated NMDAR mediated EPSCs from untransfected and neighboring MEF2-VP16 transfected Fmr1 KO neurons (scale bar = 50 pA/50 ms). F. Plotted are average mEPSC frequency, mEPSC amplitude, evoked AMPAR mediated EPSC amplitude, and paired-pulse ratio from untransfected and MEF2-VP16-ER™ transfected Fmr1 KO neurons. Group data of pharmacologically isolated, evoked, NMDAR mediated EPSC amplitudes from untransfected and neighboring MEF2-VP16 transfected Fmr1 KO neurons., G. MEF2-VP16 reduces average dendritic spine number of wildtype neurons, but not of Fmr1 KO neurons. *** p< 0.001. H. Representative images of apical dendrites from wildtype or Fmr1 KO CA1 pyramidal neurons in slice cultures transfected with GFP alone or MEF2-VP16 + GFP for 36 hours. Scale bar = 10 μm.
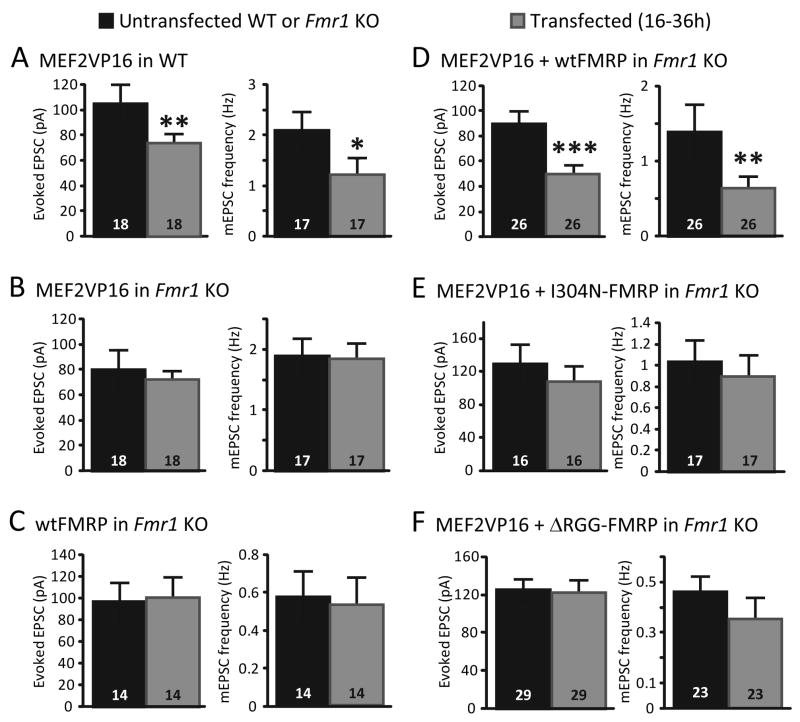
Figure 4. Acute postsynaptic coexpression of MEF2 and FMRP are required for functional synapse elimination.
A. Acute transfection of constitutively-active MEF2 (MEF2-VP16) into wildtype (WT) hippocampal slice cultures results in a decrease in synaptic transmission 16- 30 hrs post-transfection as measured by evoked EPSCs and mEPSC frequency. B. In contrast, acute transfection of MEF2-VP16 into Fmr1 KO hippocampal slice cultures has no effect on any measure of synapse function. C. Transfection of Fmr1 KO slice cultures with wildtype (wt) FMRP-GFP alone for 16–48 hours had no effect on evoked EPSCs or mEPSC frequency. D. Co-transfection of wtFMRP-GFP together with MEF2-VP16 into Fmr1 KO neurons (16–30 hrs) reduced EPSC size and mEPSC frequency. E, F. Co-transfection of I304N FMRP (E) or ΔRGG-FMRP (F) together with MEF2-VP16 (16–30 hrs) had no effect on evoked EPSCs or mEPSCs. For all experiments (A-F) mEPSC amplitudes and paired-pulse facilitation of evoked EPSCs were unaffected by transfection. *p< 0.05; **p< 0.01; *** p < 0.001.
To confirm that MEF2 activity caused a structural synapse elimination we examined the effect of MEF2-VP16 on dendritic spine number in CA1 hippocampal neurons from wildtype slice cultures. Consistent with the effects of MEF2-VP16 on synaptic function, as well as previous work, we observed that brief expression of MEF2-VP16 (24–36h) lead to a ~30% decrease in dendritic spine number (Fig. 1G,H) (Barbosa et al., 2008; Flavell et al., 2006). Together these results indicate that postsynaptic activation of MEF2 leads to a structural and functional synapse elimination in a cell autonomous manner.
To determine if FMRP is required for MEF2-induced functional synapse elimination, MEF2-VP16-ER™ was expressed in Fmr1 KO neurons. In contrast to wildtype neurons, MEF2 activation in Fmr1 KO neurons had no effect on any measure of synapse function or structure, including evoked AMPAR and NMDAR mediated EPSCs, mEPSC frequency and amplitude, paired-pulse facilitation (Fig. 1E,F; Fig. 4B) and dendritic spine number (Fig. 1G,H). This observation indicates that FMRP is required for MEF2-induced functional and structural synapse elimination.
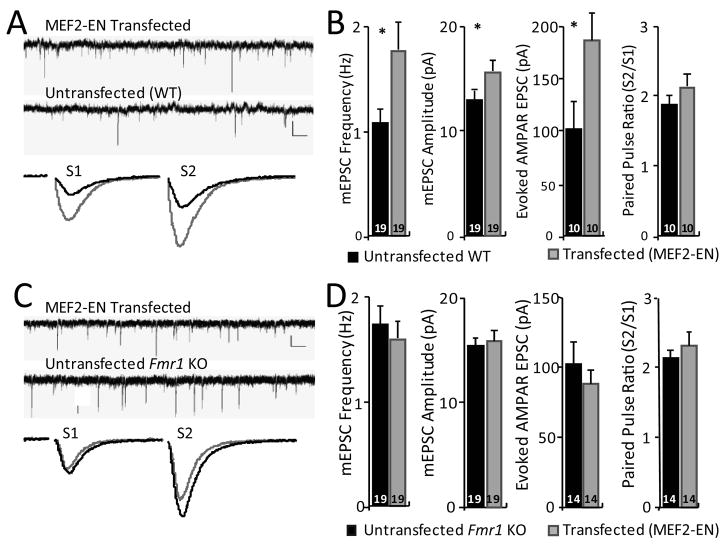
Increases in functional and structural synapse number are observed by decreasing MEF2 isoforms using RNA interference or in knockout mice (Barbosa et al., 2008; Flavell et al., 2006; Pulipparacharuvil et al., 2008). Because this effect requires neuronal activity, this suggests that tonic neuronal activity drives endogenous MEF2 transcription to suppress synapse number. To determine whether endogenous MEF2 functions to suppress synapse number in Fmr1 KO neurons, a dominant negative form of MEF2, MEF2-Engrailed (MEF2-EN), was expressed in either wildtype or Fmr1 KO slice cultures. MEF2-EN consists of the DNA-binding and dimerization domains of MEF2 fused to the potent transcriptional repressor domain of Engrailed, which results in a dominant repression of MEF2-dependent transcription (Fig. S2A,B) (Arnold et al., 2007; Shalizi et al., 2006). MEF2-EN expression in wild-type neurons resulted in a robust (~70–80%) increase in evoked EPSC amplitude and mEPSC frequency without affecting paired pulse facilitation (Fig. 2A, B). These results indicate that acute inhibition of endogenous MEF2 activity is sufficient to increase functional synapse number in wildtype neurons. In contrast, MEF2-EN transfection into Fmr1 KO neurons had no effect on any measure of synapse function or number (Fig. 2C, D). These results indicate that FMRP is required for both exogenous and endogenous MEF2 to suppress synapse number and strongly suggest that MEF2-dependent synapse regulation is dysfunctional in FXS.
Figure 2. Inhibition of endogenous MEF2 function enhances synapse number in wildtype, but not in Fmr1 KO neurons.
A, Representative traces of mEPSCs (upper; scale bar = 10 pA/500 ms.) and evoked EPSCs (lower; scale bar = 50 pA/10 ms) from a simultaneous recording from an untransfected and neighboring MEF2-Engrailed (MEF2-EN) transfected WT neuron. B. Average mEPSC frequency, mEPSC amplitude, evoked EPSC amplitude, and paired-pulse ratio from untransfected WT and MEF2-EN transfected cells. Averages are plotted + SEM and n (# of cell pairs) is indicated on each bar. * p< 0.05. C, D MEF2-EN transfection into Fmr1 KO slice cultures. C. Representative traces of mEPSCs (upper; scale bar = 10 pA/500 ms.) and evoked EPSCs (lower; scale bar = 50 pA/10 ms) from a simultaneous recording of an untransfected and neighboring MEF2-EN transfected Fmr1 KO neuron. D. Average mEPSC frequency, mEPSC amplitude, evoked EPSC amplitude, and paired-pulse ratio from untransfected and MEF2-EN transfected Fmr1 KO neurons.
MEF2-dependent transcription is normal in Fmr1-KO neurons
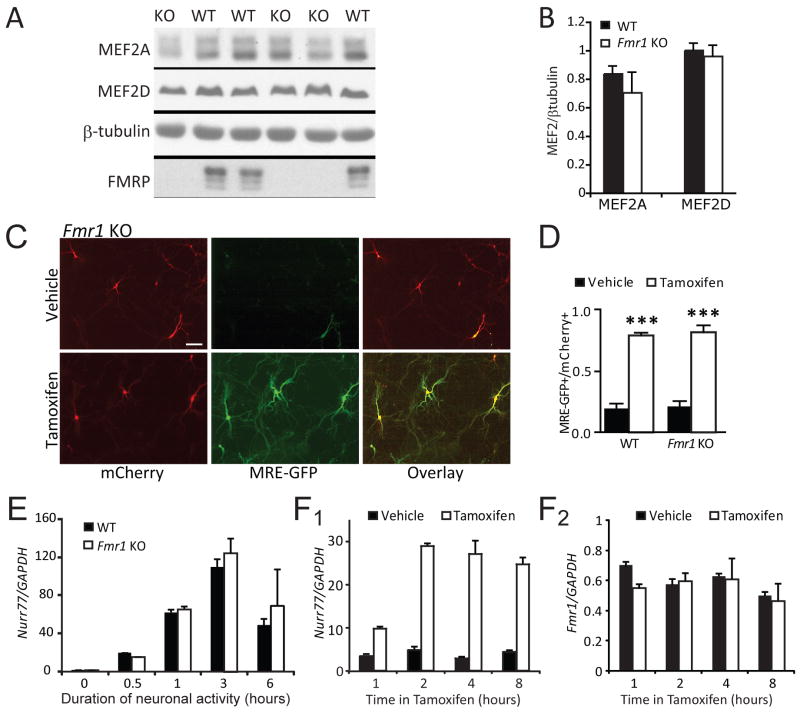
Since MEF2 fails to regulate synapse number in Fmr1 KO neurons we considered the possibility that MEF2 protein expression requires FMRP. On the contrary, expression levels of the major MEF2 isoforms in CA1 neurons, MEF2A and D, are normal in Fmr1 KO hippocampus (Fig. 3A,B). We also considered the possibility that FMRP is required for MEF2-dependent transcriptional activity. To address this possibility, dissociated neuronal hippocampal cultures from either wild-type or Fmr1-KO mice were co-transfected with MEF2-VP16-ER™ and MEF2 transcriptional reporter MRE-GFP. Induction of MEF2-VP16-ER™ activity with 4OHT induced equivalent, robust GFP expression in both genotypes (Fig. 3C, D). Similarly, the magnitude and kinetics of activity-dependent induction of the endogenous MEF2 target gene, Nurr77 (Flavell et al., 2006; Shalizi et al., 2006), is not different in Fmr1 KO slice cultures in comparison to wildtype, as determined by qRT-PCR (Fig. 3E). Taken together, these data argue that MEF2 expression and transcriptional activity is largely normal in Fmr1 KO mice and suggest that FMRP functions downstream of MEF2 transcription to regulate synapse number.
Figure 3. MEF2 levels and MEF2-induced transcription are normal in Fmr1 KO hippocampal neurons.
A Hippocampal lysates from WT and Fmr1 KO littermates (P12–14) were probed for the major MEF2 isoforms, MEF2A and MEF2D. B. Quantification of MEF2A and MEF2D levels (normalized to β-tubulin) WT and Fmr1 KO hippocampi (N = 6 mice from each genotype). C. Dissociated hippocampal neurons (7–8 days in vitro) from WT and Fmr1 KO mice were transfected with MEF2-VP16-ER™, the MEF2 transcriptional reporter, MRE-GFP, and mCherry (transfection indicator), treated 16–24hr with vehicle or 4-OHT, then imaged live. Scale bar = 100 μm. D. Group data of tamoxifen induced MRE-GFP positive WT and Fmr1 KO neurons expressed as a ratio of total number of transfected (mCherry-positive) neurons (mean±SEM; *** p < 0.001). The number of cells per condition >200 for each condition, n = 3 cultures. E. Neuronal depolarization of hippocampal slice cultures from WT and Fmr1 KO was induced for the indicated times with high K+ (55 mM KCl), isotonic media which activates MEF2 mediated transcription (Flavell et al., 2006). Quantitative real time PCR reveals similar activity-dependent induction of the MEF2 transcript Nurr77 in WT and Fmr1 KO cultures (n = 3 cultures (mice) for each time point). F. PC12 cells were transfected with MEF2-VP16-ER™ and treated with tamoxifen for the indicated times. Endogenous mRNA levels of Nurr77 and Fmr1 were determined by quantitative RT_PCR. MEF2 activation of PC12 cells effectively induces Nurr77 transcription (F1), but not Fmr1 (F2), indicating that Fmr1 is not a target gene regulated by MEF2 activation over the 8 hour time period analyzed..
As a downstream component of MEF2-dependent synapse elimination, MEF2 may directly or indirectly regulate expression of FMRP to affect synapse elimination. Several studies have examined gene promoter regions bound by MEF2 and none have observed binding of the Fmr1 promoter nor activation of FMRP expression in hippocampal neurons by MEF2. (Flavell et al., 2006; Flavell et al., 2008; Pulipparacharuvil et al., 2008). Similarly, we observed no change in Fmr1 mRNA expression in PC12 cells following activation of MEF2-VP16-ER™ by 4OHT, whereas Nurr77 mRNA was robustly induced (Fig. 3F). Therefore, these data suggest that existing FMRP may play an acute role together with MEF2 to mediate synapse elimination.
Acute postsynaptic expression of FMRP restores MEF2-dependent functional synapse elimination
To determine if FMRP plays an acute, postsynaptic role in MEF2 induced functional synapse elimination, we expressed both MEF2-VP16 and N-terminal GFP tagged wildtype FMRP (wtFMRP) in Fmr1 KO neurons (Darnell et al., 2005b; Pfeiffer and Huber, 2007). Prolonged expression (3–7 days) of wtFMRP alone in Fmr1 KO neurons has been shown to modestly suppress synapse number (~20%) (Pfeiffer and Huber, 2007). In contrast, brief periods (16–48 hr) of wtFMRP expression have no effect on synaptic function (Fig. 4C). Acute co-expression (16–30 hr) of wtFMRP together with MEF2VP16 in Fmr1 KO neurons induced robust (~40%) functional synapse elimination, as revealed by a decrease in evoked EPSC amplitude and mEPSC frequency (Fig. 4D). These results indicate that acute expression of FMRP restores MEF2-induced synapse elimination in Fmr1 KO neurons, and suggest that FMRP plays an acute role in MEF2-induced synapse elimination. Furthermore, because brief expression of wtFMRP alone does not induce synapse elimination in Fmr1 KO neurons, this result indicates that active MEF2 together with FMRP is necessary for rapid synapse elimination.
FMRP interacts with RNA through several RNA-binding motifs, including two hnRNP-K homology domains (KH domains, KH1 and KH2), and an arginine/glycine-rich RNA-binding motif (RGG box). The RGG box mediates a high affinity interaction with RNAs that form a tertiary structure termed a “G-quartet”, whereas the KH2 domain associates with a “kissing complex” RNA structure (Darnell et al., 2005a; Darnell et al., 2001; Schaeffer et al., 2001). A single point mutation in the KH2 domain of FMRP (I304N) abolishes FMRP interactions with “kissing complex” RNAs, as well as polyribosomes (Darnell et al., 2005a; Darnell et al., 2005b; Feng et al., 1997a; Laggerbauer et al., 2001). Notably, the I304N mutation occurs in a single patient with a severe form of FXS, implicating FMRP interactions with KH2 domain targets and polyribosomes in the disease (De Boulle et al., 1993). To determine if RNA interactions or translational control by FMRP is necessary for synapse elimination, we tested the ability of I304N FMRP or ΔRGG-FMRP (in which the entire RGG-box was deleted) to support MEF2-dependent synapse elimination in Fmr1 KO neurons. Although I304N FMRP and ΔRGG-FMRP were expressed similarly to wtFMRP when transfected into CA1 neurons (Pfeiffer and Huber, 2007), they failed to rescue MEF2VP16-induced synapse elimination in Fmr1 KO neurons, thus implicating FMRP-mediated translational control in synapse number regulation (Fig. 4E, F).
Our results implicate FMRP directly in a MEF2-dependent postsynaptic mechanism that regulates synapse number in a cell autonomous fashion. Synapse elimination requires the DNA binding capacity of MEF2 as well as the ability of FMRP to interact with RNA and polyribosomes. MEF2 resides mostly in the nucleus and FMRP is predominantly in the cytoplasm. FMRP shuttles in and out of the nucleus and may have the opportunity to interact with MEF2 (Feng et al., 1997b; Flavell et al., 2006; Pulipparacharuvil et al., 2008). However, we failed to observe a co-association between endogenous MEF2 and FMRP proteins in hippocampal slice cultures under several mild co-immunoprecipitation conditions (Fig. S3A). Similarly, MEF2A, 2C or 2D overexpressed in HEK293T cells did not co-IP with either exogenously-expressed GFP-wtFMRP or endogenous FMRP (Fig. S3B-D), suggesting that MEF2 and FMRP do not likely function as a complex to promote synapse elimination. More likely, FMRP regulates the processing, transport and/or translation of MEF2 generated transcripts; a scenario where FMRP and MEF2 function in the same pathway to elicit synapse elimination (Fig. S4A). Equally as plausible, FMRP and MEF2 may function in independent pathways to regulate expression of distinct transcripts that are each necessary for synapse elimination (Fig. S4B). In the future, the identification the MEF2 generated transcripts, as well as the FMRP-regulated mRNAs that mediate synapse elimination will help to differentiate between these possibilities. Because FMRP has been implicated in transport and translational control of mRNAs in the dendrite (Bassell and Warren, 2008), our results suggest that local or synaptic translation of MEF2-induced transcripts regulates synapse number and may provide a mechanism to eliminate specific subsets of synapses.
Loss of function mutations in FMR1 lead to mental retardation and autism in humans, and our data suggest that these disorders may arise from a deficit in activity- and MEF2-dependent refinement of synaptic connections. Consistent with this notion, neurons of adult Fmr1 KO mice and Fragile X patients display an excess of dendritic spines, similar to what is observed with knockout or knockdown of MEF2 isoforms (Barbosa et al., 2008; Flavell et al., 2006; Irwin et al., 2002; Irwin et al., 2001; Pulipparacharuvil et al., 2008). Interestingly, we did not observe a increase in spine number between developing Fmr1 KO and wildtype hippocampal neurons unless they were transfected with MEF2 (Fig. 1G,H). However, this is an early stage for spine formation (P12–14), and unlike functional synapses formed on the dendritic shaft, differences in spine density might not occur until later stages of development (Busetto et al., 2008; Sorra and Harris, 2000). This interpretation is consistent with previous reports of normal neocortical dendritic spine density in Fmr1 KO mice at 2–4 weeks of age (Nimchinsky et al., 2001), but excess spines in adult Fmr1 KO neurons (Galvez and Greenough, 2005). Excess dendritic spine number as a result of a deficit in MEF2 dependent synapse elimination may accumulate over the course of brain maturation and reach significant differences in the adult. Alternatively, additional mechanisms may maintain proper spine number in developing neurons independent of FMRP.
While mutations in MEF2 genes haven’t been reported in autism patients, MEF2 has been indirectly linked to autism. For example, the MEF2C gene is repressed by methyl-CpG binding protein 2 (MeCP2), the gene mutated in Rett Syndrome, an autism spectrum disorder (Chahrour et al., 2008). In addition, several activity- and MEF2-dependent transcripts, such as DIA1 (deleted in autism-1 or c3orf58), PCDH10 (protocadherin10), and Ube3A (E3 ubiquitin-protein ligase) display mutations or copy number variations in autistic patients (Glessner et al., 2009; Morrow et al., 2008; Wang et al., 2009). Therefore, dysfunction of MEF2 or MEF2-regulated transcripts may contribute to other forms of autism in addition to FXS.
EXPERIMENTAL PROCEDURES
Hippocampal Slice Cultures and Transfections
Organotypic hippocampal slice cultures were prepared from postnatal day (P) 6–7 WT or Fmr1 KO mice bred from the congenic C57BL/6 mouse strain using previously published protocols (Pfeiffer and Huber, 2007; Stoppini et al., 1991). Cultures were biolistically transfected at 3 DIV (McAllister, 2004). pcDNA1-MEF2-VP16 and pcDNA3-MEF2-VP16-ER™ and MRE-Luc constructs have been described previously (Flavell et al., 2006; Molkentin et al., 1996). MEF2-EN was provided by Dr. Eric Olson (UT Southwestern Medical Center) and has been previously described (Arnold et al., 2007; Shalizi et al., 2006). MRE-GFP reporter plasmid contains three copies of a canonical MEF2 DNA binding element upstream of a transcriptional start site and the open reading frame of EGFP. MRE-GFP was generated from the MRE-Luc plasmid (Flavell et al., 2006) by removing the open reading frame of firefly luciferase using NcoI and SacI restriction sites and subcloning the coding region of EGFP and the polyadenylation sequence of human growth hormone from pAAV-shRNA (Pulipparacharuvil et al., 2008) using the same restriction sites. All FMRP-GFP constructs were obtained from Dr. Jennifer Darnell at Rockefeller University, are driven by the human FMR1 promoter and have been described previously (Darnell et al., 2005b; Pfeiffer and Huber, 2007). wtFMRP-GFP, I304N FMRP-GFP and ΔRGG-FMRP-GFP were expressed at comparable levels as measured by fluorescence intensity of the neuron soma and dendrite (Pfeiffer and Huber, 2007).
For slice cultures transfected with MEF2-VP16-ER™, 4-hydroxytamoxifen (4OHT; 1 μM in 0.1% EtOH) was applied 24–48 hours after transfection to induce rapid nuclear transport of MEF2-VP16-ER™, and MEF2-dependent transcription. For experiments with MEF2-VP16-ER™ or MEF2-VP16, neurons were cotransfected with GFP (pC3-EGFP) or MRE-GFP. MRE-GFP allowed the visualization and confirmation of MEF2 activated transcription in individual recorded neurons. MEF2-EN was expressed for 24–48 hrs.
Electrophysiology
Simultaneous whole-cell recordings were obtained and analyzed from CA1 pyramidal neurons in slice cultures visualized using IR-DIC and GFP fluorescence to identify transfected and non-transfected neurons as described (Pfeiffer and Huber, 2007). See Supplemental Data.
Two-photon laser scanning microscopy and spine image analysis
To image dendritic spines, hippocampal slice cultures (3 DIV) were biolistically transfected with pA1-GFP (with or without MEF2-VP16) that expresses GFP and a myristolated form of GFP from two transcriptional start sites to better fill the dendritic spines. After 24–36h, apical dendrites of transfected CA1 neurons were imaged live using a Zeiss LSM 510 two-photon laser scanning microscope with an excitation wavelength of 920 nm. The two-photon excitation source was a Chameleon-Ti: sapphire standard laser. Secondary apical dendrites (150–200 μm from the cell body) were imaged with a 63X/0.9 n.a. water immersion lens (image field, 133 × 133 μm). Optical sections were taken at 1.0 μm spacing. Spine density was measured in consecutive sections using LSM510 (Zeiss) and ImageJ (NIH) software by an observer who was blind to transfection condition.
Supplementary Material
Acknowledgments
This research was supported by NIH grants 1F31NS050992 (BEP), NS045711, HD052731 (KMH), DA08227 (CWC), T32DA07290 and F32DA27265 (LNS), and grants from Autism Speaks (KMH), the Whitehall (CWC) and Simons Foundations (KMH and CWC). We would like to thank K. Loerwald and C. Hale for technical assistance; E. Kavalali for helpful discussions and E. Olson for providing the MEF2-EN construct.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busetto G, Higley MJ, Sabatini BL. Developmental presence and disappearance of postsynaptically silent synapses on dendritic spines of rat layer 2/3 pyramidal neurons. J Physiol. 2008;586:1519–1527. doi: 10.1113/jphysiol.2007.149336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran AR, Shah RD, Crair MC. Developmental homeostasis of mouse retinocollicular synapses. J Neurosci. 2007;27:1746–1755. doi: 10.1523/JNEUROSCI.4383-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005a;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005b;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997a;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997b;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet A. 2005;135:155–160. doi: 10.1002/ajmg.a.30709. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua JY, Smith SJ. Neural activity and the dynamics of central nervous system development. Nat Neurosci. 2004;7:327–332. doi: 10.1038/nn1218. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Idupulapati M, Gilbert ME, Harris JB, Chakravarti AB, Rogers EJ, Crisostomo RA, Larsen BP, Mehta A, Alcantara CJ, et al. Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am J Med Genet. 2002;111:140–146. doi: 10.1002/ajmg.10500. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel E, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Hessler NA, Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Biolistic transfection of cultured organotypic brain slices. Methods Mol Biol. 2004;245:197–206. doi: 10.1385/1-59259-649-5:197. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol Cell Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J Neurosci. 2007;27:3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232:232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. Embo J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Sorra KE, Harris KM. Overview on the structure, composition, function, development, and plasticity of hippocampal dendritic spines. Hippocampus. 2000;10:501–511. doi: 10.1002/1098-1063(2000)10:5<501::AID-HIPO1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PM, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga CJ, Walsh MF, Turrigiano GG. Temporal regulation of the expression locus of homeostatic plasticity. J Neurophysiol. 2006;96:2127–2133. doi: 10.1152/jn.00107.2006. [DOI] [PubMed] [Google Scholar]
- Wiesel TN. Postnatal development of the visual cortex and the influence of environment. Nature. 1982;299:583–591. doi: 10.1038/299583a0. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Yang G, Kwon E, Gan WB. Long-term sensory deprivation prevents dendritic spine loss in primary somatosensory cortex. Nature. 2005;436:261–265. doi: 10.1038/nature03715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.