Summary
Signaling in the most conserved branch of the endoplasmic reticulum (ER) unfolded protein response (UPR) is initiated by sequence-specific cleavage of the HAC1/XBP1 mRNA by the ER stress-induced kinase-endonuclease IRE1. We have discovered that the flavonol quercetin activates yeast IRE1’s RNase and potentiates activation by ADP, a natural activating ligand that engages the IRE1 nucleotide binding cleft. Enzyme kinetics and the structure of a co-crystal of IRE1 complexed with ADP and quercetin reveal engagement by quercetin of an unanticipated ligand binding pocket at the dimer interface of IRE1’s Kinase Extension Nuclease (KEN) domain. Analytical ultracentrifugation and crosslinking studies support the preeminence of enhanced dimer formation in quercetin’s mechanism of action. These findings hint at the existence of endogenous cytoplasmic ligands that may function alongside stress signals from the ER lumen to modulate IRE1 activity and at the potential for the development of drugs that modify UPR signaling from this unanticipated site.
INTRODUCTION
The endoplasmic reticulum (ER) unfolded protein response (UPR) couples perturbation of the protein-folding environment in the ER lumen to rectifying changes in transcription and translation (Bernales et al., 2006; Ron and Walter, 2007). The oldest and most conserved branch of the UPR is initiated by IRE1, an ER localized type-I transmembrane protein whose lumenal domain senses ER stress and transmits the signal to the cytosolic effector domain (Cox et al., 1993; Mori et al., 1993; Tirasophon et al., 1998; Wang et al., 1998). The latter possess two distinct enzymatic activities: a protein kinase, whose only known substrate is IRE1 itself and a sequence-specific endoribonuclease that cleaves the HAC1 or XBP1 mRNA in yeast and metazoans, respectively (Cox and Walter, 1996; Mori et al., 1996; Yoshida et al., 2001; Calfon et al., 2002). IRE1 recognizes an RNA stem loop structure, found twice in its substrates. Cleavage occurs at both sites, and initiates an unconventional splicing event that is completed when the two ends of the mRNA are ligated. The spliced HAC1/XBP1 mRNAs encodes a transcription factor that activates numerous UPR target genes.
There is a good correlation between IRE1 oligomerization, autophosphorylation and RNase activity (Shamu and Walter, 1996; Bertolotti et al., 2000; Liu et al., 2000). Autophosphorylation favors nucleotide binding at the kinase active site. This promotes dimerization of IRE1’s cytosolic domain (Lee et al., 2008b). Interestingly, while kinase activity is required for IRE1 function in vivo (Cox et al., 1993; Mori et al., 1993), this requirement can be bypassed by a mutation that enables non-phosphorylated IRE1 to bind an ATP competitive inhibitor at the kinase active site (Papa et al., 2003). The crystal structure of the active form of yeast IRE1 provides further clues to the coupling of kinase and nuclease activity: The cytosolic portion of IRE1 forms an extensive back-to-back dimer. Nucleotide is bound in the conventional nucleotide-binding cleft (the kinase active site) and the phosphorylated activation loop of the kinase portion is disordered in one structure (Lee et al., 2008b) and ordered in another (Korennykh et al., 2009). Together, these observations support a model whereby a lumenal stress signal initiates oligomerization in the plane of the membrane promoting (trans)-autophosphorylation of IRE1’s activation loop. The resulting conformational change favors nucleotide binding, which, in turn, stabilizes the back-to-back dimer. The dimer interface extends through the length of IRE1, juxtaposing the Kinase Extension Nuclease (KEN) domain located at the C-terminus of each IRE1 protomer, which thereby attains an active conformation for sequence-specific RNA cleavage. The RNase activity of the dimer may be further reinforced by formation of higher-order oligomers (Aragon et al., 2009; Korennykh et al., 2009)
The aforementioned model, which places nucleotide binding at the center of IRE1 activation and subordinates kinase activity to that end, suggests the possibility that ligands that bind the nucleotide binding pocket (e.g. kinase inhibitors) may activate IRE1’s endonuclease (Han et al., 2009; Korennykh et al., 2009). The desire to modulate IRE1 activity is fuelled in part by evidence that chronic ER stress (and aspects of the cellular response to it) contribute to the time and use-dependent attrition of tissues with high secretory capacity, for example, the insulin-producing beta cells of the pancreas (Kaufman, 2002; Powers et al., 2009). Furthermore, in addition to promoting the first step in XBP1 splicing, phosphorylated mammalian IRE1 is able to activate Jun N-terminal kinase (JNK), by recruiting the scaffold protein TRAF2 (Urano et al., 2000). This latter activity of phosphorylated IRE1 is believed to contribute to some of the deleterious consequences of ER stress, such as insulin resistance (Ozcan et al., 2004). Therefore, ligands that drive a wedge between IRE1 kinase and RNase activity would be of special interest. Here we report on the surprising discovery of an additional ligand-binding pocket at the dimer interface of IRE1 that can be targeted to modulate IRE1’s RNase.
RESULTS
Identification of small molecules that modulate IRE1 activity
To detect modulators of IRE1 we developed a kinetic assay for the sequence-specific RNase activity. An RNA substrate was prepared incorporating the IRE1 recognition loop from XBP1 and a destabilized stem that dissociates upon cleavage. The RNA probe was modified at the 5’ and 3’ positions with AlexaFluor647 (AF647) and Black-Hole Quencher 3 (BHQ), respectively, resulting in a quenched IRE1 substrate. Cleavage relieves the BHQ-dependent quenching of AF647 allowing fluorescence. (Figure 1A).
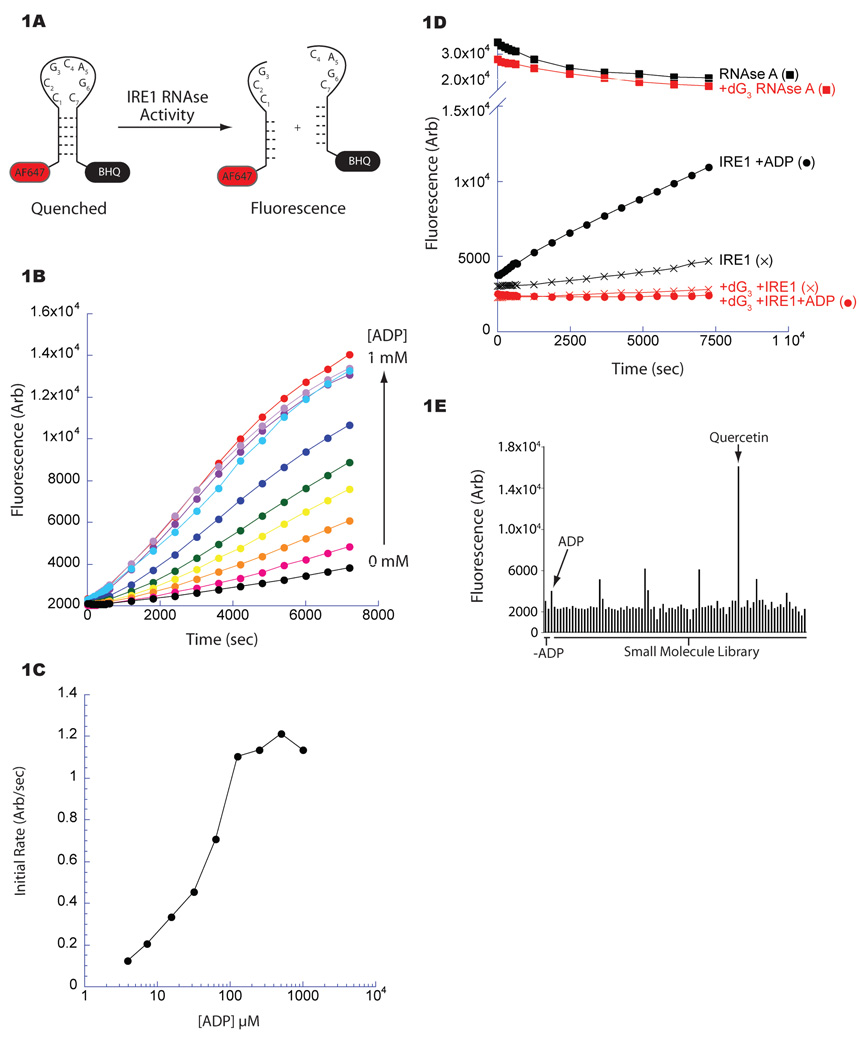
Figure 1. Small molecule screen for modulators of IRE1 RNase activity.
A. Fluorescent-based assay for yeast IRE1 (aa 658–1115) RNase activity. A stem loop RNA substrate incorporating an IRE1 endoribonuclease site (cleavage between G3-C4, labeled bases) was modified 5’ with AlexaFluor 647 (AF647) and 3’ with BlackHoleQuencher3 (BHQ). Cleavage alleviates quenching allowing fluorescence.
B. Fluorescence timecourse measuring IRE1’s RNase. The activity of IRE1 (1.0 µM) incubated with increasing concentrations of ADP was measured by the cleavage of the fluorescent substrate (25 nM) depicted in Figure 1A.
C. Quantification of the ADP-mediated activation of IRE1 RNase shown in Figure 1B. The initial rate of IRE1 RNase activity was plotted against ADP concentration demonstrating an EC50 of ~40 µM.
D. Fluorescence timecourse reporting on cleavage of the IRE1 substrate as in Figure 1A (black) or an altered substrate incorporating deoxyguanosine at position 3 (dG3; red), disrupting cleavage at that site. The maximal signal following cleavage of both substrates was determined by incubation with RNase A (squares).
E. Bar graphs depicting the activity of IRE1’s (1.0 µM) RNase in the presence of various small molecules (25 µM) from a collection of kinase inhibitors after a 10 minute reaction; the wells containing quercetin (25 µM) and ADP (2 mM) are indicated.
When purified from bacteria, the cytosolic portion of yeast IRE1 (residues 658–1115; inclusive of both the kinase and KEN domains) is phosphorylated (Sidrauski and Walter, 1997; Lee et al., 2008b; Korennykh et al., 2009), thus the assay is predicted to report on the liganddependent activation of IRE1 RNase. Cleavage of the probe by IRE1 was ADP-dependent with an EC50 of ~40 µM, consistent with the previously reported binding constant of ADP to IRE1 (KD = 20 µM) (Lee et al., 2008b) (Figure 1B & 1C). Removal of the 2’ hydroxyl from the 5’ residue at the predicted cleavage site (G3→dG3 of IRE1’s recognition loop) blocked cleavage, attesting to the sequence-specificity of the assay (Figure 1D).
IRE1 binds nucleotide in its conventional kinase active site cleft herein referred to as the nucleotide binding site. Therefore we searched for IRE1 modulators among a collection of small molecules that are known to bind the active site of various kinases. This exercise led to the identification of several small molecules that modulated IRE1’s RNase; the naturally occurring flavonol, quercetin, a well known ATP competitive kinase inhibitor (e.g. Sicheri et al., 1997; Walker et al., 2000), stood out as a particularly potent activator in this assay (Figure 1E).
Quercetin activates IRE1 through a site distinct from the nucleotide-binding site
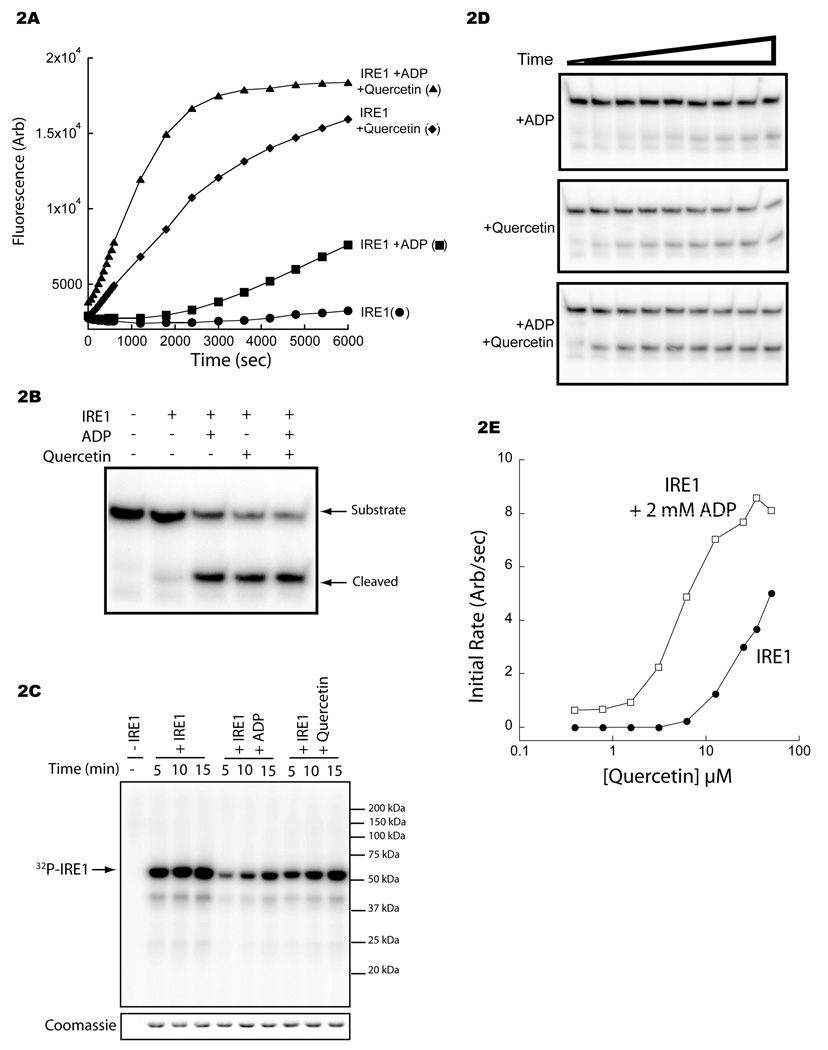
At a concentration of 30 µM, quercetin promoted ~10 fold more RNase activity than saturating concentrations of ADP (2 mM; Figure 1B & Figure 2A), without altering the specificity of IRE1’s RNase (Figure 2B and Supplemental Figure 1A).
Figure 2. Quercetin potentiates ADP-mediated IRE1 RNase activity.
A. Fluorescence timecourse of substrate (25 nM) cleavage by IRE1 (0.5 µM), alone (circles) and in the presence of ADP (2 mM, squares), quercetin (25 µM, diamonds) or both (triangles).
B. Autoradiograph showing the cleavage of a 32P-labeled IRE1 substrate (20 nM), of identical sequence to that depicted in Figure 1A, by IRE1 (0.5 µM) incubated with ADP (2 mM) or quercetin (25 µM) following a 2 hour reaction. The full-length substrate and cleavage product are indicated.
C. Autoradiograph of IRE1 (aa 658–1115) (0.5 µM) incubated with [32P]γ-ATP for the indicated time in the presence of ADP (30 µM) or quercetin (30 µM). A Coomassie stain of the same gel is shown below.
D. Autoradiograph depicting the time-dependent cleavage of a 32P-labeled IRE1 substrate (20 nM; as in Figure 2B) by IRE1 (0.5 µM) in the presence of ADP (2 mM), quercetin (25 µM) or both.
E. Plot of the initial rate of IRE1 RNase activity as a function of quercetin concentration in the absence (filled circles) and presence (open squares) of ADP (2 mM).
(see also Supplemental Figure 1).
To determine if quercetin’s activity correlated with its affinity towards the nucleotide-binding site, we compared ADP and quercetin’s ability to compete with ATP and block IRE1 autophosphorylation. Quercetin was the weaker inhibitor, indicating a lower affinity for the nucleotide-binding site (Figure 2C). These findings predict that if quercetin activates IRE1 exclusively by allosteric effects mediated from the nucleotide-binding site, the enhanced RNase activity of quercetin-bound IRE1 would be inhibited by an excess of ADP; as the weaker activator, but more avid ligand (ADP), displaces the more potent but less avid quercetin from a common binding site. Surprisingly the opposite was observed; addition of saturating amount of ADP to quercetin-activated IRE1 enhanced RNase activity (Figure 2A and 2D). Furthermore, chelating divalent cations by adding EDTA strongly attenuated ADP’s effect, indicating that potentiation is observed when ADP engages the nucleotide-binding site in complex with Mg+2;Supplemental figure 1B).
Analysis of the reaction’s initial rate revealed that in the presence of saturating concentrations of ADP (2 mM), the EC50 for quercetin-mediated activation of IRE1’s RNase was diminished from >20 µM (in the absence of ADP) to 6 µM in its presence (Figure 2E); in the absence of ADP, an accurate EC50 could not be determined, as the analysis was limited by quercetin’s solubility. Together these observations are consistent with simultaneous engagement of ADP and quercetin and imply the existence of a second site for ligand binding to IRE1.
A co-crystal structure of IRE1, ADP and quercetin defines an unanticipated ligand-binding pocket
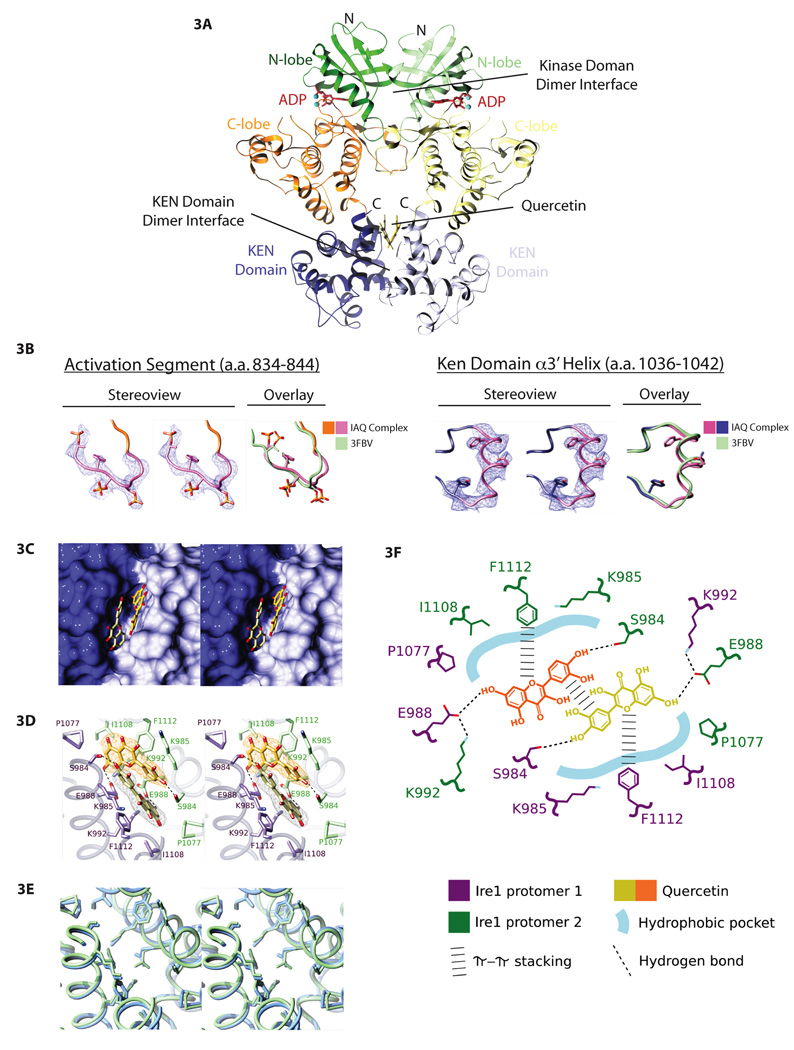
We solved the crystal structure of IRE1 in the presence of both quercetin and ADP (PDB: 3LI0), using the same construct of yeast IRE1 (658–1115) that had previously been crystallized with ADP alone (Lee et al., 2008b).
IRE1:ADP:quercetin crystals diffracted to better than 3.2 Å resolution (Supplemental Table 1) revealing a protein dimer defined by two discontinuous contact surfaces formed by the kinase domain and KEN domain (Figure 3A). The structure of IRE1 in the IRE1:ADP:quercetin complex demonstrated a tri-lobed architecture nearly identical to that of the previously reported IRE1:ADP complex (Supplemental Figure 2A) with two notable exceptions: Two regions that are disordered in the IRE1:ADP binary complex (PDB: 2RIO) are now visible as ordered structural elements in the IRE1:ADP:quercetin crystal structure, namely amino acids 837 to 844 corresponding to the N-terminal portion of the activation loop and amino acids 1036 to 1042 corresponding to a flexible region in the KEN domain thought to contain putative nuclease specificity determinants (Figure 3B). The structure of these regions corresponds well to an alternative crystal structure of the IRE1 cytosolic domain (PDB: 3FBV, Korennykh et al., 2009).
Figure 3. The crystal structure of IRE1–bound by quercetin and ADP reveals a second ligand binding site, the Q-site.
A. Structure of IRE1 (658–1115, Δ869–892) crystallized in the presence of both ADP and quercetin shown in ribbon format (PDB: 3LI0). The kinase domain N- and C-terminal lobes of each protomer in the dimer are green and orange, respectively, and the KEN domains are blue. The kinase and the KEN domain dimer interfaces are indicated. ADP and quercetin are shown in a ball-and-stick representation.
B. Electron density (blue-wire mesh) and tube representation of regions of the IRE1:ADP:quercetin ternary complex that are unstructured in the IRE1:ADP dimer (PDB: 2RIO) are shown in stereo view (two left panels) and superimposed on the structure of the same segments in PBD: 3FBV. Left – Amino acids 837–844 including phosphorylated residues S840, S841, and T844 in the activation segment of IRE1 are shown with side chains displayed in a ball and stick format. Right – Amino acids 1036–1042 from the α3’ helix of the KEN domain are shown with side chain residues in ball and stick format. Both structured regions show significant overlap with the previous crystal structure of oligomeric IRE1 (PDB – 3FBV).
C. Stereo view of the two-fold symmetric quercetin binding pocket (Q-site). The solvent accessible surface of the KEN domain is shown in light blue and dark blue for the two protomers. Quercetin is shown in a ball-and-stick representation.
D. Structure of the Q-site with the residues from the two protomers (purple and green) that interact with quercetin (shown as ball-and-stick in the same view as Figure 3E). Unbiased sigma A-weighted Fo-Fc electron density (see Supplementary Figure 3 for details) for each quercetin molecule is shown as orange or gray wire-mesh.
E. A comparison of the Q-site from IRE1:ADP:quercetin (PDB: 3LI0, blue) and IRE1:ADP (PDB: 2RIO, green) shown in a stereo view and depicted as tubes. The residues that line this pocket are shown in ball-and-stick format (the quercetin ligand has been removed from the IRE1:ADP:quercetin structure).
F. Schematic depicting the spatial arrangement and interactions of IRE1 and quercetin that define quercetin binding to the Q-site.
(also see Supplemental Table 1 and Supplemental Figure 2.)
Apart from ADP at the nucleotide binding site of IRE1 (which is indistinguishable between the IRE1:ADP and IRE1:ADP:quercetin crystals, Supplemental Figure 2B), the IRE1:ADP:quercetin complex also demonstrated a unique density at the dimeric interface of the KEN domains attributed to two quercetin molecules (Figure 3A, 3C). We refer to this deep solvent exposed symmetric binding pocket as the Q-site. The electron density for quercetin in the Q-site is highly asymmetric and we were able to unambiguously dock the larger bi-cyclic benzopyran ring and smaller monocyclic phenyl ring of quercetin into the wider and narrower part of the quercetin electron density, respectively (Figure 3D). The similarity at the Q-site between the structures of IRE1:ADP:quercetin and IRE1:ADP (Figure 3E) suggests that this site is a preformed pocket generated through dimerization.
The Q-site is lined by residues S984, K985, E988, K992, P1077, I1108 and F1112, with the quercetin pair interacting with residues from both IRE1 protomers (Figure 3D). This site is predicted to be largely conserved in mammalian IRE1 (Supplemental Figure 2C & 2D) and is large enough to accommodate potential endogenous ligands the size of a sterol molecule (Supplemental Figure 2E & 2F). The primary contacts that dictate quercetin binding are mediated by 1) pi-pi stacking between F1112 and the bi-cyclic benzopyran ring of quercetin; 2) a hydrophobic interaction between K985 and the monocylic quercetin ring (which is rotated 30 degrees relative to the benzopyran ring); and 3) hydrogen bonds between the 4’-hydroxyl of quercetin and the S984 sidechain. Quercetin also engages the other protomer in the IRE1 dimer through formation of a hydrogen bond between the 7-hydroxyl and the carboxylate moiety of E988, stabilizing the dimeric interface between the two protomers (Figure 3D). The two quercetin molecules in the Q-site are symmetrically arranged about the two-fold axis of the IRE1 dimer bridging and strengthening the interaction between the two IRE1 protomers through propagation of the pi-pi stacking in the pocket across the KEN domain dimer interface (Figure 3F).
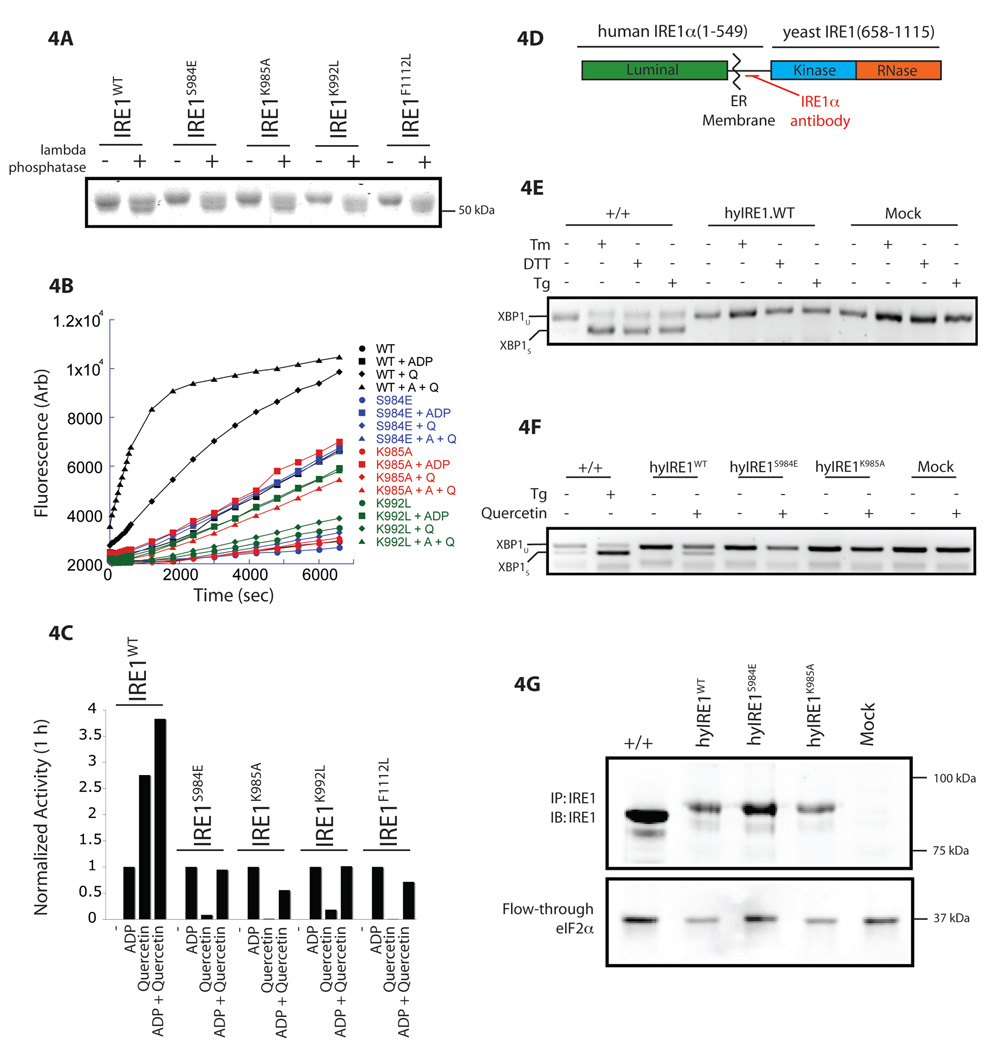
To test the importance of the contacts predicted by the crystal structure, we examined several mutations in residues predicted to interact with the bound ligand (Figure 3F). Like the wildtype, IRE1S984E, IRE1K985A, IRE1K992L and IRE1F1112L mutants were well expressed in E. coli and purified as soluble phosphorylated proteins (as evidenced by enhanced mobility following incubation with lambda phosphatase, Figure 4A) and exhibited ADP-mediated RNase activity (Figure 4B–C). All four mutations abolished quercetin-mediated activation of IRE1’s RNase (Figure 4B–C), validating the importance of the contacts revealed in the crystal structure to quercetin-mediated activation and supporting the crystal-based model of the ligand’s disposition in the Q-site.
Figure 4. Mutation of residues lining the Q-site interfere with quercetin-mediated activation of IRE1 RNase.
A. Coomassie stained SDS-PAGE of wildtype and mutant yeast IRE1 (aa 658–1115) variants before and after incubation with lambda phosphatase.
B. Timecourse of IRE1 RNase activity of IRE1WT (black), IRE1S984E (blue), IRE1K985A (red), and IRE1K992L (green) incubated alone, with ADP (+A; 2 mM), quercetin (+Q; 25 µM) or both (+A +Q). The concentration of each enzyme was adjusted to approximate the activity of IRE1WT in the presence of ADP.
C. Bar graph comparing IRE1 RNase activity of IRE1WT, IRE1S984E, IRE1K985A, IRE1K992L, and IRE1F1112L measured as in Figure 4B. The activity of the different enzymes, defined by the fluorescent signal following 1 h treatment, in the presence of ADP was normalized to 1.
D. Domain structure of the human-yeast IRE1 chimera (hyIRE1). The rabbit serum used to detect the endogenous and chimeric proteins recognizes the juxta-membrane region as indicated.
E. RT-PCR analysis of XBP1 mRNA purified from wild-type MEFs (+/+) or IRE1 knockout cells transduced with empty vector (Mock) or hyIRE1WT exposed to tunicamycin (Tm; 2.5 µg/mL; 4h), DTT (2 mM; 1 h) or thapsigargin (Tg; 0.5 µM; 1h). The position of the unspliced (XBP1U) and spliced (XBP1S) product is indicated.
F. RT-PCR analysis of XBP1 mRNA purified from wild-type MEFs (+/+) or IRE1 knockout cells transduced with empty vector (Mock), hyIRE1WT, hyIRE1S984E, or hyIRE1K985A and exposed to 450 µM quercetin for 2 h or thapsigargin (Tg; 0.5 µM for 2h), as indicated.
G. Immunoblot of endogenous IRE1 or the hyIRE1 chimeras immunoprecipitated from lysates of the cells shown in Figure 4F. The hyIRE1 chimera migrates slower than endogenous IRE1 due to the larger size of the yeast kinase and endonuclease domains. The anti-eIF2α immunoblot from the flow-through of the immunoprecipitation reaction (lower panel) serves as a loading control.
(also see Supplemental Figure 3).
Quercetin’s pleiotropic effects in mammalian cells, which include induction of ER stress (Ito et al., 1999), obscure analysis of possible interactions with endogenous mammalian IRE1 in vivo, whereas inertness in yeast, suggesting poor bioavailability, precluded analysis in that species (data not shown). In an effort to circumvent these difficulties, we stably transfected IRE1 knockout cells (Calfon et al., 2002) with a chimeric IRE1 construct consisting of residues 1–549 of human IRE1α (encoding the lumenal, transmembrane and juxta-membrane domains) and residues 658–1115 of yeast IRE1 (the kinase and endonuclease domains) (hyIRE1; Figure 4D). Fortunately, the chimeric protein proved unresponsive to ER stress alone (Figure 4E) and thus could be exploited to evaluate the Q-site dependent activation of hyIRE1 RNase by quercetin in vivo. Exposure of hyIRE1-rescued knockout cells to quercetin promoted hyIRE1-dependent splicing of endogenous XBP1 (Figure 4F) and mutations that blocked quercetin-mediated activation of yeast IRE1 in vitro also blocked activation in vivo (Figure 4F & 4G). While this observation does not indicate that Q-site activation by quercetin is sufficient to promote XBP1 splicing, it strongly suggests that engagement of the Q-site is required for quercetin-mediated activation.
The crystal structure of IRE1 in complex with two other activators (JAK inhibitor I (PDB: 3LI1) and CDK1/2 inhibitor III (PDB: 3LI2), also indentified from a library of kinase inhibitors) showed that these ligands engage the conventional nucleotide binding site and, like ADP, activated the RNase of both the wildtype and quercetin-unresponsive K985A mutant IRE1 (Supplemental Figure 3). Furthermore, no density was observed at the Q-site in either co-crystal structure or in the complex of IRE1 with a different activator that binds the nucleotide site, APJ1999 (data not shown and PDB 3FBV). Together these findings point to the unusual mode of IRE1 activation through the Q-site by quercetin.
In the crystal structure quercetin is modeled with the 3’-hydroxyl facing the bulk solvent (Figure 3C, 3D), but the resolution of the X-ray data cannot unambiguously rule out an alternative orientation whereby the monocyclic phenyl ring is rotated 180 degrees (Supplemental Movie 1). To identify the most probable orientation of the quercetin 3’-hydroxyl group (syn or anti relative to the 3 and 3’ hydroxyl moieties) and to further probe the interaction between quercetin and the Q-site, we compared the ability of structurally diverse flavonols to activate IRE1’s RNase in vitro (Figure 5A).
Figure 5. The hierarchy of IRE1 activation by flavonols supports a model of ligand binding in the syn orientation of the prime ring.
A. Chemical structures of flavonol-based analogs of quercetin. Quercetin and other relevant flavonols are shown with the 3- and 3’-moieties in the syn orientation; except morin, which is shown in the anti conformation due to clash between the 3- and 2’-hydroxyls.
B. Timecourse of RNase activity of IRE1 (0.5 µM) incubated in the presence of various flavonols (25 µM). The activity of IRE1 in the absence of flavonol is indicated by the x symbols.
C. Timecourse of RNase activity of IRE1 (0.5 µM) incubated in the presence of both ADP (2 mM) and flavonols (25 µM). The activity of IRE1 in the absence of flavonol is indicated by the x symbols.
D. Autoradiograph of IRE1 (0.5 µM) incubated with [32P]γ-ATP in the presence of the known kinase inhibitor staurosporine (25 µM, a positive control for inhibition) or the flavonols (25 µM), following either 10 or 20 min reaction, as indicated. A Coomassie stain of the 20 min gel is shown below. The asterisk indicates the presence of an activity in the sample containing myrecetin that reproducibly degrades IRE1.
E. RT-PCR analysis of XBP1 mRNA purified from wild-type MEF (+/+) or IRE1 knockout cells transduced with hyIRE1WT following exposure to thapsigargin (Tg; 0.5 µM for 2 h) or flavonols (450 µM for 2 h), as indicated.
F. RT-PCR analysis of XBP1 mRNA purified from wild-type MEF (+/+) or IRE1 knockout cells transduced with hyIRE1WT, hyIRE1S984, hyIRE1K985A or empty vector (Mock) following exposure to thapsigargin (Tg, 0.5 µM for 2h) or Luteolin (450 µM for 2 h), as indicated.
(also see Supplemental Movie 1).
Among the flavonols tested, quercetin was the most potent activator of IRE1 RNase in vitro, both in the absence (Figure 5B) and presence of saturating concentrations of ADP (Figure 5C). Kampferol, a quercetin derivative lacking the 3’-hydroxyl, was next in potency. Kampferol is a weak inhibitor of IRE1 autokinase activity (Figure 5D), suggesting that its ability to activate IRE1’s RNase is derived mainly through binding the Q-site. The weak potentiation with ADP of isorhamnetin and morin, luteolin and apigenin (relative to quercetin) supports quercetin binding in the syn-orientation between its 3- and 3’-hydroxyl moieties, which likely defines the angular rotation of the two ring systems of quercetin.
The structure activity relationship in vivo was similar to that observed in vitro, but produced some surprises: Luteolin and galangin were strong activators of XBP1 splicing in IRE1 knockout cells expressing hyIRE1 (Figure 5E). The potent activation of XBP1 splicing by luteolin was dependent on binding the Q-site, as splicing was strongly attenuated in cells expressing Q-site mutant hyIRE1 chimeras (Figure 5F). The basis of the observed differences in the relative activity of flavonols in vitro and in vivo remains to be determined.
Quercetin increases the population of IRE1 dimers in vitro
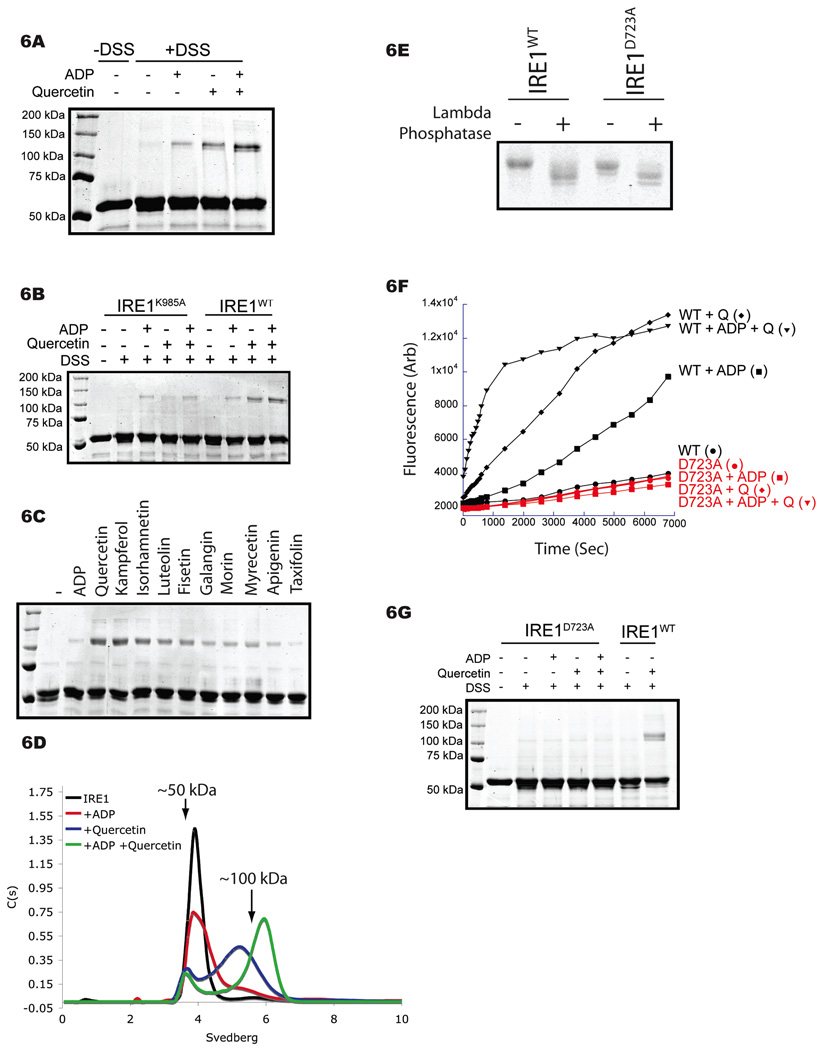
As the Q-site is located at the dimer interface of the KEN domain and as dimerization is a pre-requisite for RNase activity (Lee et al., 2008b; Korennykh et al., 2009), we sought to determine if quercetin binding affects dimer stability. On SDS-PAGE IRE1 (658–1115) has the mobility predicted of a monomer (53 kDa), which is unaltered by the crosslinker disuccinimidyl suberate (DSS). Upon addition of ADP, a faint slower migrating, DSS-dependent band that corresponds in mobility to a predicted dimer was observed. The intensity of this slower migrating band was increased by incubation with quercetin and was increased further upon addition of both quercetin and ADP (Figure 6A). Quercetin’s effects on IRE1 crosslinking were abolished by the Q-site mutant IRE1K985A, although ADP-dependent crosslinking was still observed (Figure 6B). The hierarchy at which the various flavonols activated IRE1’s RNase correlated with their ability to promote IRE1 crosslinking (Figure 6C), further demonstrating that quercetin-mediated crosslinking is dependent on binding to the Q-site identified in the crystal.
Figure 6. Quercetin increases the population of IRE1 dimers.
A. Coomassie stained SDS-PAGE of yeast IRE1 (aa 658–1115, 5 µM) incubated with ADP (2 mM), quercetin (25 µM) or both in the absence (−DSS) or presence of 200 µM disuccinimidyl suberate (+DSS), an irreversible chemical cross-linker.
B. Coomassie stained SDS-PAGE of IRE1K985A and IRE1WT incubated with ADP (2 mM), quercetin (25 µM) or both in the presence of 200 µM DSS.
C. Comassie stained SDS-PAGE of IRE1 (5 µM) incubated with ADP (2 mM) and the flavonols (25 µM) in the presence of DSS (200 µM).
D. Sedimentation distribution of yeast IRE1 (aa 658–1115, 5 µM; black) in the presence of ADP (2 mM; red), quercetin (25 µM; blue) or both (green), as determined by sedimentation velocity analytical ultracentrifugation. Approximate molecular weights are indicated.
E. Coomassie stained SDS-PAGE of IRE1WT and IRE1D723A incubated without or with lambda phosphatase.
F. Fluorescence timecourse of RNase activity of IRE1WT (black) and IRE1D723A (red) in the presence of ADP (2 mM), quercetin (Q; 25 µM) or both.
G. Coomassie stained SDS-PAGE of IRE1WT and IRE1D723A (5 µM) incubated with ADP (2 mM), quercetin (25 µM) or both in the presence of 200 µM DSS.
Quercetin’s effect on the oligomeric state of IRE1 was further explored by sedimentation velocity analytical ultracentrifugation (AUC). Non-liganded IRE1 sedimented as a single ~4S species, corresponding to the predicted molecular weight of ~50 kDa. In the presence of ADP, IRE1 sedimented predominantly as a 4S monomer (50kDa) with a small amount of a larger species (5.3S), consistent with the previously demonstrated ADP-dependent dimerization of IRE (Lee et al., 2008b). Quercetin alone increased the population of the higher molecular weight state and addition of ADP increased this further (Figure 6D), corresponding well with both enzymatic activity (Figure 2A) and DSS-dependent crosslinking (Figure 6B). Significant populations of larger species were not identified by AUC, indicating that in the conditions used here IRE1 oligomers consisted primarily of dimers. The observed differences in the sedimentation coefficient of IRE1 oligomers in the presence of ADP, quercetin or both is consistent with the formation of a more stable dimer by the ternary complex, which presumably undergoes less rapid equilibration with the monomer during sedimentation (Brown et al., 2008).
To further evaluate dimerization’s role in quercetin-dependent IRE1 activation, we assessed quercetin’s ability to activate an IRE1 variant with a well-characterized mutation in the dimer interface, D723A, located ~30Å from the Q-site. As reported previously, IRE1D723A was purified from bacteria as an active protein kinase in a yield similar to that of wildtype IRE1 (Figure 6E). Neither ADP, nor quercetin nor a combination thereof activated IRE1D723A (Figure 6F). Furthermore, neither ligand promoted DSS-induced crosslinking of the mutant protein (Figure 6G). These findings indicate that quercetin-mediated activation of IRE1’s RNase entails dimerization at the same interface required for ADP-mediated activation, as suggested by the co-crystal structure.
Quercetin-binding at the Q-site modifies the nucleotide binding site
Dimerization of IRE1’s kinase and endonuclease domain, critical to activation of the RNase, is dependent on engagement of the nucleotide binding site by a suitable ligand. Under normal circumstances autophosphorylation must precede such ligand binding, however this requirement can be bypassed by a mutation that alters the architecture of the nucleotide binding site (Papa et al., 2003). Given this plasticity, we sought to determine if quercetin-mediated activation of IRE1 also depends on autophosphorylation.
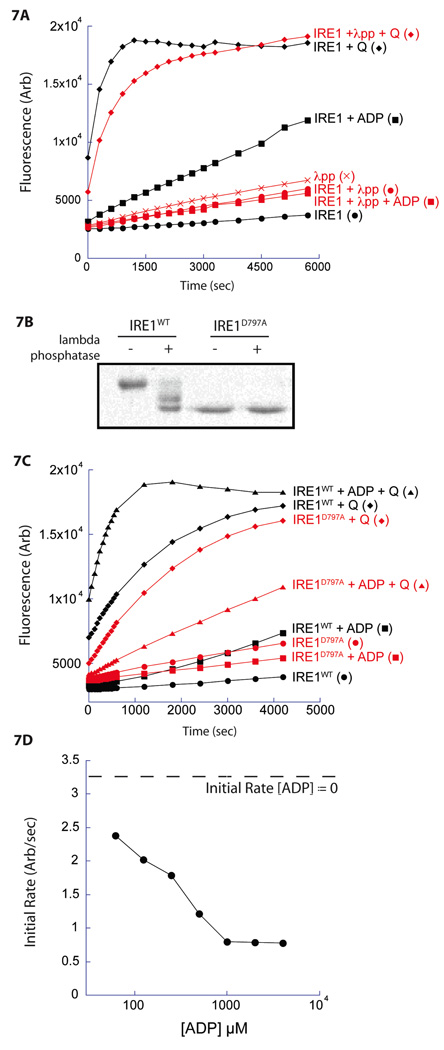
As expected, dephosphorylation with lambda phosphatase markedly diminished ADP-mediated RNase activity. Surprisingly, the dephosphorylated IRE1 remained strongly responsive to quercetin (Figure 7A). However, as dephosphorylation with lambda phosphatase was incomplete (Figure 7B), we further explored the relationship between quercetin-mediated activation and phosphorylation with a mutation that abolishes kinase activity without (directly) altering the structure of the nucleotide binding site.
Figure 7. The nucleotide binding site and the quercetin binding pocket interact to regulate IRE1 RNase.
A. Timecourse of IRE1 RNase activity incubated without or with lambda phosphatase (λpp) in the presence of ADP (2 mM) and quercetin (Q; 25 µM).
B. Comassie-stained SDS-PAGE of IRE1WT and IRE1D797A incubated with or without lambda phosphatase.
C. Fluorescence timecourse of RNase activity of IRE1WT (filled symbols) and IRE1D797A (open symbols) incubated with ADP (2 mM), quercetin (Q; 25 µM) or both.
D. Quantification of RNase activity for IRE1D797A incubated with quercetin (25 µM) and increasing concentrations of ADP. The initial rate of IRE1 RNase activity is plotted against ADP concentration (the initial rate of IRE1D797A incubated with quercetin (25 µM) alone is shown by the dashed line).
(also see Supplemental Figure 7).
D797A alters a residue essential for de-protonation of the kinase substrate and abolishes all measureable kinase activity (Lee et al., 2008b). Compared with the wildtype, bacterially-expressed, IRE1D797A exhibits enhanced mobility on SDS-PAGE that is unaltered by treatment with lambda phosphatase (Figure 7B), consistent with an unphosphorylated form of the enzyme. While IRE1D797A had no detectable ADP-mediated RNase activity (as previously noted, Lee et al., 2008b), addition of quercetin substantially activates its RNase (Figure 7C). Such activation was Q-site dependent, as it was absent in the IRE1D797A,K985A double mutant (Supplemental Figure 4A & 4B).
Isothermal titration calorimetry had previously indicated that the IRE1D797A is unable to bind ADP (Lee et al., 2008b), remarkably, addition of ADP attenuated the quercetin-mediated activation of IRE1D797A (Figure 7D), suggesting that both accessibility and functionality of the nucleotide binding site of the unphosphorylated IRE1D797A had been altered by binding of ligand to the Q-site.
DISCUSSION
The discovery of a ligand-binding site at the dimer interface of the kinase-endonuclease domain of yeast IRE1 provides an interesting perspective on features previously shown to regulate the enzyme. The new finding suggests the existence of hitherto unanticipated modes of physiological regulation of IRE1’s endonuclease activity as well as features of the enzyme that may be exploited for pharmacological manipulation of the UPR.
The crystal structure of IRE1 with ADP and quercetin reveals two sites occupied by different ligands: ADP in the nucleotide binding site (the kinase active site) and a very unusual arrangement of a pair of quercetin molecules nestled at the back-to-back dimer interface of the KEN domain engaging what we refer to as the Q-site. The two quercetin molecules interact broadly with one another and make numerous symmetrical contacts with residues from both IRE1 protomers.
Quercetin is known to engage the nucleotide-binding site of diverse kinases and likely does so in the case of IRE1. Nonetheless, the observation that mutations in the Q-site abolish all RNase activation indicates the functional dominance of the Q-site in IRE1 activation. The suggestion that quercetin promotes RNase activity first and foremost by stabilizing the back-to-back active dimeric configuration of the enzyme is further supported by the enhanced population of dimers observed by analytical ultracentrifugation and by the enhanced crosslinkability of IRE1 protomers in the presence of quercetin.
The preeminence of dimerization in quercetin’s mechanism of action fits well the prevailing notions on how ER stress modulates IRE1 activity: Lumenal signals initiated by dissociation of an inhibitory ligand (Bertolotti et al., 2000) (Liu et al., 2000) or by direct binding of misfolded proteins to IRE1’s lumenal domain (Credle et al., 2005) promote interaction between IRE1 protomers in the plane of the ER membrane. The resulting trans-autophosphorylation (Shamu and Walter, 1996) enhances affinity for nucleotide (likely ADP) (Sidrauski and Walter, 1997; Papa et al., 2003) whose engagement at the nucleotide binding site promotes conformational change(s) that favor back-to-back dimerization of the cytoplasmic effector domain (Lee et al., 2008b). The dimer may be further incorporated into higher order structures that favor cooperativity (Aragon et al., 2009; Korennykh et al., 2009). Higher order oligomers were not observed in this study, perhaps because the construct used lacked the 32 N-terminal residues required for higher order oligomers (present in Korennykh et al., 2009). Thus, it seems likely that quercetin activates IRE1 by promoting the dimerization step in the cascade described above.
The structure of IRE1’s cytoplasmic domain dimer in the IRE1:ADP (PDB: 2RIO), IRE1:APJ1999 (PDB: 3FBV) and IRE1:ADP:quercetin crystals (this study; PDB: 3LI0) is nearly identical with notable further structuring in both the kinase activation loop and the RNA binding site in the IRE1:ADP:quercetin structure. The structure of these two region in the ternary IRE1:ADP:quercetin complex is similar to that observed previously in IRE1 that assembled into large oligomers (Korennykh et al., 2009), suggesting that quercetin binding mimics the impact of oligomerization and potentially alters enzymatic activity. Unfortunately, we are unable to detect (or exclude) allosteric regulation at the RNase site, as we are unable to measure IRE1’s affinity for RNA (Km) or its enzymatic activity in the presence of saturating concentrations of RNA substrate (Vmax, data not shown).
Quercetin’s effect on the activity of the phosphorylation-defective IRE1D797A mutant suggests a complex interplay between phosphorylation state of the activation loop, ligand engagement at the Q- and nucleotide binding sites and RNase activity: IRE1D797A, which lacks detectable autokinase activity, cannot bind nucleotide and is refractory to activation by ADP (Lee et al., 2008b). Nonetheless the mutant is activated by quercetin and, as activation of IRE1D797A is abolished by a concomitant K985A mutation, the mode of ligand binding is likely similar in the wildtype and IRE1D797A mutant enzyme. Remarkably, addition of ADP to quercetin bound IRE1D797A attenuates its RNase activity. Positive cooperativity between quercetin and ADP binding, driven by quercetin-mediated dimerization, may explain the responsiveness of quercetin-bound IRE1D797A to ADP. But the inhibitory nature of the response suggests that the phosphorylation status of the activation loop profoundly affects the coupling of nucleotide binding to RNase activity and reveals hitherto unanticipated complexity in regulation of IRE1’s outputs.
It is tempting to speculate that the Q-site may engage endogenous ligand(s) and that perhaps quercetin itself or related flavonols, which are produced in plants may represent such ligands for plant IRE1s. Alternatively, the hydrophobic nature of the Q-site may provide an important pathway-specific feedback component to the known links between phospholipid metabolism and yeast IRE1 (Nikawa and Yamashita, 1992) (Cox et al., 1997). Furthermore, modeling suggests that mammalian IRE1α also possesses a pocket analogous to the Q-site that may be accessed by quercetin-like compounds (Supplemental Figure 2C–2D). Therefore, interaction between products of lipid or sterol metabolism and mammalian IRE1 at the Q-site site may help explain the emerging links between lipid metabolism and the UPR in mammals (Sriburi et al., 2004) (Lee et al., 2008a).
The findings described here also suggest a hitherto unanticipated mode for accessing the UPR pharmacologically independent of the nucleotide binding site (that is common to many kinases). Drugs that may bind a homologous Q-site in mammalian IRE1 and recapitulate quercetin activation of IRE1’s RNase could serve as potent modulators of the protein-folding environment in the ER. This may affect so-called proteostasis networks and alter the fate of misfolding-prone medically-important mutant proteins (Powers et al., 2009). Conversely, drugs that access the Q-site to inhibit XBP1 splicing could be used to neuter the IRE1 branch of the UPR with beneficial effects in fighting cancers, like multiple myeloma, which rely on the pathway for their survival.
It has recently been suggested that animal cell IRE1 can degrade mRNA promiscuously (Hollien and Weissman, 2006) and that such degradation contributes to cell death during ER stress. The regulation of this promiscuous IRE1 dependent RNAse activity has been linked to occupancy of the nucleotide binding site and to IRE1’s phosphorylation status (Han et al., 2009; Hollien et al., 2009). Given the evidence of allosteric regulation of IRE1’s activity from the Q-site, it is interesting to speculate on the possibility that ligands of mammalian IRE1 may have selective effects on XBP1 splicing and generalized RNA degradation and thus bias the response in a beneficial manner. Similarly, an activator of XBP1 splicing that works selectively through the Q-site could drive a wedge between potentially salubrious signaling from IRE1 to XBP1 and potentially damaging IRE1 effector functions linked to its kinase activity (e.g. JNK activation).
Experimental Procedures
Plasmids and reagents
Quercetin, ADP, and flavonols were purchased from Sigma-Aldrich. The initial small molecule screen was performed with the Kinase Inhibitor Library (BioMol International). RNA probes with the sequence 5’-CAUGUCCGCAGCGCAUG-3’ were purchased from Invitrogen either with or without modifications on the 5’ (AlexaFluor647) or 3’ (Black Hole Quencher 3). IRE1(658–1115) was cloned into the pGEX.Smt3 vector (derived from H6.Smt3, a kind gift of Chris Lima). All mutations in IRE1 were prepared by site-directed mutagenesis and sequenced to confirm incorporation of the appropriate mutation.
Purification of IRE1(658–1115)
BL21 E. coli expressing GST.Smt3.IRE1(658–1115) were induced with 1 mM IPTG and grown overnight at 18 deg C. Bacterial lysates were prepared by disruption in an EmulsiFlex-C3 (Avestin, Inc.) and cleared by centrifugation. GST.Smt3.IRE1(658–1115) was purified on a GSTrap 4B (GE Healthcare) affinity column and eluted with 40 mM glutathione (Sigma). The protein was further purified on a Mono Q (GE Healthcare) anion exchange column using an Akta FPLC (GE Healthcare). IRE1(658–1115) was cleaved from GST.Smt3 by incubation with 2 µg of Ulp1 per mg of IRE1(658–1115) overnight at 4 deg C. The cleaved protein was gel filtered into 20 mM Hepes pH 7.5, 200 mM NaCl on a Superdex 75 column (GE Healthcare) and frozen at −80 deg C in 10% glycerol at a concentration of ~10 µM.
Biochemical Assays for IRE1(658–1115)
IRE1 RNase activity was measured by incubation of purified IRE1(658–1115) 0.5–1µM with 25 nM of the quenched RNA probe at room temperature in a 384 well low-volume round bottom plate (Corning) with a final reaction volume of 20 µL. IRE1 reaction buffer (20 mM Hepes pH 7.5 50 mM KOAc 1 mM MgOAc and 1 mM DTT) was used for all RNase activity measurements. Time-dependent fluorescence was measured on a TecanF500 (Tecan US) using an excitation filter of 612 nm (bandpass 10 nm) and emission filter of 670 (bandpass 25 nm). An analogous protocol was followed to measure the IRE1-dependent cleavage of 20 nM of 32P-labeled RNA substrate, prepared by 5’ end-labeling with T4 polynucleotide kinase, Following the reaction the products were separated by denaturing acrylamide gel electrophoresis.
IRE1 autophosphorylation was measured by incubation of 0.5 µM IRE1(658–1115) with 0.1 mCi [32P]γ-ATP (MP Biomedicals) for the indicated time in 20 mM Hepes pH 7.5 50 mM MgCl2 2 mM MnCl2 1 mM DTT at room temperature. Incorporation of 32P was followed by SDS-PAGE and radiography.
Crosslinking was performed by incubation of 5 µM IRE1(658–1115) with 200 µM disuccinimidyl suberate (DSS; Pierce) for 30 min at room temperature in IRE1 reaction buffer. The reaction was quenched by the addition of 50 mM Tris pH 7.5. The samples were then boiled in Laemmili buffer and separated by SDS-PAGE.
Crystallization of IRE1
Bacterial expressed IRE1(658–1115) lacking residues 869–892 was purified as previously described (Lee et al., 2008b). Crystals of IRE1 were grown in the presence of Mg2+-ADP and quercetin or in the presence of JAK inhibitor 1 or Cdk1/2 inhibitor III in a hanging drop. Ire1:ADP:quercetin crystals flash-frozen in cryo-protectant (10% PEG 8K, 50 mM Na-Cacodylate pH 6.5, 300 mM KCl, 100 mM SrOAc and 30 % glycerol) and Ire1:JAK inhibitor I or Ire1:Cdk1/2 inhibitor III crystals flash frozen in cryo-protectant (1.6M LiSO4, 50 mM hepes pH 7.1, 20 mM MgCl2 and 30% glycerol) were used for data collection at the NE-CAT 24-ID-C beamline at the Advanced Photon Source at the Argonne National Laboratory. Data processing was performed using MOSFLM (Leslie, 2006) and SCALA (Evans, 2006). Molecule replacement was performed using Phaser (McCoy et al., 2007). Initial coordinates and refinement restraints for ADP and quercetin were obtained from HIC-up server. The final model was obtained using iterative cycles of manual building in Coot (Emsley and Cowtan, 2004)and automated refinement using Refmac5 (Murshudov et al., 1997) and CNS (Brunger et al., 1998). Model validation was performed using PROCHECK (Laskowski et al., 1993). Structural images presented in figures were prepared in Chimera (Pettersen et al., 2004). The statistical parameters for IRE1:ADP:quercetin (PDB: 3LI0), IRE1:JAK I inhibitor (PDB: 3LI1) and IRE1:Cdk1/2 inhibitor III (PDB: 3LI2) are provided in Supplemental table 1.
Quercetin mediated XBP1 spicing in mammalian cells by human-yeast chimeric IRE1
Cells lacking both IRE1α and IRE1β (Calfon et al., 2002) were transduced with GFP-marked retroviruses expressing human-yeast IRE1 chimeric proteins. Transduced fluorescent cells were FAC sorted and tested for expression of the chimeric protein by immunoprecipitation followed by immunoblotting with a poly-clonal serum directed to the cytosolic juxtamembrane region of mammalian IRE1α, common to the endogenous IRE1α (of wildtype MEFs) or the chimeric proteins (Bertolotti et al., 2000). Transduced cells were exposed to the indicated concentrations of flavonols, or tunicamycin, thapsigargin or dithiothreitol (DTT) and RNA was procured and XBP1 splicing measured by a ratio-metric reverse transcriptase-PCR assay.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tim Cardozo and Hong Wang (NYU), Tom Dever and Madusudan Dey (NICHD), Jeff Kelly (Scripps), Spencer Williams (U. Melbourne) and members of the Sicheri Lab for fruitful discussions and Chris Lima (MSKCC) for the Ulp1 and Smt3 expression plasmids, Andy Ryan and Matt Shoulders (Scripps) for their help in docking cholesterol to the IRE1 Q-site, the staff of the Northeastern Collaborative Access Team (NE-CAT) at the Advanced Photon Source (APS) at Argonne National Laboratories, where diffraction data were collected and Steffan Knapp (Structural Genomics Consortia, Oxford) for assistance with small molecule screening. Supported by grants DK047119, DK075311 and ES08681 to DR, F32-ES014775 to RLW, and Canadian Institutes for Health Research grant MOP-84370 to FS. KPKL is a recipient of a Canadian Institutes for Health Research Canada Graduate Scholarship and DR is a Wellcome Trust Principal Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot L, Harding H, Ron D. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr Protoc Immunol Chapter 18. 2008:15. doi: 10.1002/0471142735.im1815s81. Unit 18. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Ito T, Warnken SP, May WS. Protein synthesis inhibition by flavonoids: roles of eukaryotic initiation factor 2alpha kinases. Biochem. Biophys. Res. Commun. 1999;265:589–594. doi: 10.1006/bbrc.1999.1727. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R, MacArthur M, DS M, JM T. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008a;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KPK, Dey M, Neculai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in non-conventional RNA splicing. Cell. 2008b;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress-response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein- response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Papa FR, Zhang C, Shokat K, Walter P. Bypassing a Kinase Activity with an ATP-Competitive Drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological Chemical Approaches to Diseases of Proteostasis Deficiency. Annu. Rev. Biochem. 2009 doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: A Link Between the Unfolded Protein Response, Lipid Biosynthesis and Biogenesis of the Endoplasmic Reticulum. J. Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding H, Ron D. Coupling of Stress in the Endoplasmic Reticulum to Activation of JNK Protein Kinases by Transmembrane Protein Kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.