Abstract
Our understanding of the molecular genetic basis of two common neurodegenerative dementias, Alzheimer’s disease (AD) and frontotemporal lobar degeneration (FTLD) has greatly advanced in recent years. Progranulin mutations were identified as a major cause of FTLD and a potential susceptibility factor for other forms of dementia. In addition, through copy-number analyses of previously identified disease genes and the study of microRNA regulation in dementia, new evidence emerged to support the view that subtle variability in the expression of known disease proteins may increase the risk for sporadic forms of dementia. Finally, in late-onset AD populations, the first genome-wide association studies were performed and novel potential AD susceptibility genes reported. These exciting findings provide novel insights into the disease mechanisms underlying dementia and hold promise for the development of potential treatments.
Introduction
A brief overview of the neurodegenerative dementias
Dementia or the significant loss of intellectual abilities such as memory capacity, severe enough to interfere with social or occupational functioning, is one of world’s most devastating and burdensome health conditions. It is estimated that 24.3 million people have dementia today, with 4.6 million new cases annually [1]. Moreover, as a result of our aging population, the number of people affected with dementia is expected to double every 20 years. Alzheimer’s disease (AD) is by far the most frequent form of dementia comprising 50-70% of all cases and affecting 40% of individuals older than 85 years [2]. A definite diagnosis of AD requires the presence of extraneuronal β-amyloid plaques and intraneuronal neurofibrillary tangles composed of hyperphosphorylated tau in the degenerating brain [3]. Dementia is also present as a common clinical feature in a host of related neurodegenerative diseases (Table 1). Lewy-body disease (LBD) is considered the second commonest cause of dementia in patients older than 65 years [4]; however, in presenile dementia patients frontotemporal lobar degeneration (FTLD) is the second most common diagnosis after AD [5]. FTLD patients are clinically characterized by personality changes and disinhibited behavior, often combined with a gradual and progressive language dysfunction. Memory impairment is typically preserved in the early phase of disease, which distinguishes them from patients with AD. Pathologically, about 40% of FTLD patients present with neuronal and/or glial tau aggregates (FTLD-tau), whereas the majority of FTLD patients show ubiquitin-immunoreactive cytoplasmic (NCI) and intranuclear inclusions (NII) composed of hyperphosphorylated and carboxy-terminal truncated fragments of the nuclear protein TAR DNA-binding protein 43 (TDP-43), characteristic of FTLD-U. Up to four subtypes of FTLD-U have been delineated that are based on the distribution of NCIs, dystrophic neurites and the presence of NIIs (ref). Despite well-defined clinical diagnostic criteria, the differential diagnosis of neurodegenerative dementias is often challenging. This is particularly true for AD and FTLD which have been suggested to belong to a spectrum of neurodegenerative diseases [6].
Table 1. Clinical and pathological features of neurodegenerative diseases associated with dementia.
| Neurodegenerative Disease | Main clinical features | Most affected brain regions | Characteristic pathology (major disease protein) |
|---|---|---|---|
| Alzheimer disease | Memory loss, cortical dementia (anomia, agnosia, apraxia) |
Cerebral cortex, hippocampus, amygdala, basal forebrain, brainstem (locus ceruleus) |
Senile plaques (β-amyloid); Neurofibrillary tangles and neuropil threads (tau) |
| Dementia with Lewy bodies (DLB) and Parkinson disease dementia (PDD) |
Subcortical dementia, parkinsonism, recurrent visual hallucinations, sleep disorders, fluctuation |
Limbic cortex, basal forebrain, amygdala, brainstem (substantia nigra, locus ceruleus) |
Lewy bodies and Lewy neurites (α-synuclein); senile plaques (β-amyloid) |
| Frontotemporal lobar degeneration | Cortical dementia, behavior and personality changes, language dysfunction; parkinsonism or motor neuron disease in some |
Frontal and temporal cortex, hippocampus, amygdala, basal ganglia (striatum) |
Neuronal cytoplasmic & intranuclear inclusions and dystrophic neurites (TDP-43) |
| Pick bodies (tau); ballooned neurons (neurofilament) | |||
| Progressive supranuclear palsy | Subcortical dementia, parkinsonism, vertical gaze palsy, axial dystonia, bulbar signs, |
Basal ganglia (striatum & pallidum), subthalamic nucleus, brainstem (substantia nigra), cerebellum |
Neurofibrillary tangles and tufted astrocytes (tau) |
| Corticobasal degeneration | Cortical/subcortical dementia; asymmetric rigidity, apraxia and dystonia; aphasia and frontal dementia in some |
Cerebral cortex, basal ganglia (striatum & pallidum) and brainstem (substantia nigra) |
Neurofibrillary tangles, neuropil threads and astrocytic plaques (tau); ballooned neurons (neurofilament) |
| Amyotrophic lateral sclerosis | Spasticity, muscle weakness and atrophy, bulbar signs; subcortical dementia in some |
Motor cortex, brainstem and spinal cord motor neurons |
Neuronal cytoplasmic, dystrophic neurites and intranuclear inclusions (TDP-43) |
| Huntington’s disease | Chorea, psychiatric problems; subcortical dementia in some |
Cerebral cortex, basal ganglia (striatum) |
Intranuclear inclusions and cytoplasmic aggregates (huntingtin) |
| Transmissible spongiform encephalopathies |
Rapidly progressive cortical dementia, myoclonus, ataxia, visual problems, psychiatric problems; insomnia |
Cerebral cortex, basal ganglia, thalamus, cerebellum |
Spongiform degeneration, amyloid plaques (prion) |
In recent years, significant progress has been made in understanding the molecular genetics and underlying pathological substrates of AD and FTLD, which will be the focus of the current review. For additional information on LBD and other Parkinson-related dementias we refer to recent reviews by Bonifati et al. [7] and Ferman and Boeve [8].
Genetic aspects of AD and FTLD
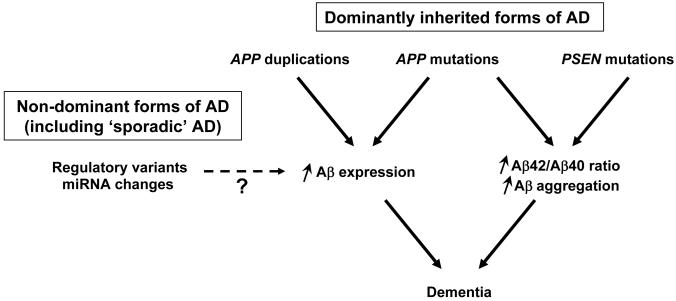
AD is a genetically complex and heterogeneous disorder [9]. Less than 10% of AD patients have an early-onset of disease (before the age of 65 years), and about 60% of these patients are familial, with an autosomal dominant inheritance of the disease in approximately 13% of patients [10]. In autosomal dominant AD families, rare mutations in the amyloid precursor protein gene (APP) and the presenilin 1 (PSEN1) and 2 (PSEN2) genes have been identified [11-13]. The presenilins are components of the proteolytic γ-secretase complex that together with β-secretase (BACE 1) generates β-amyloid from APP [14]. Two major β-amyloid species of either 40 (Aβ40) or 42 (Aβ42) amino acids are produced with Aβ40 being the more common and Aβ42 being the more fibrillogenic and neurotoxic species. The identification of mutations in the genes encoding both the substrate and the key enzyme for the generation of β-amyloid has led to major advances in the characterization of the pathophysiology of AD and provides the main support for the amyloid cascade hypothesis which has dominated the AD field for over 15 years [15]. According to this hypothesis, the central event in AD is the imbalance between the production, maturation and clearance of β-amyloid in brain leading to neuronal degeneration and dementia (Figure 1). This original hypothesis has undergone alterations over time [16], primarily in the description of the nature of pathogenic β-amyloid species that is proposed to initiate deleterious events and cause AD, and it should be mentioned that additional disease mechanisms have also been proposed [17, 18].
Figure 1. The amyloid cascade hypothesis.
Autosomal dominant familial forms of Alzheimer’s disease (AD) mainly result from mutations in PSEN1 and to a smaller extent from mutations in PSEN2 and APP, including APP duplications. PSEN mutations lead to aberrant γ-secretase cleavage of APP resulting in an overproduction of the more fibrillogenic Aβ42 and an increased Aβ42/Aβ40 ratio. All missense mutations in APP are located at or near the three proteolytic cleavage sites of APP and depending on their location result in (i) increased production of Aβ40 and Aβ42, (ii) increased Aβ42/Aβ40 ratio, or (ii) enhanced fibrillogenic potential of mutant Aβ, accompanied by vascular tropism responsible for cerebral amyloid angiopathy (CAA). The essence of the amyloid cascade is that the increased production or decreased clearance of Aβ peptides result in the aggregation and accumulation of Aβ, which in a yet uncertain pathogenic form, triggers a number of downstream deleterious events, finally leading to neuronal death. Recently described APP duplications in AD patients also fit this model. Although it has not been finally demonstrated, it is likely that APP duplication leads to enhanced Aβ expression, which is phenotypically associated with AD and CAA in patients. Finally, this model suggests that in sporadic forms of AD slight increases in APP expression, mediated either by genetic variants in the promoter or 3′ untranslated regions of APP or by micro RNA changes, could constitute risk factors for late-onset AD.
Aggregation of late-onset AD also occurs in families, though 75-85% of late-onset AD is sporadic [9]. The disease in these patients is expected to result from the interaction of multiple susceptibility genes and unknown environmental factors. So far, the ε4 allele of the apolipoprotein E (APOE) gene has been the only consistently replicated genetic risk factor for late-onset AD [19]. Meta-analyses showed a 3-times increased risk to develop AD in individuals carrying one copy of the APOE ε4 allele and a 15-times increased risk for APOE ε4 homozygote individuals [20]. The APOE ε4 allele further modifies the onset of disease, shifting it to an earlier age [21] Several mechanisms have been proposed to explain APOE ε4 detrimental effects in AD (for review, see [22]). Through its fundamental role in lipid transports, APOE is implicated in numerous cellular pathways linked to AD, including cholesterol redistribution, oxidative stress, neurite outgrowth, tau phosphorylation and β-amyloid clearance and aggregation. It was demonstrated that the β-amyloid plaque load is increased in APOE ε4 allele carriers [23] and that the APOE ε4 allele enhances fibrillar Aβ burden in cognitively normal older people [24].
In contrast to AD, FTLD patients usually present first symptoms in their 50s and 60s, with a positive family history of dementia in as much as 50% of patients [5]. In the past decade, several different genes and chromosomal loci have been associated with FTLD [25]. Mutations in the microtubule associated protein tau (MAPT) and progranulin (GRN) genes were identified as important causes of FTLD, explaining 10-25% of familial FTLD patients and 5-10% of all FTLD [25-27]. Mutations in the genes encoding charged multi-vesicular body protein (CHMP2B) and valosin-containing protein (VCP) on 9p21-12 were further reported as rare causes of FTLD [28, 29]. Several families with FTLD and motor neuron disease (MND) have also shown linkage to 9p21-13 but to a gene other than VCP, which still awaits identification [30, 31].
The detection of rare disease causing mutations in the autosomal dominant dementia genes unequivocally shaped our current understanding of the pathogenic mechanisms underlying AD and FTLD. However, despite these major advances, the cause of the disease in the majority of familial dementia patients and in the common sporadic forms largely remained unknown. Here, we review the novel insights into the molecular genetics of dementia; we discuss the identification of mutations in GRN as a novel cause of FTLD and a potential risk gene for other dementias, and fascinating new data suggesting that genetic variability in the expression of known disease proteins may contribute to the risk for the common sporadic forms of dementia and may cause disease in previously unexplained dementia families. We specifically discuss the importance of copy-number variants (CNVs) and the regulation of gene expression by micro RNAs (miRNAs), two novel disease mechanisms implicated in dementia. Finally, we summarize the recent progress made in the identification of genetic risk factors for AD and FTLD.
PGRN and TDP43: new players in dementia
The most exciting recent breakthrough in the search for novel causal dementia genes was undoubtedly the identification of loss-of-function mutations in GRN as a major cause of FTLD-U [26, 27]. The identification of GRN mutations had long been hampered by its genomic location on chromosome 17q21, only 1.7Mb centromeric of the other major FTLD gene, MAPT, whose role in the development of FTLD-U was unclear. Only after exhaustive candidate-gene sequencing in 17q21-linked FTLD-U families, mutations in GRN could be identified.
GRN is a member of the epithelin family of growth factors and is described in the literature by various synonyms including acrogranin, epithelin precursor, proepithelin and PC cell-derived growth factor [32]. It is a secreted precursor protein composed of a signal peptide and 7.5 tandem repeats of a rare 12 cysteinyl motif [33, 34]. As a result of proteolytic cleavage of GRN by extracellular proteases, a family of active ~6KDa peptides (granulins A to G and paragranulin) are formed that each contain 10-12 highly conserved cysteine residues which are folded into four stacked β-hairpins stabilized by disulphide bridges critical for biological function [35]. Both GRN and the granulins are widely expressed and have been implicated in a range of biological processes including development, wound repair, inflammation and tumorigenesis [36, 37]. At present, the role of GRN and the granulins in brain remains unknown; however, recent in vitro findings suggest that GRN may act as a neurotrophic factor [38].
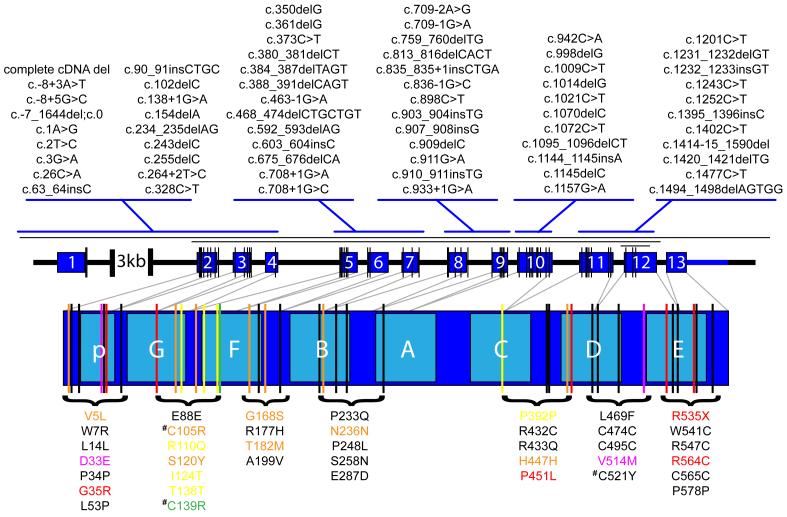
In less than three years, 66 different pathogenic GRN mutations have been reported in 199 families (FTD mutation database) [39]. Mutations are found in all GRN exons, except in the most 3′ exon 13, and include different types of mutations, such as nonsense (N=14) and splice-site (N=11) mutations as well as small insertions and deletions leading to a shift in the normal reading frame (N=34) (Figure 2). All GRN mutations are expected to lead to the loss of 50% functional GRN, suggesting a haploinsufficiency disease mechanism [40]. Loss-of-function mutations are not typically responsible for dominantly inherited diseases, however, soon after the identification of the first GRN mutations, expression analyses in mutation carriers showed a 50% reduction in GRN transcript due to nonsense-mediated mRNA decay (NMD) as well as a 30-35% reduction of full-length GRN protein, without detectable levels of truncated GRN species [26]. The haploinsufficiency disease mechanism has since been validated by the identification of mutations directly affecting the Met1 translation initiation codon (N=3), which prevent translation of the mutant transcript, and more recently by the identification of larger GRN deletions (N=3), including a complete heterozygous GRN locus deletion [41]. Finally, the p.A9D mutation located in the hydrophobic core of the signal peptide segregated with FTLD-U in the autosomal dominant family HDDD2 [42]. This mutation showed evidence of cytoplasmic missorting with extremely low expression in cell culture along with significantly reduced levels of mutant GRN mRNA in brain [43].
Figure 2. Spectrum of GRN mutations in dementia.
Schematic representation of the genomic structure of progranulin (GRN) and the mRNA encoding the GRN protein. The dark blue numbered boxes in the genomic structure indicate non-coding exon 1 and coding exons 2-13. The light blue lettered boxes in the GRN protein refer to the individual granulin domains. Mutations in GRN were recently reported as a novel cause of dementia. Loss-of-function mutations in GRN result in frontotemporal lobar degeneration with TDP-43 positive inclusions (FTLD-U). A total of 66 different loss-of-function mutations scattered over all GRN exons except exon 13, have been reported. These mutations are indicated with vertical black lines on the GRN exons and listed above the exons with their cDNA numbering relative to the largest GRN transcript (GenBank accession number NM_002087.2). One complete and two partial GRN deletions have also been identified as indicated with horizontal black lines above the GRN genomic structure. An additional 39 patient specific mutations with unknown pathogenic significance were identified in neurodegenerative disease patients. These include 28 missense mutations, 10 silent mutations and one nonsense mutations in GRN exon 13. These mutations are indicated with vertical lines on the GRN protein relative to the individual granulin domains and listed below the GRN protein with the protein numbering relative to GenPept accession number NP_002078.1. Each of the mutations is color coded to indicate that the mutations were identified in patients with a clinical diagnosis of FTLD (black), FTLD and ALS (orange), ALS (yellow), AD (red), AD and FTLD (green) or AD and PD (purple). Missense mutations affecting conserved cysteine residues within the granulin peptides are indicated with a hash symbol.
The sequencing analyses of GRN in an ever-increasing number of dementia patients and controls also identified numerous missense and silent mutations with unknown pathogenicity (Figure 2). Thus far, 39 patient-specific genetic variants were observed; however, none of these segregated with disease in extended dementia families and the disease mechanism remains poorly understood. In-vitro analyses of missense mutations p.P248L and p.R432C demonstrated that they were expressed as immature proteins, but inefficiently transported through and partially degraded within the secretory pathway resulting in a significant reduction of secreted GRN [43]. An additional three missense mutations (p.C105R, p.C139R and p.C521Y) are predicted to affect highly conserved cysteine residues within the granulin peptides, which may disrupt the disulphide bonds necessary to maintain the granulin quaternary structure and function. Indeed, two independent studies detected reduced GRN levels in plasma of a patient and in serum of a patient and an asymptomatic carrier of the p.C139R mutation affecting granulin F [44, 45]. Together it is expected that at least some missense mutations may cause disease through a (partial) loss of GRN function, while others may function as susceptibility alleles, potentially by making neurons more vulnerable to neurodegeneration through subtle decreases in neurotrophic support.
A limited number of studies have looked at the frequency of GRN mutations in other neurodegenerative diseases, including AD [39]. GRN loss-of-function mutations, similar to those identified in FTLD-U, were not identified in population-based mutation screenings of AD, amyotrophic lateral sclerosis (ALS) or Parkinson disease (PD). One possible exception is the nonsense mutation, p.R535X, identified in a clinically diagnosed AD patient; however, cDNA analyses predicted that this mutation would escape NMD [46]. In contrast, numerous potentially pathogenic missense mutations were identified in AD, ALS and PD patients (Figure 2). This is an interesting observation which suggests that a partial loss of GRN function could also contribute to the neurodegenerative disease process in dementias other than FTLD. In support of this hypothesis, genetic association studies showed significant association of a haplotype spanning the coding GRN region in a Belgian late-onset AD case-control population [47], which was partially confirmed in a Finnish AD population, although in this case the effect was male-specific [48]. In ALS, common GRN variants (rs9897526, rs34424835, and rs850713) and a haplotype including these variants were significantly associated with a reduction in age at onset and a shorter survival after onset of ALS in a Belgian ALS population [49]. The association of GRN variants and survival after disease onset was further replicated in a Dutch ALS population [49]. Additional mutation and association studies in extended AD, ALS and PD populations should shed light on the importance of GRN in the development of these related neurodegenerative disorders.
Soon after the discovery of GRN mutations, the nuclear protein TDP-43 was identified as the major disease protein in all ubiquitin-positive inclusions in FTLD-U patients as well as in the ubiquitinated inclusions in the lower motor neurons in sporadic patients with ALS [50]. It was further shown that all GRN mutation carriers have a common FTLD-U subtype, characterized by TDP-43 immunoreactive NCIs, short, thin neurites in layer II of the cortex and lentiform NIIs [51-53]. This FTLD-U subtype is referred to as Type 1 by Mackenzie et al. [51] and Type 3 by Sampathu et al. [53]. One study provided a possible link between the loss of functional GRN and TDP-43 pathology, by showing that decreased GRN levels can induce caspase-dependent accumulation of TDP-43 fragments in-vitro [54]; however this finding was not confirmed by a second study using both human cell lines and zebrafish [43].
TDP-43 is a highly conserved protein, expressed by a variety of tissues and cell types including neurons, with proposed functions that include the regulation of transcription, alternative splicing and the transport and stabilization of mRNA [55]. TDP-43 has also been implicated in miRNA biogenesis [56]. Through these functions, TDP-43 may potentially be involved in the pathophysiology of FTLD-U and ALS, as well as in other neurodegenerative dementias, including AD and Lewy-body related diseases, were TDP43-immunoreactivity was also reported in a subset of patients [57-59]. Initial mutation screenings of the TARDBP gene, encoding TDP-43, in FTLD and AD populations, did not support a direct role for TARDBP in the etiology of dementia [60]. However, TARDBP missense mutations were identified as a cause of sporadic and familial ALS, supporting a direct role for TDP-43 in neurodegeneration [61, 62]. More recently, the spectrum of phenotypes associated with TARDBP mutations expanded to include FTLD-ALS, when two genealogically unrelated French patients carrying the p.G295S mutation were reported who each presented with clinical FTLD, two years prior to the development of ALS symptoms [63]. Also, an Italian patient carrying p.G294V developed clinical AD three years prior to the onset of ALS [64]. These findings suggest that TARDBP mutations may also be a rare cause of dementia.
Finally, mutations in the FUS/TLS (fused in sarcoma/translocation in liposarcoma) gene, encoding another RNA associated protein with structural and functional similarities to TDP-43, were recently identified in patients with familial ALS. Whether mutations in FUS/TLS are also involved in the etiology of dementia has not yet been determined.
Copy-number alterations as a cause of dementia
In the past few years, genome-scanning array technologies and comparative DNA-sequence analyses have revealed an unexpected abundance of submicroscopic DNA rearrangements in normal individuals [65, 66]. These genomic rearrangements were collectively termed copy number variations (CNVs) and defined as “DNA segments of at least 1kb in size, for which copy number differences have been observed in the comparison of two or more genomes” [67]. According to the database of genomic variants (http://projects.tcag.ca/variation/), almost 20,000 CNVs overlapping a total of 7,000 genes have already been discovered, illustrating that CNVs represent a substantial proportion of the total genetic variability in human populations.
The identification of duplications and triplications of the α-synuclein gene in autosomal dominant Parkinson’s disease (PD) provided the first evidence for the involvement of rare CNVs in neurodegenerative diseases [68, 69]. The first hint for a possible role in dementia came from the observation that presence of an additional copy of the APP gene on chromosome 21q21 in patients with Down syndrome, leads to overproduction of APP and deposition of Aβ peptide in amyloid plaques and the development of early-onset AD associated with cerebral amyloid angiopathy (CAA) [70]. In 2006, an extensive screen for APP copy-number mutations finally resulted in the identification of duplications at the APP locus in 5 families with autosomal dominant early-onset AD associated with CAA [71]. Despite variable sizes of the genomic duplications in these families, ranging from 0.58 to 6.37Mb and involving 5 to 12 genes, the phenotypes of patients were similar, without any obvious signs of Down syndrome. These results were further corroborated by another study, which demonstrated that APP duplication is sufficient to cause early-onset AD with CAA [72].
These data are of importance for several reasons. First, they provide further evidence that increased gene dosage can be involved in the etiology of neurodegenerative disorders caused by peptide or protein accumulation, emphasizing the importance of copy-number analyses in molecular genetic studies of dementia. Second, mechanistically, APP duplication in early-onset AD constitutes a strong support to the amyloid cascade hypothesis (Figure 1). It demonstrates that increased APP gene dosage may initiate the cascade of events leading to β-amyloid plaques and neurofibrillary tangles, although it remains to be evaluated to what extent APP duplication leads to protein overexpression. Finally, it suggests that even a small increase in APP expression (<50% increase in expression), perhaps resulting from alterations in the 5′ or 3′ regulatory elements of APP, could constitute a risk factor for late-onset AD. In fact, APP promoter analyses in a Dutch and a Belgian early-onset AD population and an extended Belgian late-onset AD population, revealed a number of APP promoter mutations with an in-vitro 1.2-1.8-fold neuron-specific increase in APP transcriptional activities [73, 74]. Although the association of rare APP promoter mutations with AD could not be confirmed in a French AD case-control population, genetic variant rs463946 located in the APP upstream regulatory region showed evidence of association with AD in two independent French AD case-control series [75]. Also, Lv et al. [76] demonstrated that two SNPs (rs466433 and rs364048) in linkage disequilibrium with rs463946, were associated with AD in Chinese Han populations.
In addition to APP duplications, a number of partial and complete gene deletions have been reported in autosomal dominant AD and FTLD. The first report described genomic deletions encompassing PSEN1 exon 9 in patients presenting with AD associated with cotton wool plaques and spastic paraparesis [77]. More recently, deletions of GRN and MAPT genes were reported in FTLD. In GRN, a complete GRN locus deletion was described in a Belgian FTLD patient without known family history of dementia [41], followed by a second French family with two affected siblings carrying partial GRN deletions encompassing exons 1-11 [78]. In this family, one patient presented with FTLD-U, while the other was diagnosed with clinical PD, further illustrating the phenotypic variability associated with GRN mutations. Although previously hypothesized, the identification of GRN deletions established haploinsufficiency as the sole mechanism of GRN mutations necessary to initiate the disease process. This was further supported by the observation of strongly reduced GRN expression levels in plasma in all studied GRN loss-of-function mutation carriers, including a GRN deletion carrier [44].
Lastly, a partial MAPT genomic deletion encompassing exons 6 to 9 was found in an FTLD patient [79]. This genomic deletion resulted in a truncated tau protein that (i) loses its ability to bind microtubules and (ii) acquires the ability to bind microtubule-associated protein 1B (MAP-1B), another axonal MAP. Of note, complete genomic deletions of MAPT are not associated with dementia but were previously identified in individuals with mental retardation [80-82].
These studies demonstrate that CNVs leading to protein overexpression, haploinsufficiency or to a gene product with modified functional properties may contribute to various phenotypes and mechanisms leading to AD and FTLD. Collectively, these data highlight the importance of well-regulated expression of neurodegenerative disease proteins, not only as a cause of dominantly inherited disease in previously unexplained families but also as a risk factor for the more common sporadic forms of dementia.
Altered micro RNA regulation in dementia
Increasing evidence suggests that miRNAs may be a contributing factor in neurodegeneration and could potentially influence dementia risk. miRNAs are a class of small, endogenous, non-coding RNA molecules that serve as posttranscriptional regulators of gene expression [83]. They are thought to directly promote degradation of target mRNAs or suppress translation of corresponding protein via non-perfect base-pairing with the target mRNAs [84]. The current release of miRBase (http://microrna.sanger.ac.uk/), the primary repository and database resource for miRNA data maintained by the Sanger Institute, contains entries for 533 human miRNAs; however, it is predicted that there are as many as 1,000 miRNAs in the human genome that likely regulate 30% of all human transcripts. Many miRNAs are tissue-specific and/or temporally regulated in their expression. Interestingly, compared to other organs, human brain expresses an exceptionally diverse spectrum of distinct miRNAs and at somewhat higher levels [85].
The most popular approach to identify specific miRNAs implicated in dementias has been miRNA microarray profiling in brain tissue samples derived from patients and controls. Using miRNA expression profiling in temporal cortex samples from a well characterized clinicopathological series of elderly subjects, either non-demented without AD pathology, non-demented with early AD pathology, mild cognitive impairment (MCI) with moderate AD pathology and AD, Wang et al. identified miR-107 to be specifically decreased early in the course of AD [86]. In situ hybridization further demonstrated that miR-107 is strongly expressed in neurons with a characteristic laminar expression pattern, which suggested a specific decrease of miR-107 in cortical layers with abundant AD pathology. Computational analyses predicted BACE1 mRNA as a target of miR-107 and correlative mRNA expression studies confirmed that subjects with lower miR-107 levels (generally presenting with more AD pathology) showed higher BACE1 mRNA levels. An independent miRNA profiling study by Hebert and colleagues using temporal cortex of AD cases and age-matched controls further confirmed the importance of BACE1 regulation by miRNAs and identified the miR-29a/b-1 cluster as a potential major suppressor of BACE1 protein expression [87]. The miR-29a/b-1 cluster was significantly and specifically downregulated in AD patients and correlated with increased expression of BACE1 protein in AD brain. They also showed a tight correlation of miR-29a/b-1 and BACE1 expression during brain development and in isolated primary cells. Further evidence came from Boissonneault and colleagues who showed that miR-298 and miR-328 regulate BACE1 protein expression in mouse cultured neuronal cells [88]. Finally, in vitro evidence suggested that brain expressed miR-106a can also directly regulate APP protein levels through translational repression of APP mRNA [89]. Together these studies provide strong support for a regulatory role of miRNAs in BACE1 expression, with possible implications for sporadic AD. Changes in miRNA expression may well contribute to the increased BACE1 expression observed in sporadic AD patients, providing a novel link between sporadic AD and the amyloid cascade hypothesis (Figure 1) [90]. Interestingly, additional BACE1 expression regulation by non-coding RNAs was recently reported when a natural antisense transcript for BACE1 was identified [91]. The levels of BACE1 antisense transcripts were elevated in AD patients and in APP transgenic mice and alterations in BACE1 antisense RNA concentrations impacted Aβ40 and Aβ42 production [91].
Recently the first miRNA array on pooled miRNA samples of AD cases and controls was performed, revealing yet another miRNA elevated in AD brain, miR-146a, whose expression is regulated by the transcription factor NF-κB [92]. The authors showed that the NF-κB sensitive miR-146a was able to downregulate complement factor H, an important repressor of the inflammatory response in the brain, suggesting a novel regulatory pathway which may contribute to inflammatory pathology in AD. Unfortunately, each miRNA profiling study in AD has thus far identified a different set of differentially expressed miRNAs, which underlines the challenge of using human brain samples with intrinsic significant biological variation. Confirmation of these findings in independent AD patient and control populations will therefore be critical.
In contrast to AD, miRNA array profiling studies have yet to be performed in other dementia populations. However, an important hint towards a possible involvement of miRNA in the development of FTLD was recently provided by a combination of genetic and biochemical studies. In a series of pathologically confirmed FTLD-U patients, it was shown that carriers homozygous for the T-allele of rs5848, located in a miR-659 binding site in the 3′ untranslated region of GRN, have a 3.2-fold increased risk to develop FTLD-U compared to homozygous C-allele carriers [93]. In vitro studies confirmed that miR-659 can regulate GRN expression, with miR-659 binding more efficiently to the high risk T-allele of rs5848 resulting in augmented translational inhibition of GRN. These results are consistent with the loss-of-function disease mechanism of GRN mutations and further emphasize the role of gene dosage effects in neurodegenerative disorders. Moreover, these findings suggest a larger contribution of GRN to the development of FTLD then initially anticipated. Follow-up studies in additional FTLD-U patient-control populations should now be performed to confirm these findings.
The widespread role of miRNAs in human diseases, now including dementias, has suggested that miRNAs might be viable targets for therapeutic intervention. An important future challenge associated with miRNA therapeutics for dementia will be the efficient and targeted delivery of miRNAs or antisense oligonucleotides to the brain [94].
Finally, Cogwell and colleagues discovered that miRNAs can be detected in the cerebrospinal fluid (CSF) and importantly that the expression pattern of miRNAs are altered in AD patients compared to controls [95]. This study provides initial hope that miRNAs could provide accessible biomarkers to aid clinical diagnosis in the future.
Novel genetic risk factors in dementia
Since the identification of the three autosomal dominant early-onset AD genes, APP, PSEN1 and PSEN2, well over a decade ago, no novel causal AD genes and few AD loci have been reported [9]. This can be explained, in part, by the low prevalence of familial AD (less than 1% of the total number of AD patients have another family member with AD) and the resulting scarcity of extended AD families with sufficient power for genetic linkage studies. In recent years, the focus of AD genetic studies has therefore shifted towards association studies aimed at the identification of genetic risk factors, which has led to the identification of several potential AD risk genes.
One popular way to identify new susceptibility genes for AD has been to perform association studies in functional candidate genes, selected based on current knowledge and hypotheses of the physiopathological mechanisms underlying AD. In this view, the autosomal dominant dementia genes (APP, PSEN1, MAPT, PRNP) and genes encoding for proteins involved in APP cleavage (e.g. BACE-1, nicastrin, presenilin enhancer-2) or β-amyloid degradation (e.g. insulin degrading enzyme, neprilysin) have been studied. Thus far, these studies produced mostly inconsistent results and additional analyses of potential disease variants, including functional studies, will be essential to distinguish false positives from true genetic risk factors. One interesting finding is the identification of positive association of AD with a subtype of the extended H1 MAPT haplotype, which has been suggested to be more efficient in driving MAPT gene expression than the H2 haplotype [96, 97]. Despite the lack of MAPT mutations in AD, these findings suggest that tau dysfunction may contribute to AD risk. Another important candidate gene that was recently reported to be associated with AD is SORL1, a neuronal sorting receptor involved in recycling of APP from the cell surface via the endocytic pathways. Two distinct clusters of intronic SNPs, one towards the 5′ end and one towards the 3′ end of SORL1, were significantly associated with late-onset AD in multiple case-control populations [98]. Since exonic functional variants could not be identified, it was hypothesized that intronic regulatory sequences within SORL1 might govern cell type-specific or tissue-specific expression of SORL1. Lastly, novel candidate genes have also been characterized through study of expression profiles in AD brains. This strategy enabled the identification of the calcium homeostasis modulator 1 gene (CALHM1), located on chromosome 10q24.33, which encodes a multipass transmembrane glycoprotein that controls cytosolic Ca2+ concentrations and β-amyloid levels. Dreses-Werringloer et al. found that the coding p.P86L variant in CALHM1 increases β-amyloid levels by interfering with the CALHM1-mediated Ca2+ permeability and was associated with late-onset AD in four case-control series [99]; however, independent follow-up studies have not been able to confirm these findings [100-102]. For a continuously updated list of the genes most strongly associated with AD based on meta-analyses of all published studies, we refer to the AlzGene database (http://www.alzforum.org/res/com/gen/alzgene/default.asp)[103].
The completion of the Human Genome Project, combined with advances in high-throughput and high-density genotyping technology, has led to the emergence of genome wide association studies (GWAS). For GWAS, contrary to candidate gene studies, no a priori knowledge about the disease is needed. These studies currently represent the most promising and powerful tool to raise new susceptibility genes. Although the GWAS field is still in its relative infancy, eight GWAS have already been completed for AD and a number of novel susceptibility genes have been proposed (Table 2). However, caution is warranted when interpreting the reported findings. First, with the exception of APOE, none of the reported novel genes overlapped between studies. This may be explained by limited sample sizes, slight differences in study design and the stringent corrections for multiple testing inherent to these types of studies. In an attempt to overcome this last hurdle, Feulner et al. performed an initial analysis of their GWAS data only including the current top 10 candidate genes according to the AlzGene database [104]. Using this approach, all four analyzed genes that were previously identified through GWAS (GAB2, PGBD1, PCK1 and LMNA) showed nominally significant association, compared to only two genes (MAPT and SORL1) identified through candidate gene studies. Second, since all GWAS studies were performed within the last two years, few independent replication studies have been reported and it is likely that some genes may not replicate. The only extensively studied gene identified by GWAS to date is GAB2, encoding a scaffolding protein implicated in numerous growth and differentiation signaling pathways [105]. Both positive and negative association studies for GAB2 have been reported and a current meta-analysis of all published studies suggests a significant association of GAB2 with AD [103]. Interestingly, some studies reported a stronger risk in those individuals who also carried an APOEε4 allele [105, 106]. In the future, independent replication and validation of GAB2 and other newly identified AD candidate genes in multiple populations combined with functional genomic analyses will be essential.
Table 2. GWAS performed in late-onset AD.
| GWAS | Study Design | Sample origin | Number of samples in GWAS (patients/controls) |
Genotyping Platform | Genes or SNPs most associated with late-onset AD |
Additional notes | Refs |
|---|---|---|---|---|---|---|---|
| Carrasquillo et al. | Clinical and neuropathological case-control |
USA | 844/1255 | Illumina HumanHap300 | APOE, PCDH11X | First AD candidate gene identified on chromosome X |
[111] |
| Beecham et al. | Clinical case-control | USA | 492/496 | Illumina HumanHap300 | APOE, FAM113B | Associated SNPs were analyzed in a previous GWAS [105] using imputation data |
[112] |
| Feulner et al. | Clinical case-control | Germany | 491/479 | Illumina HumanHap500 | APOE a | Only top 10 genes from AlzGene were analyzed |
[104] |
| Abraham et al. | Clinical case-control | UK | 1082/1239 | Illumina HumanHap300 and HumanHap240S |
APOE, LRAT | DNA pooling strategy was used |
[113] |
| Betram et al. | Family based | USA | 941/404 | Affymetrix GeneChip Human Mapping 500K Array |
APOE, ATXN1, CD33, rs11159647 at 14q31.2 |
First study using family-based method in AD field |
[114] |
| Li et al. | Clinical case-control | Canada and UK |
753/736 | Affymetrix GeneChip Human Mapping 500K Array |
APOE, GOLM1, rs9886784 at 9p24.3, rs10519262 at 15q21.2 |
[115] | |
| Grupe et al. | Clinical case-control | USA and UK | 380/396 | Celera Functional SNPs |
APOE, GALP, TNK1,
PCK1 |
GWAS performed with functional SNPs |
[116] |
| Coon et al. and Reiman et al. |
Clinical and neuropathological case-control |
USA and The Netherlands |
664(446)/422(290) | Affymetrix GeneChip Human Mapping 500K Array |
APOE, GAB2 |
GAB2 association only identified in APOEε4+ carriers |
[105, 117] |
Nominally significant association was also found for GAB2, PGBD1, PCK1, LMNA, MAPT and SORL1 but p-values were not corrected for multiple testing.
AD = Alzheimer’s disease; APOE = Apolipoprotein E; PCDH11X = protocadherin 11, x-linked; FAM113B= family with sequence similarity 113, member B; LRAT=lecithin retinol acyltransferase; ATXN1= Ataxin 1, CD33= CD33 molecule; GOLM1= golgi membrane protein 1; GALP = galanin-like peptide precursor; TNK1 = non-receptor tyrosine kinase 1; PCK1= phosphoenolpyruvate carboxykinase 1; GAB2 = GRB2-associated binding protein.
In contrast to AD, candidate-gene association studies in clinical FTLD populations have produced little success in recent years. In part this may reflect the heterogeneous composition of a clinical FTLD cohort, which is predicted to include both tau-positive and FTLD-U patients as well as a subset of patients with other pathological conditions. Clinicopathological studies have suggested that as much as 15-30% of patients with a clinical diagnosis of FTD have AD at autopsy [107-109]. It is expected that future studies, focused on pathologically more homogenous FTLD subpopulations, could lead to the identification of novel genetic risk factors involved in the various pathways underlying FTLD.
Concluding Remarks
Most of our current mechanistic insight into the pathogenesis of AD and FTLD has resulted from the molecular genetic dissection of the rare autosomal dominant dementia families and the identification of mutations in APP, PSEN1, PSEN2 and MAPT. The recent discovery of loss-of-function mutations in GRN as an important cause of FTLD-U identified yet another key player in these neurodegenerative disease processes [26, 27]. Given the presumed role of GRN in neuronal survival and the identification of possible (partial) GRN loss-of-function mutations in other dementias, including AD, a wider role for GRN as a genetic risk factor for neurodegeneration was suggested [40]. It is the hope that future studies using GRN animal models will shed light on the molecular pathway by which GRN maintains neuronal fitness, which could lead to identification of novel therapeutic targets [110].
Despite major progress in unraveling the etiology of the rare Mendelian forms of AD and FTLD, the currently known disease genes explain less than half of the familial dementia patients and only a minority of the apparently sporadic patients. However, recent interest in copy-number alterations led to the identification of genomic duplications of APP in AD and genomic deletions of GRN and MAPT in FTLD, explaining the disease in a number of previously unresolved families [41, 71]. Furthermore, a subtle change in expression of the known disease proteins was suggested to contribute to the risk for the common sporadic forms of dementia: APP promoter variants with an expected ~1.5-fold increase in APP transcriptional activities were identified in AD patients [74], while genetic variability in the 3′ untranslated region of GRN was associated with a 3-times increased risk to develop FTLD-U, as a result of partially reduced levels of GRN [93]. In addition, a new and emerging role for miRNAs in the development of AD was suggested, including a specific down-regulation in AD of miRNAs regulating BACE1, which correlated with an increase in BACE 1 expression in AD brain [86, 87]. Together these findings support a much larger role for the currently known disease genes in the etiology of dementia than initially anticipated. In addition, they hold the promise that therapeutic strategies designed to target the monogenic forms of the disease will also proof effective in sporadic dementia patients.
Finally, through GWAS, a novel opportunity of identifying dementia risk genes and associated disease pathways has emerged with several potential AD risk genes already reported (Table 2). Further validation and replication in appropriately large patient populations combined with functional characterization of these genes could lead to a better understanding of the pathophysiology underlying dementia and open new therapeutic possibilities for AD and FTLD.
Acknowledgements
We would like to thank the families who contributed samples that were critically important to past, present and future research. We also thank Richard Crook for preparation of Figure 2 and Dr. Dominique Campion for careful reading of the manuscript. Research in the authors’ laboratory was supported by the NIH (Mayo Clinic ADRC grant P50 AG16574), the Pacific Alzheimer Research Foundation and the Association for Frontotemporal dementia (AFTD).
GLOSSARY
- Nonsense-mediated decay (NMD):
Nonsense mediated decay is a eukaryotic quality control mechanism that selectively degrades mRNA species harboring premature termination (nonsense) codons to prevent the expression of truncated or erroneous proteins.
- Haploinsuffiency:
Haploinsufficiency refers to a situation where an individual who is heterozygous for a certain gene mutation, is clinically affected because 50% of the level of gene function is not sufficient to assure normal function.
- Cerebral amyloid angiopathy (CAA):
Cerebral amyloid angiopathy, also known as congophilic angiopathy, is a disease of small blood vessels in the brain in which deposits of β-amyloid in the blood vessel walls may lead to stroke, brain hemorrhage, or dementia. CAA is a common feature in AD patients with APP duplications and APP missense mutations in the α-secretase cleavage site.
- Linkage disequilibrium:
In population genetics, the term linkage disequilibrium refers to a situation where a particular allele at one locus is found together on the same chromosome with a specific allele at a second locus - more often than expected if the loci were segregating independently in the population.
REFERENCES
- 1.Ferri CP, et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366:2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breteler MM, et al. Epidemiology of Alzheimer’s disease. Epidemiol Rev. 1992;14:59–82. doi: 10.1093/oxfordjournals.epirev.a036092. [DOI] [PubMed] [Google Scholar]
- 3.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 4.McKeith I, et al. Dementia with Lewy bodies. Lancet Neurol. 2004;3:19–28. doi: 10.1016/s1474-4422(03)00619-7. [DOI] [PubMed] [Google Scholar]
- 5.Graff-Radford NR, Woodruff BK. Frontotemporal dementia. Semin Neurol. 2007;27:48–57. doi: 10.1055/s-2006-956755. [DOI] [PubMed] [Google Scholar]
- 6.van der Zee J, et al. Invited article: the Alzheimer disease-frontotemporal lobar degeneration spectrum. Neurology. 2008;71:1191–1197. doi: 10.1212/01.wnl.0000327523.52537.86. [DOI] [PubMed] [Google Scholar]
- 7.Bonifati V. Recent advances in the genetics of dementia with lewy bodies. Curr Neurol Neurosci Rep. 2008;8:187–189. doi: 10.1007/s11910-008-0030-1. [DOI] [PubMed] [Google Scholar]
- 8.Ferman TJ, Boeve BF. Dementia with Lewy bodies. Neurol Clin. 2007;25:741–760. vii. doi: 10.1016/j.ncl.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird TD. Genetic aspects of Alzheimer disease. Genet Med. 2008;10:231–239. doi: 10.1097/GIM.0b013e31816b64dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campion D, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goate A, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 12.Levy-Lahad E, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 13.Sherrington R, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 14.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 15.Hardy J. Amyloid, the presenilins and Alzheimer’s disease. Trends Neurosci. 1997;20:154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 16.Pimplikar SW. Reassessing the amyloid cascade hypothesis of Alzheimer’s disease. Int J Biochem Cell Biol. 2008 doi: 10.1016/j.biocel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bezprozvanny I, Mattson MP. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci. 2008;31:454–463. doi: 10.1016/j.tins.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Small SA, Duff K. Linking Abeta and tau in late-onset Alzheimer’s disease: a dual pathway hypothesis. Neuron. 2008;60:534–542. doi: 10.1016/j.neuron.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strittmatter WJ, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrer LA, et al. APOE and Alzheimer Disease Meta Analysis Consortium Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. Jama. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 21.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 22.Mahley RW, et al. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc Natl Acad Sci U S A. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drzezga A, et al. Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology. 2009;72:1487–1494. doi: 10.1212/WNL.0b013e3181a2e8d0. [DOI] [PubMed] [Google Scholar]
- 24.Reiman EM, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rademakers R, Hutton M. The genetics of frontotemporal lobar degeneration. Curr Neurol Neurosci Rep. 2007;7:434–442. doi: 10.1007/s11910-007-0067-6. [DOI] [PubMed] [Google Scholar]
- 26.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 27.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 28.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 29.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 30.Morita M, et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 31.Vance C, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 32.Bateman A, Bennett HP. Granulins: the structure and function of an emerging family of growth factors. J Endocrinol. 1998;158:145–151. doi: 10.1677/joe.0.1580145. [DOI] [PubMed] [Google Scholar]
- 33.Bhandari V, et al. Isolation and sequence of the granulin precursor cDNA from human bone marrow reveals tandem cysteine-rich granulin domains. Proc Natl Acad Sci U S A. 1992;89:1715–1719. doi: 10.1073/pnas.89.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plowman GD, et al. The epithelin precursor encodes two proteins with opposing activities on epithelial cell growth. J Biol Chem. 1992;267:13073–13078. [PubMed] [Google Scholar]
- 35.Hrabal R, et al. The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat Struct Biol. 1996;3:747–752. doi: 10.1038/nsb0996-747. [DOI] [PubMed] [Google Scholar]
- 36.He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med. 2003;81:600–612. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- 37.He Z, et al. Progranulin is a mediator of the wound response. Nat Med. 2003;9:225–229. doi: 10.1038/nm816. [DOI] [PubMed] [Google Scholar]
- 38.Van Damme P, et al. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol. 2008;181:37–41. doi: 10.1083/jcb.200712039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gijselinck I, et al. Granulin mutations associated with frontotemporal lobar degeneration and related disorders: an update. Hum Mutat. 2008;29:1373–1386. doi: 10.1002/humu.20785. [DOI] [PubMed] [Google Scholar]
- 40.Cruts M, Van Broeckhoven C. Loss of progranulin function in frontotemporal lobar degeneration. Trends Genet. 2008;24:186–194. doi: 10.1016/j.tig.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Gijselinck I, et al. Progranulin locus deletion in frontotemporal dementia. Hum Mutat. 2008;29:53–58. doi: 10.1002/humu.20651. [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee O, et al. HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann Neurol. 2006;60:314–322. doi: 10.1002/ana.20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shankaran SS, et al. FTLD-U linked missense mutations in the progranulin gene reduce progranulin production and secretion. J Biol Chem. 2007 doi: 10.1074/jbc.M705115200. [DOI] [PubMed] [Google Scholar]
- 44.Finch N, et al. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 2009 doi: 10.1093/brain/awn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sleegers K, et al. Serum biomarker for progranulin-associated frontotemporal lobar degeneration. Ann Neurol. 2009 doi: 10.1002/ana.21621. [DOI] [PubMed] [Google Scholar]
- 46.Brouwers N, et al. Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch Neurol. 2007;64:1436–1446. doi: 10.1001/archneur.64.10.1436. [DOI] [PubMed] [Google Scholar]
- 47.Brouwers N, et al. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology. 2008;71:656–664. doi: 10.1212/01.wnl.0000319688.89790.7a. [DOI] [PubMed] [Google Scholar]
- 48.Viswanathan J, et al. An association study between granulin gene polymorphisms and Alzheimer’s disease in Finnish population. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30889. [DOI] [PubMed] [Google Scholar]
- 49.Sleegers K, et al. Progranulin genetic variability contributes to amyotrophic lateral sclerosis. Neurology. 2008 doi: 10.1212/01.wnl.0000289191.54852.75. [DOI] [PubMed] [Google Scholar]
- 50.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 51.Mackenzie IR, et al. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–549. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackenzie IR, et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain. 2006;129:3081–3090. doi: 10.1093/brain/awl271. [DOI] [PubMed] [Google Scholar]
- 53.Sampathu DM, et al. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol. 2006;169:1343–1352. doi: 10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang YJ, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 56.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 57.Amador-Ortiz C, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol. 2007;61:435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higashi S, et al. Concurrence of TDP-43, tau and alpha-synuclein pathology in brains of Alzheimer’s disease and dementia with Lewy bodies. Brain Res. 2007;1184C:284–294. doi: 10.1016/j.brainres.2007.09.048. [DOI] [PubMed] [Google Scholar]
- 59.Nakashima-Yasuda H, et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol (Berl) 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- 60.Mackenzie IR, Rademakers R. The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr Opin Neurol. 2008;21:693–700. doi: 10.1097/WCO.0b013e3283168d1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008 doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 62.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benajiba L, et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- 64.Corrado L, et al. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Hum Mutat. 2009;30:688–694. doi: 10.1002/humu.20950. [DOI] [PubMed] [Google Scholar]
- 65.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 66.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feuk L, et al. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 68.Chartier-Harlin MC, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 69.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 70.Wisniewski KE, et al. Alzheimer’s disease in Down’s syndrome: clinicopathologic studies. Neurology. 1985;35:957–961. doi: 10.1212/wnl.35.7.957. [DOI] [PubMed] [Google Scholar]
- 71.Rovelet-Lecrux A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 72.Sleegers K, et al. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–2983. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 73.Brouwers N, et al. Genetic risk and transcriptional variability of amyloid precursor protein in Alzheimer’s disease. Brain. 2006;129:2984–2991. doi: 10.1093/brain/awl212. [DOI] [PubMed] [Google Scholar]
- 74.Theuns J, et al. Promoter mutations that increase amyloid precursor-protein expression are associated with Alzheimer disease. Am J Hum Genet. 2006;78:936–946. doi: 10.1086/504044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guyant-Marechal L, et al. Variations in the APP gene promoter region and risk of Alzheimer disease. Neurology. 2007;68:684–687. doi: 10.1212/01.wnl.0000255938.33739.46. [DOI] [PubMed] [Google Scholar]
- 76.Lv H, et al. Promoter polymorphisms which modulate APP expression may increase susceptibility to Alzheimer’s disease. Neurobiol Aging. 2008;29:194–202. doi: 10.1016/j.neurobiolaging.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Crook R, et al. A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin 1. Nat Med. 1998;4:452–455. doi: 10.1038/nm0498-452. [DOI] [PubMed] [Google Scholar]
- 78.Rovelet-Lecrux A, et al. Deletion of the progranulin gene in patients with frontotemporal lobar degeneration or Parkinson disease. Neurobiol Dis. 2008;31:41–45. doi: 10.1016/j.nbd.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Rovelet-Lecrux A.e.a. Partial Deletion of the MAPT Gene: A Novel Mechanism of FTDP-17. Human Mutation. 2009 doi: 10.1002/humu.20979. in press. [DOI] [PubMed] [Google Scholar]
- 80.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 81.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 82.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 83.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 84.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 85.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang WX, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hebert SS, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boissonneault V, et al. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patel N, et al. MicroRNAs can regulate human APP levels. Mol Neurodegener. 2008;3:10. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fukumoto H, et al. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- 91.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lukiw WJ, et al. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rademakers R, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol Med. 2006;12:99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 95.Cogswell JP, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 96.Myers AJ, et al. The H1c haplotype at the MAPT locus is associated with Alzheimer’s disease. Hum Mol Genet. 2005;14:2399–2404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 97.Myers AJ, et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25:561–570. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 98.Rogaeva E, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dreses-Werringloer U, et al. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer’s disease risk. Cell. 2008;133:1149–1161. doi: 10.1016/j.cell.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bertram L, et al. No association between CALHM1 and Alzheimer’s disease risk. Cell. 2008;135:993–994. doi: 10.1016/j.cell.2008.11.030. author reply 994-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Minster RL, et al. No association between CALHM1 variation and risk of Alzheimer disease. Hum Mutat. 2009 doi: 10.1002/humu.20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sleegers K, et al. No association between CALHM1 and risk for Alzheimer dementia in a Belgian population. Hum Mutat. 2009 doi: 10.1002/humu.20990. [DOI] [PubMed] [Google Scholar]
- 103.Bertram L, et al. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 104.Feulner TM, et al. Examination of the current top candidate genes for AD in a genome-wide association study. Mol Psychiatry. 2009 doi: 10.1038/mp.2008.141. [DOI] [PubMed] [Google Scholar]
- 105.Reiman EM, et al. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sleegers K, et al. Common variation in GRB-associated Binding Protein 2 (GAB2) and increased risk for Alzheimer dementia. Hum Mutat. 2009;30:E338–344. doi: 10.1002/humu.20909. [DOI] [PubMed] [Google Scholar]
- 107.Forman MS, et al. Frontotemporal dementia: clinicopathological correlations. Ann Neurol. 2006;59:952–962. doi: 10.1002/ana.20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Knibb JA, et al. Clinical and pathological characterization of progressive aphasia. Ann Neurol. 2006;59:156–165. doi: 10.1002/ana.20700. [DOI] [PubMed] [Google Scholar]
- 109.Knopman DS, et al. Antemortem diagnosis of frontotemporal lobar degeneration. Ann Neurol. 2005;57:480–488. doi: 10.1002/ana.20425. [DOI] [PubMed] [Google Scholar]
- 110.Vossel KA, Miller BL. New approaches to the treatment of frontotemporal lobar degeneration. Curr Opin Neurol. 2008;21:708–716. doi: 10.1097/WCO.0b013e328318444d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carrasquillo MM, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer’s disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beecham GW, et al. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. 2009;84:35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abraham R, et al. A genome-wide association study for late-onset Alzheimer’s disease using DNA pooling. BMC Med Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bertram L, et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li H, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 116.Grupe A, et al. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 117.Coon KD, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]