Abstract
Deletion of mouse preprotachykinin-A (PPTA), which encodes mainly for neuropeptide substance P, has been shown to protect against lung injury and mortality in sepsis. This study explored microarray-based differential gene expression profiles in mouse lung tissue 8 h after inducing microbial sepsis and the effect of PPTA gene deletion. A range of genes differentially expressed (more than two-fold) in microarray analysis was assessed, comparing wild-type and PPTA-knockout septic mice with their respective sham controls, and the data were further validated. Genetic deletion of substance P resulted in a significantly different expression profile of genes involved in inflammation and immunomodulation after the induction of sepsis, compared with wild-type mice. Interestingly, apart from the various proinflammatory mediators, the antiinflammatory cytokine interleukin-1 receptor antagonist gene (IL1RN) was also elevated much more in PPTA−/− septic mice. In addition, semiquantitative RT-PCR analysis supported the microarray data. The microarray data imply that the elevated levels of inflammatory gene expression in the early stages of sepsis in PPTA-knockout mice are possibly aimed to resolve the infection without excessive immunosuppression. As scientists are divided over the effects of pro- and antiinflammatory mediators in sepsis, it seems prudent to define the status depending on a complete genome profile. This is the first report exploring pulmonary gene expression profiles using microarray analysis in PPTA-knockout mice subjected to cecal ligation and puncture-induced sepsis and providing additional biological insight into the protection received against lung injury and mortality.
INTRODUCTION
Sepsis is a serious clinical condition caused by an imbalance in the inflammatory response of the body against overwhelming bacterial infection. The initial proinflammatory response to eliminate pathogens is followed by an excessive production of antiinflammatory mediators that also contributes to the patho-physiology of sepsis (1–3). Mechanisms of the antiinflammatory response have not been clearly understood so far (2). A balance between anti- and proinflammatory responses is needed to clear the invading microorganisms without damaging the host in sepsis (4–6).
Sepsis is generally caused by mixed infection (7), and multiple mediators have been reported to be involved in the development of sepsis (4,8). Substance P (SP), a preprotachykinin-A (PPTA) gene product, is an immunoregulatory neuropeptide implicated in various inflammatory diseases. Previously we have shown that PPTA-knockout mice are protected against polymicrobial sepsis (9) and lipopolysaccharide (LPS)-induced endotoxemia (10). Deletion of PPTA in those mice significantly attenuated inflammation and damage in the lungs (9,10).
Gene expression profiling using microarray is a novel approach. The aim is to comprehensively analyze various mediators that are differentially expressed in a disease state and visualize the genetic network (11). This global approach could eventually aid in better understanding of the complete mechanism involved in the pathogenesis of a disease. With the help of microarray analysis, researchers can search for diagnostic and therapeutic markers. However, there are few reports of using microarrays to study sepsis in vivo (12). In the present study, we sought to evaluate the genome-wide tissue-specific differential expression pattern after the induction of polymicrobial sepsis in PPTA-knockout mice, with particular focus on chemokines and cytokines. The cecal ligation and puncture (CLP) technique that we used to induce sepsis in mice is a reliable and clinically relevant model of polymicrobial sepsis. It reproduces many of the inflammatory and pathological sequences found in patients. The microarray data were further supported with semiquantitative RT-PCR and enzyme-linked immunosorbent assay (ELISA). The results indicated that the deletion of PPTA resulted in a significantly different expression profile of inflammatory and immunomodulatory mediators in the early stages of sepsis, compared with wild-type mice.
MATERIALS AND METHODS
Materials
TRIzol® reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), RNeasy® mini kit (Qiagen, Valencia, CA, USA) and GelRedTM (Biotium, Hayward, CA, USA) were used for RNA isolation and quantification. GeneChip® Mouse Genome 430 2.0 array, GeneChip® Hybridization Wash and Stain kit, One-cycle Target labeling and control reagents (containing in vitro transcription [IVT] labeling kit, One-cycle cDNA Synthesis kit, Sample Cleanup module, Poly-A RNA Control kit, Hybridization Controls) were purchased from Affymetrix (Santa Clara, CA, USA). NanoDrop® ND1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used for RNA quantification. PCR was performed in a MyCyclerTM thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) was used for cDNA synthesis. Primers were synthesized by 1st Base, Singapore. GeneChip® Fluidics Station 450, GeneChip Hybridization Oven 640, and GeneChip Scanner 3000 at the DSO were used for data collection. Pentobarbitone was purchased from Jurox (Rutherford, NSW, Australia).
Animal Model of Polymicrobial Sepsis
Animal experiments were performed in accordance with the DSO National Laboratories Animal Care and Use Committee, Singapore guidelines, which follow the established International Guiding Principles for Animal Research. Mice were maintained at a controlled temperature (21–24°C) and lighting (12-h light/dark cycle) and fed with standard laboratory diet and drinking water, provided ad libitum. PPTA−/− (with Balb/c genetic background) and wild-type Balb/c male mice (25–30 g) were randomly divided into sham or CLP experimental groups (n > 6 in each group). Polymicrobial sepsis was induced by CLP as explained earlier (13–15). The same surgical procedure except for the cecal ligation and puncture was carried out on sham-operated animals. The animals were killed 8 h after surgery by an intraperitoneal injection of pentobarbitone. Samples of lung were snap-frozen in liquid nitrogen and stored at −80°C for subsequent RNA isolation.
RNA Isolation and Quantification
Total RNA was isolated from the lung of PPTA−/− and wild-type mice subjected to sham and CLP surgery using TRIzol reagent according to the manufacturer’s protocol. RNeasy mini kit was used to clean up the total RNA after extraction. The quantity of extracted RNA was determined by spectrophotometric analysis (NanoDrop ND1000). RNA samples used for microarray hybridization and RT-PCR experiments were of highest purity, with A260/A280 ratios 1.9–2.1. The integrity of RNA was assessed by 1% wt/vol denaturing agarose gel electrophoresis using GelRed dye. The RNA sample was stored at −80°C until microarray analysis or RT-PCR.
Microarray Experiments
GeneChips were prepared individually for each of the mouse lung samples from the four groups (n = 3 for each group; n = 12 total) according to Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, double-stranded cDNA was synthesized from total RNA extracted from the mouse lung; IVT and One-cycle Target labeling were done to obtain biotin-labeled cRNA from the cDNA, which was purified and fragmented before hybridization to the arrays. Biotin-labeled RNA fragments (“target”) were hybridized to the oligonucleotide probes on the array and stained with streptavidin–phycoerythrin conjugate using the GeneChip fluidics station. Fluorescent hybridization signal was detected by the GeneChip Scanner 3000, enabled for high-resolution scanning. The amount of light emitted at 570 nm was proportional to the bound target at each location on the probe array. GeneChip Operating Software with Affymetrix Microarray Suite 5.0 algorithm was used to define the probe cells and compute intensity for each cell. Each complete probe array was imaged and stored in a separate data file.
Microarray Data Analysis
Expression Console™ software was used to enable probe set summarization and verify if the hybridization results and initial data were of sufficient quality for secondary analysis. By use of Gene-Spring™ 7.3 software (Agilent Technologies, Santa Clara, CA, USA), differentially expressed genes that showed a fold change of ≥ 2 against sham group were selected. Differentially expressed genes were further grouped based on Gene Ontology-Biological Process (Database for Annotation, Visualization, and Integrated Discovery [DAVID] Bioinformatics Resources 2008, National Institute of Allergy and Infectious Diseases [NIAID], NIH, Gene function classification Tool [http://niaid.abcc.ncifcrf.gov/]). Representation of specific inflammatory and immunoregulatory pathways among the differentially expressed genes was analyzed with Pathway Studio® software (Ariadne Genomics, Rockville, MD, USA), version 5.0. The software uses information available in the current literature to identify common pathways, targets or regulators that are associated with the altered genes to generate biological interaction networks. Microarray expression data was imported into Pathway Studio to graphically represent all known relationships and potential interactions between the differentially expressed genes. A biological network pathway was proposed according to Pathway Studio definitions regarding gene expression, interactions and regulations.
The microarray data discussed in this publication have been deposited in the Gene Expression Omnibus (GEO) site (http://www.ncbi.nlm.nih.gov/geo), accession number GSE15379.
Semiquantitative RT-PCR Analysis
Total RNA from lung (n > 6 for each group) was extracted using TRIzol reagent as per the manufacturer’s protocol. RNA (1 μg) was reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad) at 25°C for 5 min, 42°C for 30 min, followed by 85°C for 5 min. The cDNA was used as a template for PCR amplification by iQ Supermix (Bio-Rad). The primer sequences for detection of interleukin (IL)-1β, C-C chemokine ligand 2 (CCL-2), C-X-C chemokine ligand 2 (CXCL-2), IL-1 receptor antagonist (IL-1ra), CCL-3, CCL-4, interferon-inducible protein 10 (IP-10), serum amyloid A3 (SAA3), chemokine (C-C motif) receptor 2 (CCR-2), CCR-5 and 18S gene, optimal annealing temperature, and optimal cycles are shown in Table 1. PCR amplification was carried out in a MyCycler (Bio-Rad). The reaction mixture was first subjected to 95°C for 3–5 min, followed by an optimal cycle of amplification (denaturation, annealing and elongation) and a final extension at 72°C for 5–7 min. PCR products were analyzed on 1.5% wt/vol agarose gel containing 0.1 μL/mL GelRed. Densitometry results from PCR products were normalized to the mouse 18S band densities as a control.
Table 1.
Primer sequences and optimal conditions used in PCR analysis.
| Gene | Sense primer sequence (5′–3′) | Antisense primer sequence (5′–3′) | Amplification conditions | Amplification cycles, n |
|---|---|---|---|---|
| SAA3 | AGC CTT CCA TTG CCA TCA TTC TT | AGT ATC TTT TAG GCA GGC CAG CA | 94°C 1min; 58°C 1min; 74°C 1min | 25 |
| IP10 | GTGTTGACATCATTGCCACG | GCTTACAGTACAGAGCTAGG | 95°C 30 sec; 60°C 30 sec; 45°C 45 sec | 30 |
| CCL-2 | CCCCACTCACCTGCTGCTACT | CACTGTCACACTGGTCACTCC | 95°C 50 sec; 64°C 50 sec; 72°C 1 min | 36 |
| CCL-3 | ACTGCCCTTGCTGTTCTTCTCT | AGGCATTCAGTTCCAGGTCAGTGA | 95°C 30 sec; 61°C 30 sec; 72°C 30 sec | 33 |
| CCL-4 | CCCTCTCTCTCCTCTTGCTCGT | TTCAACTCCAAGTCACTCATGTACTCA | 94°C 30 sec; 55°C 30 sec; 72°C 1 min | 32 |
| CXCL-2 | GCTGTCAATGCCTGAAGACC | TAGTTCCCAACTCACCCTCTC | 95°C 50 sec; 65°C 50 sec; 72°C 1 min | 36 |
| IL-1β | AAGGAGAACCAAGCAACGAC | GAGATTGAGCTGTCTGCTCA | 95°C 50 sec; 63°C 50 sec; 72°C 1 min | 34 |
| CCR-2 | CACGAAGTATCCAAGAGCTT | CATGCTCTTCAGCTTTTTAC | 94°C 30 sec; 58°C 45 sec; 72°C 70 sec | 35 |
| CCR-5 | TTCCCTGTCATCGCTTGCTCT | CGGATGGAGATGCCGATTTT | 94°C 1 min; 60°C 1 min; 72°C 2 min | 40 |
| IL-1ra | GACCCTGCAAGATGCAAGCC | CAGGACGGTCAGCCTCTAGT | 95°C 20 sec; 51°C 20 sec; 72°C 20 sec | 36 |
| 18S | GTA ACC CGT TGA ACC CCA TT | CCA TCC AAT CGG TAG TAG CG | 95°C 30 sec; 59°C 30 sec; 72°C 30 sec | 24 |
ELISA Analysis
For the measurement of cytokine IL-1ra, ELISA kit from R&D Systems (Minneapolis, MN, USA) was used according to the manufacturer’s instructions employing a quantitative sandwich enzyme immunoassay. The lower limit of detection was 31.25 pg/mL. Briefly, standards and samples were pipetted into the microplate wells precoated with mouse IL-1ra–specific polyclonal antibody and incubated for 2 h. After washing away unbound substances, an enzyme-linked mouse IL-1ra–specific polyclonal antibody was added to the wells and incubated for 2 h. After a wash, a substrate solution was added to the wells and incubated for 30 min to yield a blue product that turned yellow when the stop solution was added. The optical density of each well was determined using a microplate reader set to 450 nm. The intensity of the color measured was proportional to the amount of mouse IL-1ra bound in the initial step. The sample values were then read off the standard curve and corrected for the DNA content of the lung tissue. Lung IL-1ra was expressed as pg/μg of DNA and plasma as pg/mL.
Statistical Analysis
Differentially expressed genes were analyzed by Student unpaired t test (P < 0.05) for array data to compare differences between the medians of the groups. For RT-PCR and ELISA data, values were expressed as mean ± SEM, and significant difference between groups was evaluated by one-way ANOVA, followed by post hoc Tukey test. P < 0.05 was considered statistically significant.
RESULTS
Microarray Quality Control
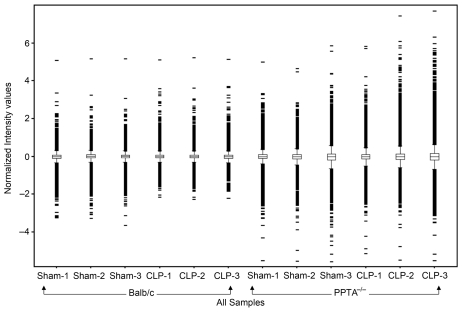
Each of the Affymetrix high-density oligonucleotide arrays that we used for lung expression profiling had 45,000 probe sets to analyze the expression level of more than 39,000 transcripts and variants from about 34,000 well-characterized mouse genes. As the high cost of genomic investigations was a limitation, we used three arrays for each group according to Minimum Information about a Microarray Experiment (MIAME) guidelines (16). All 12 arrays passed the quality controls included within the arrays such as hybridization controls, poly-A controls, normalization control set and housekeeping/control genes. Illustration of the box-whisker plot was done to examine the distribution of data and ensure proper dynamic range (Figure 1). Data distributions were assessed based on the highest intensity value at saturation. Appropriately, similar distribution of data was considered to be of high technical quality for further analysis. The expression of genes was compared between sham controls and CLP-induced sepsis groups among wild-type and PPTA−/− mice. We focused on genes that were either consistently increased or decreased in all sets of experiments. Only those genes whose expressions changed by two-fold or more were taken as significant (P < 0.05). These differentially expressed genes were further annotated based on their known biological functions using DAVID. Many of the genes were present in more than one functional class.
Figure 1.
The box-whisker plot of distribution of normalized intensity values for each sample. The boxes represent interquartile range, with 75th percentile at the top and 25th percentile at the bottom. The line in the middle of the box represents the 50th percentile (median). Whiskers represent the rest of the distribution, with intensity values beyond 1.5 times the interquartile range shown in red. Whiskers represent the rest of the distribution, with intensity values beyond 1.5 times the interquartile range. The x axis represents individual microarray, and the y axis represents the normalized intensity values.
Inflammatory Gene Profile of Wild-Type Septic Mice
Table 2 lists a majority of the genes altered by CLP in wild-type and PPTA−/− mice, grouped under the biological process gene ontology classes of inflammatory response, chemotaxis, leukocyte activation, response to bacterium, regulation of cellular process, signal transduction and cytokine- and chemokine-mediated signaling pathway. A few others of mixed or unknown functions are grouped separately, as they are also reported in the literature to be differentially expressed in sepsis. As expected, CLP-induced sepsis in wild-type mice resulted in a significant increase in many of the inflammatory response and chemotaxis genes compared with sham surgery. Some of the chemokine superfamily members that were upregulated significantly include C-C chemokines (CCL-2, CCL-3, CCL-4, CCL-9) and C-X-C chemokines (CXCL-1, CXCL-2, CXCL-10). In addition, SAA3, IL-1β, CCR-1, toll-like receptor 2, s100 calcium binding protein a8 (S100A8) and matrix metallopeptidase (MMP) 8 and 9 were also increased by more than two-fold in wild-type septic mice compared with sham-treated mice. It was interesting to note that the antiinflammatory cytokine IL-1ra also showed a greater than sixfold increase after CLP in these mice.
Table 2.
Differentially expressed genes in wild-type and PPTA−/− mice 8 h after CLP-induced sepsis, compared with sham-operated mice.
| Fold increase/decreasea |
||
|---|---|---|
| Gene | Wild-type CLP | PPTA−/− CLP |
| Inflammatory response | ||
| Serum amyloid A 3b | 4.21 | 54.52 |
| Chemokine (C-C motif) ligand 2b | 5.52 | 54.37 |
| Fc receptor, IgE, high affinity I, γ polypeptide | 2.30 | 7.15 |
| Chemokine (C-X-C motif) ligand 10b | 5.04 | 12.58 |
| Chemokine (C-C motif) receptor 1 | 4.66 | 15.52 |
| Chemokine (C-X-C motif) ligand 1 | 5.35 | 37.74 |
| Interleukin-1 receptor antagonistb | 6.23 | 125.25 |
| Chemokine (C-X-C motif) ligand 2b | 107.41 | 114.76 |
| CD14 antigen | 7.93 | 13.70 |
| Toll-like receptor 13 | 3.72 | 5.52 |
| Chemokine (C-C motif) ligand 4b | 3.55 | 17.59 |
| Interleukin-1βb | 6.77 | 8.85 |
| Selectin, platelet | 2.57 | 12.03 |
| Toll-like receptor 2 | 4.09 | 6.09 |
| Complement component 5a receptor 1 | 3.19 | 6.50 |
| Tumor necrosis factor receptor superfamily, member 1b | 2.15 | 8.14 |
| CD28 antigen | 1.87 | 2.56 |
| Chemokine (C-C motif) ligand 3b | 13.05 | 73.16 |
| Toll-like receptor 6 | 2.18 | 2.68 |
| Tumor necrosis factor receptor superfamily, member 1a | –c | 2.09 |
| Mitogen-activated protein kinase kinase 3 | – | 2.12 |
| Oxidized low-density lipoprotein (lectin-like) receptor 1 | – | 5.51 |
| Orosomucoid 1 | – | 34.00 |
| Signal transducer and activator of transcription 3 | – | 2.63 |
| Chemokine (C-C motif) receptor 2b | – | 5.26 |
| Chemokine (C-C motif) ligand 22 | – | 2.83 |
| Fc receptor, IgG, low affinity IIb | – | 9.90 |
| Fc receptor, IgG, low affinity III | – | 6.59 |
| Toll-like receptor 5 | – | −2.24d |
| Toll-like receptor 1 | – | 3.53 |
| CD44 antigen | – | 3.62 |
| Signal transducer and activator of transcription 5a | – | 2.44 |
| Phospholipase a2, group VII (platelet-activating factor acetylhydrolase, plasma) | – | 7.59 |
| Chemotaxis | ||
| Chemokine (C-C motif) ligand 9 | 2.27 | 10.09 |
| s100 calcium binding protein A8 (calgranulin A) | 2.68 | – |
| Formyl peptide receptor 1 | 9.01 | 48.18 |
| Chemokine (C-C motif) ligand 17 | – | 8.49 |
| Chemokine (C-C motif) receptor 5b | – | 14.45 |
| Cysteine-rich protein 61 | – | −5.25 |
| Integrin β 2 | – | 3.68 |
| Interleukin-16 | – | 3.71 |
| Chemokine (C-C motif) ligand 6 | – | 3.63 |
| Leukocyte activation | ||
| CD80 antigen | 3.19 | 2.60 |
| CD52 antigen | 1.92 | 2.27 |
| Interleukin-12a | – | 2.61 |
| Growth arrest and DNA-damage-inducible 45 γ | – | 4.21 |
| fas (TNF receptor superfamily member) | – | 4.82 |
| CD40 antigen | – | 2.10 |
| Response to wounding | ||
| Coagulation factor XIII, a1 subunit | 2.12 | 4.17 |
| Gap junction membrane channel protein α 1 | – | 3.25 |
| Coagulation factor X | – | 17.55 |
| Coagulation factor VII | – | 2.43 |
| Fibrinogen, γ polypeptide | – | 11.57 |
| Response to bacterium | ||
| Cathelicidin antimicrobial peptide | 5.33 | 40.93 |
| Peptidoglycan recognition protein 1 | 4.44 | 2.26 |
| Lipopolysaccharide-binding protein | – | 2.46 |
| Regulation of cellular process | ||
| Protein tyrosine phosphatase, nonreceptor type 11 | −2.25 | – |
| Matrix metallopeptidase 9 | 3.48 | 1.95 |
| Mitogen-activated protein kinase kinase kinase 8 | 4.01 | 8.26 |
| Caspase 4, apoptosis-related cysteine peptidase | – | 8.62 |
| Heat shock protein 1A | – | −7.87 |
| Interferon regulatory factor 1 | – | 3.25 |
| Interferon regulatory factor 7 | – | 2.63 |
| Signal transduction | ||
| Interleukin-7 receptor | – | 9.81 |
| G protein–coupled receptor 35 | 2.77 | 6.49 |
| Rho family GTPase 1 | 3.24 | 8.83 |
| Interleukin-1 receptor, type II | 8.97 | 18.83 |
| G protein–coupled receptor 27 | 2.24 | 3.49 |
| Interleukin-10 receptor, α | 2.06 | 2.53 |
| Regulator of G protein signaling 1 | 4.64 | 8.55 |
| Suppressor of cytokine signaling 3 | 4.73 | 10.67 |
| Regulator of G protein signaling 2 | 2.36 | – |
| Janus kinase 2 | – | 2.41 |
| Interleukin-17 receptor | – | 2.61 |
| Mitogen-activated protein kinase kinase kinase kinase 2 | – | −15.93 |
| Phospholipase C, δ 3 | – | −2.55 |
| Chemokine (C-X-C motif) receptor 6 | – | 2.29 |
| Peroxisome proliferator activator receptor δ | – | 2.27 |
| Sphingosine kinase 1 | – | 6.67 |
| Suppressor of cytokine signaling 2 | – | 2.27 |
| Interleukin-4 receptor, α | – | 4.38 |
| Interleukin-8 receptor, β | – | 3.05 |
| Neuropilin 1 | – | −2.64 |
| Metallothionein 1 | 2.68 | 5.58 |
| Plasminogen activator, urokinase receptor | 2.47 | 4.42 |
| Phosphatidylinositol 3-kinase catalytic delta polypeptide | – | 3.39 |
| Mitogen-activated protein kinase kinase kinase kinase 5 | – | −2.69 |
| Mitogen-activated protein kinase kinase kinase 1 | – | 2.83 |
| Mitogen-activated protein kinase kinase kinase 6 | – | 9.08 |
| Mitogen-activated protein kinase 8 | – | −4.66 |
| Response to other organism | ||
| Neutrophilic granule protein | 4.70 | 27.44 |
| Response to fungus | ||
| Pentraxin-related gene | – | 11.74 |
| Cytokine- and chemokine-mediated signaling pathway | ||
| Colony-stimulating factor 2 receptor, β 1, low-affinity (granulocyte-macrophage) | 2.86 | 10.45 |
| Leukocyte immunoglobulin-like receptor, subfamily b (with TM and ITIM domains), member 3 | 2.57 | 6.62 |
| Colony-stimulating factor 2 receptor, β 2, low-affinity (granulocyte-macrophage) | – | 4.57 |
| Other | ||
| Lipocalin 2 | 2.66 | 11.97 |
| Arginase type II | 2.10 | 5.74 |
| Interleukin-1 family, member 9 | 5.18 | 19.02 |
| Matrix metallopeptidase 8 | 9.61 | 12.30 |
| Interferon-induced transmembrane protein 6 | 5.61 | 34.46 |
| s100 calcium binding protein A9 (calgranulin B) | 3.19 | 2.92 |
| Inhibitor of DNA binding 2 | – | 3.91 |
| Cytokine-inducible SH2-containing protein | – | 2.01 |
| Secretory leukocyte peptidase inhibitor | – | 3.76 |
| Amphiregulin | – | 3.59 |
| Interferon-induced transmembrane protein 2 | – | 2.51 |
| Mitogen-activated protein kinase kinase kinase 4 | – | −3.23 |
| Mitogen-activated protein kinase-activated protein kinase 3 | – | 2.36 |
| Mitogen-activated protein kinase kinase kinase 14 | – | 2.60 |
| Natriuretic peptide receptor 3 | – | 3.12 |
Values represent fold changes in gene expression in wild-type and PPTA−/− mice subjected to CLP-induced sepsis, compared with corresponding sham-operated mice. All the fold changes analyzed by GeneSpring 7.3 software were statistically significant (P < 0.05) compared with sham control.
Genes validated by RT-PCR.
Absence of a value (−) indicates that the fold change was less than two.
Negative values represent decreased mRNA levels compared with respective sham group values.
Inflammatory Gene Profile of PPTA−/−Septic Mice Differs
PPTA−/− mice also exhibited high expression of inflammatory response and chemotaxis genes 8 h after CLP compared with sham mice. The fold increase was significantly higher than that in wild-type mice, especially for the chemokines CCL-2, CCL-3, CCL-4, CCL-9, CXCL-1, CXCL-2 and CXCL-10. Similarly, the expression of SAA3, IL-1β, CCR-1, toll-like receptor 2 and MMP-8 increased significantly in PPTA−/− mice 8 h after CLP compared with the corresponding fold increase in wild-type mice. Unlike in wild-type mice, S100A8 and MMP-9 in PPTA−/− mice did not show more than two-fold increase after CLP. Further, CCL-6, CCL-17, CCL-22, CCR-2, CCR-5, toll-like receptor 1 and phosphatidylinositol 3-kinase catalytic delta polypeptide were expressed more in PPTA−/− mice after the induction of sepsis; this effect was not seen in wild-type mice after CLP. We also observed a much higher increase of IL-1ra gene IL1RN in PPTA−/− mice 8 h after CLP compared with the wild-type mice. Last, there was a significant downregulation of toll-like receptor 5 and heat shock protein 1A only in PPTA−/− mice after the induction of sepsis.
Semiquantitative RT-PCR Data Supports the Microarray Results
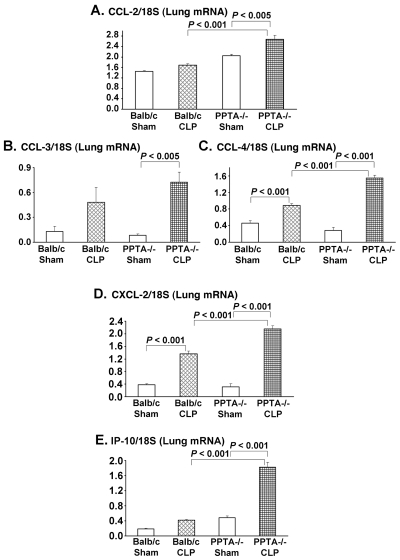
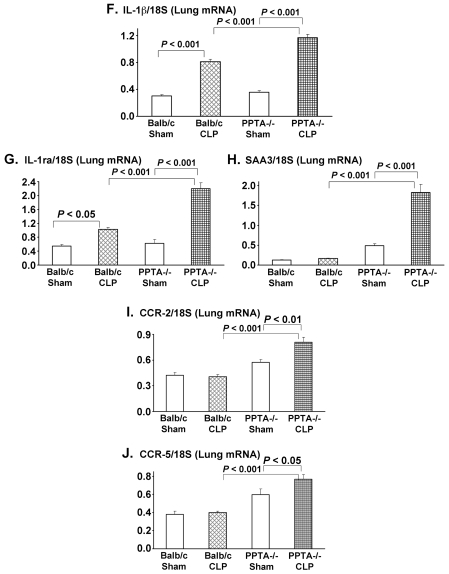
To further validate the microarray results, RT-PCR was carried out. Ten genes were selected whose expression altered by more than two-fold in septic mice. RNA from individual mouse lung was evaluated for these genes by RT-PCR. Eight h after CLP-induced sepsis, pulmonary gene expression levels of CCL-2, CCL-3, CCL-4, CXCL-2, IP-10 (CXCL-10), IL-1 β, IL-1ra and SAA3 were increased compared with the corresponding levels in sham-operated mice (Figure 2A–H). The elevated mRNA levels were observed in both the wild-type and PPTA−/− mice after sepsis, with a significantly higher increase in the latter. There was no significant change in the expression levels of chemokine receptors CCR-2 and CCR-5 in septic wild-type mice (Figure 2I, J). However, a significant upregulation was observed for both CCR-2 and CCR-5 in PPTA−/− mice 8 h after CLP (CCR-2 P < 0.01 and CCR-5 P < 0.05 versus sham) (see Figure 2I, J). The mRNA levels evaluated by RT-PCR showed a similar trend to the microarray gene expression data for the chosen 10 genes.
Figure 2.
Pulmonary inflammatory and chemotaxis gene transcripts in wild-type and PPTA−/− mice. A, CCL-2; B, CCL-3; C, CCL-4; D, CXCL-2; E, IP-10; F, IL-1β; G, IL-1ra; H, SAA3; I, CCR-2; J, CCR-5. mRNA levels of these genes in lung were evaluated 8 h after CLP or sham surgery in wild-type and PPTA−/− mice by semiquantitative RT-PCR analysis (expressed as a ratio of band density of the gene to 18S) as described in Materials and Methods. Mouse 18S was used as a control. Results were expressed as mean ± SEM (n = 6–9 mice per group). P values were determined by one-way ANOVA, followed by post hoc Tukey test.
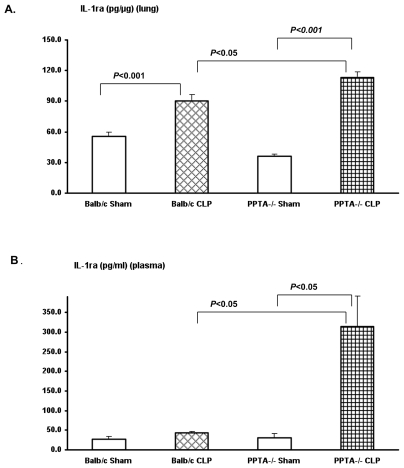
IL-1ra Protein Levels Significantly Elevated in PPTA−/− Mice after Sepsis
We also measured the protein levels of antiinflammatory cytokine IL-1ra in lung and plasma by ELISA. IL-1ra level was upregulated after CLP-induced sepsis, with a pattern similar to the gene expression (Figure 3). The increase in lung (Figure 3A) and plasma (Figure 3B) IL-1ra levels was significantly higher in PPTA−/− mice after the induction of sepsis (lung P < 0.001 and plasma P < 0.05 versus sham) compared with the corresponding increase in wild-type mice.
Figure 3.
Protein levels of IL-1ra in wild-type and PPTA−/− mice in lung (A) and plasma (B). IL-1ra levels in lung and plasma were measured 8 h after CLP or sham surgery in wild-type and PPTA−/− mice by ELISA as described in Materials and Methods. Results were expressed as mean ± SEM (n = 6–12 mice per group). P values were determined by one-way ANOVA, followed by post hoc Tukey test.
Pathway Analysis of Differentially Expressed Genes
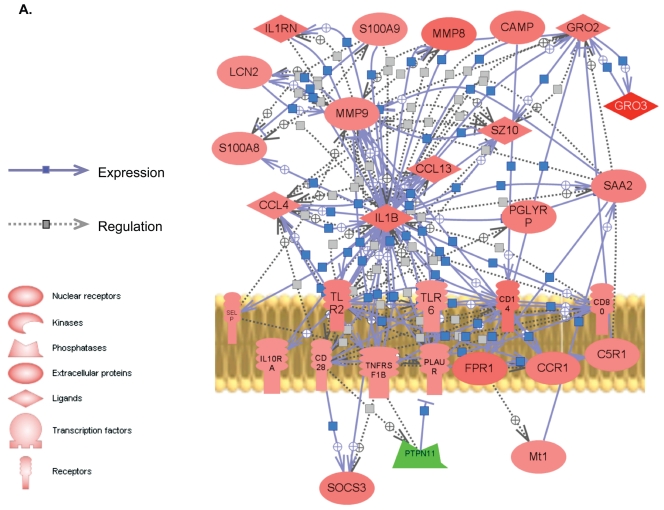
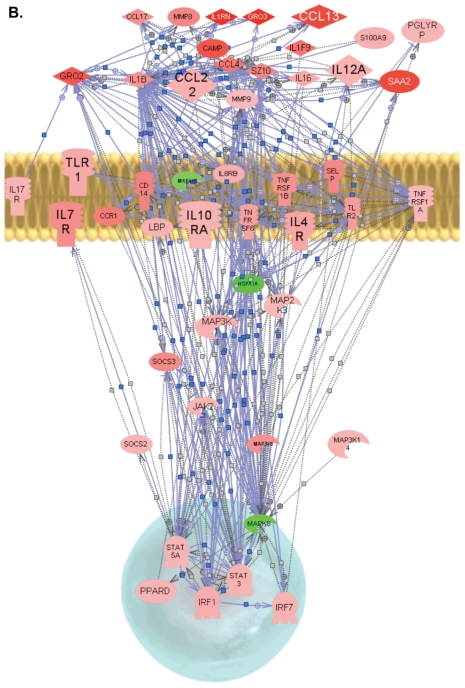
PathwayStudio software was used to identify a possible gene network in wild-type and PPTA−/− mice subjected to CLP-induced sepsis using the genes listed in Table 2. Genes were linked to each other based on the published literature (Figure 4A, B). Most of the differentially expressed genes were found to be connected, and the few unconnected genes were excluded from the figure. Our pathway analysis highlighted a significantly higher number of links and interactions and higher expressions in PPTA−/− mice among known inflammatory and immune response genes compared with the wild-type mice (see Figure 4A, B).
Figure 4.
Proposed biological network of differentially expressed genes in sepsis. By use of PathwayStudio, a pathway network was built for microarray data based on previously reported interactions in the literature. The links and nodes represent literature-based findings. Some of the prominent, differentially expressed genes shared in inflammatory and immune response in sepsis were used to create the network. (A), Wild-type septic mice. (B), PPTA−/− septic mice. + depicts positive regulation/expression; line ending with | depicts negative regulation/expression. Up- and downregulated genes are indicated in red and green, respectively. IL1RN, interleukin-1 receptor antagonist; S100A9, S100 calcium binding protein A9 (calgranulin B); CAMP, cathelicidin antimicrobial peptide; GRO2, growth-related oncogene 2 (CXCL2); GRO3, growth-related oncogene 3 (CXCL3); PGLYRP, peptidoglycan recognition protein; TLR, toll-like receptor; LBP, lipopolysaccharide-binding protein; HSP1A, heat shock protein 1A.
DISCUSSION
This is the first report exploring pulmonary gene expression profiles using microarray analysis in PPTA−/− mice subjected to CLP-induced sepsis. It has been shown previously that the pathogenesis of sepsis was delayed in PPTA−/− mice and that the gene deletion protected against lung injury in sepsis (9). SP is also implicated in other inflammatory conditions such as acute pancreatitis (17) and endotoxemia (10). To better understand the molecular mechanisms of polymicrobial sepsis and associated lung damage in PPTA−/− mice, we employed microarray analysis of pulmonary gene expression. Our main focus was on lung injury, as respiratory failure is one of the main causes of mortality in sepsis. As the lung myeloperoxidase (MPO) activity was highest 8 hours after CLP (data not shown), lung tissue was collected after killing the mice at this time point.
It is well known that the inflammatory response is an important part of sepsis. During the course of polymicrobial sepsis, a range of pro- and antiinflammatory cytokine and chemokine genes is upregulated (18), as is further evident from our pathway analysis. Although the inflammatory mediators activate leukocyte trafficking to the site of infection to fight the invading pathogens, excessive inflammation could be damaging (7). At the same time, the host also produces counterbalancing antiinflammatory mediators (7) that, if produced in excess, can cause immunosuppression and fatalities (1). Concurrent overexpression of both proinflammatory and antiinflammatory cytokines has been reported in the early phase of lethal sepsis (19). So far there is a lack of consensus on whether to suppress or boost immunity or to do each at different times, as clinical trials aimed at the inflammatory axis in severe sepsis have failed (7).
Levels of chemokines such as CXCL-2, CCL-3 and CCL-2 are reported to be increased 8 hours after CLP-induced severe sepsis in mice (20). Peritoneal CCL-6 (21) and CCL-22 (22,23) concentrations were also elevated in the CLP mouse model of sepsis. Our microarray analysis showed a significantly higher expression of a few of the inflammatory genes in PPTA−/− mice after sepsis compared with the wild-type mice. Although gene expression of CCL-2, CCL-3, CCL-4, CCL-9, CXCL-1, CXCL-2 and CXCL-10 was increased in wild-type mice after CLP, which was as expected, the increase was more significant in PPTA−/− mice 8 hours after sepsis. In spite of the elevated gene expression of proinflammatory chemokines, PPTA−/− mice are protected against CLP-induced sepsis (9). The precise mechanism of protection is not clear yet. Interestingly, CCL-3, CCL-6 and CXCL-10 have been demonstrated to be protective in sepsis-induced injury and mortality in a murine CLP model (21,23–25). Macrophage-derived chemokine (MDC) (CCL-22) also protected mice against CLP-induced death (26). We found a significant increase in CCL-6, CCL-17 and CCL-22 exclusively in PPTA−/− mice after the induction of sepsis. It is possible that these chemokines provide some protection against pathogens and damage in PPTA−/− mice after CLP. Pathway analysis further provides an insight into the complex network of interactions among these proinflammatory chemokines. The role of chemokine receptors in sepsis is not very clear (21).
Cytokines are considered both friend and foe in sepsis (5). Successful treatment of sepsis involves cytokine modulation and restriction of the systemic reactions to the inflammatory foci only. It should be noted, however, that only a small amount of cytokines can defend host, and a complete blockade may damage the host (27). Our microarray analysis showed that IL-1β increased significantly in both wild-type and PPTA−/− septic mice compared with sham-operated mice. However, the antiinflammatory cytokine IL-1ra also showed a significant increase after CLP in these mice, and the expression was much higher in PPTA−/− mice than in wild-type mice. Elevated IL-1ra gene expression in wild-type mice after sepsis is consistent with a literature report that IL-1ra mRNA level increased (by 10-fold) 3–6 hours after CLP and was sustained for 18 hours (18). A recent clinical trial of neonatal-onset multisystem inflammatory disease showed a promising improvement with IL-1ra treatment (6,28). It is believed that antiinflammatory strategies applied early in patients with a hyperinflammatory immune response may be lifesaving (5). PPTA−/− mice exhibited high expression of IL-1ra early in sepsis along with the hyperinflammatory state, which could partly explain the improvement in survival. High levels of blood IL-1ra are found in septic patients and in animal models of sepsis, although the role of IL-1ra in immunosuppression is not clearly defined (2).
Among the genes whose expression was found to be affected by PPTA−/− gene deletion, SAA3 was increased in wild-type and PPTA−/− septic mice compared with sham-operated mice, with a higher increase in the knockout mice, probably providing more antibacterial immunity (7). The increase in S100A8 in wild-type mice after sepsis was absent in PPTA−/− septic mice. PPTA−/− mice showed much higher fold increase of cathelicidins (surfactants containing antibacterial substances) after sepsis, compared with the corresponding wild-type mice (29). Similarly, a more prominent upregulation of MMP-8 gene was observed in PPTA−/− mice after sepsis, possibly providing antiinflammatory protection and neutrophil apoptosis (30). Furthermore, PPTA−/− mice did not show more than two-fold increase in MMP-9 after CLP, unlike the wild-type mice, which implies some beneficial antiinflammatory effect (31). However, PPTA−/− mice had elevated phospho-inositide 3-kinase (PI3K) gene expression after CLP and a significant downregulation of heat shock protein 1A (Figure 4B) associated with beneficial effects (32,33).
The RT-PCR data showed a similar expression of all 10 genes evaluated when compared with microarray results. Although the fold change between the sham control and CLP-induced sepsis mice determined by RT-PCR differed from that found on microarray, the trend of the response was similar between the two methods. To evaluate if transcription and translation are coordinately regulated, IL-1ra was quantified in plasma and tissue by ELISA. In line with literature reports of elevated circulating IL-1ra in sepsis (34), and consistent with our microarray and RT-PCR data, we found a significant increase in IL-1ra protein levels in both wild-type and PPTA−/− mice 8 hours after sepsis. Thus it seems prudent to define the inflammatory status on the basis of complete plasma profile and genome data, even if grouping into traditional immunologic categories becomes difficult (19).
Failed clinical trials have shown that simply blocking the inflammatory response may result in an impaired resolution of infection. Instead of targeting any individual mediator that reflects the functional status of sepsis, inflammation should be treated at the right time in the right place. In this perspective our study provides a valuable molecular fingerprint of PPTA−/− mice that are protected against mortality in CLP-induced sepsis. For the first time, we report the change in pulmonary gene expression of a large set of inflammatory mediators in PPTA−/−septic mice using microarray analysis. Elevated levels of pro- and antiinflammatory gene expression in the early stages of sepsis may help in resolving the infection without excessive immunosuppression. Additional antimicrobial mediators that were observed in our study would further support in maintaining this precarious balance of inflammatory forces. In conclusion, in this study we have shown the gene profile following sepsis and the effect of PPTA gene deletion. This study will help define the mechanisms by which PPTA gene products contribute to lung injury in sepsis.
ACKNOWLEDGMENTS
This work was supported by research grants from National Medical Research Council (NMRC) grant R-184-000-111-213 and Joint Applied R&D Co-operation Programme (JPP) grants R-184-000-144-123 and R-184-000-144-232. We thank Allan Basbaum (University of California, San Francisco, CA, USA) for the gift of PPTA−/− mice. We also thank Michelle Chong (Research Instruments), Yeo Su Yin Dawn (DSO), Shoon Mei Leng (Department of Pharmacology, National University of Singapore), and Parvathi Rajagopal (DSO) for the technical assistance.
Footnotes
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Online address: http://www.molmed.org
REFERENCES
- 1.Ashare AL, et al. Anti-inflammatory response is associated with mortality and severity of infection in sepsis. Am J Physiol Lung Cell Mol Physiol. 2005;288:L633–40. doi: 10.1152/ajplung.00231.2004. [DOI] [PubMed] [Google Scholar]
- 2.Reddy RC, Chen GH, Tekchandani PK, Standiford TJ. Sepsis-induced immunosuppression: from bad to worse. Immunol Res. 2001;24:273–87. doi: 10.1385/IR:24:3:273. [DOI] [PubMed] [Google Scholar]
- 3.Van der Poll T, Deventer SJ. Cytokines and anticytokines in the pathogenesis of sepsis. Infect Dis Clin North Am. 1999;13:413–26. doi: 10.1016/s0891-5520(05)70083-0. [DOI] [PubMed] [Google Scholar]
- 4.Okazaki Y, Matsukawa A. Pathophysiology of sepsis and recent patents on the diagnosis, treatment and prophylaxis for sepsis. Recent Pat Inflamm Allergy Drug Discov. 2009;3:26–32. doi: 10.2174/187221309787158416. [DOI] [PubMed] [Google Scholar]
- 5.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–50. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 6.Remick DG. Pathophysiology of sepsis. Am J Pathol. 2007;170:1435–44. doi: 10.2353/ajpath.2007.060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sriskandan S, Altmann DM. The immunology of sepsis. J Pathol. 2008;214:211–23. doi: 10.1002/path.2274. [DOI] [PubMed] [Google Scholar]
- 8.Marshall JC, et al. Measures, markers, and mediators: toward a staging system for clinical sepsis. Crit. Care Med; A report of the Fifth Toronto Sepsis Roundtable; Toronto, Ontario, Canada. October –25–26, 2000; 2003. pp. 1560–67. [DOI] [PubMed] [Google Scholar]
- 9.Puneet P, et al. Preprotachykinin-A gene products are key mediators of lung injury in polymicrobial sepsis. J Immunol. 2006;176:3813–20. doi: 10.4049/jimmunol.176.6.3813. [DOI] [PubMed] [Google Scholar]
- 10.Ng SW, Zhang H, Hegde A, Bhatia M. Role of preprotachykinin-A gene products on multiple organ injury in LPS-induced endotoxemia. J Leukoc Biol. 2008;83:288–95. doi: 10.1189/jlb.0807575. [DOI] [PubMed] [Google Scholar]
- 11.Schulze A, Downward J. Navigating gene expression using microarrays: a technology review. Nat Cell Biol. 2001;3:E190–5. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 12.Chung TP, et al. Molecular diagnostics in sepsis: from bedside to bench. J Am Coll Surg. 2006;203:585–98. doi: 10.1016/j.jamcollsurg.2006.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayala A, Herdon CD, Lehman DL, Ayala CA, Chaudry IH. Differential induction of apoptosis in lymphoid tissues during sepsis: variation in onset, frequency, and the nature of the mediators. Blood. 1996;87:4261–75. [PubMed] [Google Scholar]
- 14.Zhou M, Chaudry IH, Wang P. The small intestine is an important source of adrenomedullin release during polymicrobial sepsis. Am J Physiol. 2001;281:R654–60. doi: 10.1152/ajpregu.2001.281.2.R654. [DOI] [PubMed] [Google Scholar]
- 15.Hegde A, Zhang H, Moochhala SM, Bhatia M. Neurokinin-1 receptor antagonist treatment protects mice against lung injury in polymicrobial sepsis. J Leukoc Biol. 2007;82:678–85. doi: 10.1189/jlb.0407217. [DOI] [PubMed] [Google Scholar]
- 16.Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 17.Bhatia M, Slavin J, Cao Y, Basbaum AI, Neoptolemos JP. Preprotachykinin-A gene deletion protects mice against acute pancreatitis and associated lung injury. Am J Physiol. 2003;284:G830–36. doi: 10.1152/ajpgi.00140.2002. [DOI] [PubMed] [Google Scholar]
- 18.Salkowski CA, Detore G, Franks A, Falk MC, Vogel SN. Pulmonary and hepatic gene expression following cecal ligation and puncture: monophosphoryl lipid A prophylaxis attenuates sepsis-induced cytokine and chemokine expression and neutrophil infiltration. Infect Immun. 1998;66:3569–78. doi: 10.1128/iai.66.8.3569-3578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osuchowski MF, Welch K, Siddiqui J, Remick DG. Circulating cytokine/inhibitor profiles reshape the understanding of the SIRS/CARS continuum in sepsis and predict mortality. J Immunol. 2006;177:1967–74. doi: 10.4049/jimmunol.177.3.1967. [DOI] [PubMed] [Google Scholar]
- 20.Walley KR, Lukacs NW, Standiford TJ, Strieter RM, Kunkel SL. Elevated levels of macrophage inflammatory protein 2 in severe murine peritonitis increase neutrophil recruitment and mortality. Infect Immun. 1997;65:3847–51. doi: 10.1128/iai.65.9.3847-3851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ness TL, et al. CCR1 and CC chemokine ligand 5 interactions exacerbate innate immune responses during sepsis. J Immunol. 2004;173:6938–48. doi: 10.4049/jimmunol.173.11.6938. [DOI] [PubMed] [Google Scholar]
- 22.Matsukawa A, et al. Pivotal role of the CC chemokine, macrophage-derived chemokine, in the innate immune response. J Immunol. 2000;164:5362–8. doi: 10.4049/jimmunol.164.10.5362. [DOI] [PubMed] [Google Scholar]
- 23.Ness TL, Hogaboam CM, Strieter RM, Kunkel SL. Immunomodulatory role of CXCR2 during experimental septic peritonitis. J Immunol. 2003;171:3775–84. doi: 10.4049/jimmunol.171.7.3775. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi H, et al. An essential role of macrophage inflammatory protein 1α/CCL3 on the expression of host’s innate immunities against infectious complications. J Leukoc Biol. 2002;72:1190–7. [PubMed] [Google Scholar]
- 25.Yang D, et al. Many chemokines including CCL20/MIP-3α display antimicrobial activity. J Leukoc Biol. 2003;74:448–55. doi: 10.1189/jlb.0103024. [DOI] [PubMed] [Google Scholar]
- 26.Matsukawa A, et al. Pivotal role of the CC chemokine, macrophage-derived chemokine, in the innate immune response. J Immunol. 2000;164:5362–8. doi: 10.4049/jimmunol.164.10.5362. [DOI] [PubMed] [Google Scholar]
- 27.Kato T, et al. Interleukin 10 reduces mortality from severe peritonitis in mice. Antimicrob Agents Chemother. 1995;39:1336–40. doi: 10.1128/aac.39.6.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldbach-Mansky R, et al. Neonatal-onset multisystem inflammatory disease responsive to interleukin-1beta inhibition. N Engl J Med. 2006;355:581–92. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- 30.Gueders MM, et al. Matrix metallopro-teinase-8 deficiency promotes granulocytic allergen-induced airway inflammation. J Immunol. 2005;175:2589–97. doi: 10.4049/jimmunol.175.4.2589. [DOI] [PubMed] [Google Scholar]
- 31.Grimm T, et al. Inhibition of NF-kappaB activation and MMP-9 secretion by plasma of human volunteers after ingestion of maritime pine bark extract (Pycnogenol) J. Inflamm. (Lond.) 2006;3:1. doi: 10.1186/1476-9255-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cinel I, Opal SM. Molecular biology of inflammation and sepsis: a primer. Crit Care Med. 2009;37:291–304. doi: 10.1097/CCM.0b013e31819267fb. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Voegeli TS, Liu PP, Noble EG, Currie RW. Heat shock paradox and a new role of heat shock proteins and their receptors as anti-inflammation targets. Inflamm. Allergy Drug Targets. 2007;6:91–100. doi: 10.2174/187152807780832274. [DOI] [PubMed] [Google Scholar]
- 34.Cavaillon JM, Adib-Conquy M, Fitting C, Adrie C, Payen D. Cytokine cascade in sepsis. Scand J Infect Dis. 2003;35:535–44. doi: 10.1080/00365540310015935. [DOI] [PubMed] [Google Scholar]