Abstract
AIM: To investigate the association between peroxisome proliferator-activated receptor-γ (PPAR-γ) gene polymorphism 34 C>G and colorectal cancer (CRC), a meta-analysis review was performed in this report.
METHODS: A systematic literature search and selection of eligible relevant studies were carried out. Nine independent studies with a total number of 4533 cases and 6483 controls were included in the meta-analysis on the association between polymorphism 34 C>G and CRC.
RESULTS: There was no evidence for the association between PPAR-γ 34 C>G and CRC if all of the subjects in the nine studies were included. However, CG + GG showed a marginally significant difference from CC (OR = 0.84, 95% CI: 0.69-1.01, P = 0.07) in random-effect model. Stratified meta-analysis indicated that PPAR-γ 34 C>G was associated with colon cancer (OR = 0.8, 95% CI: 0.65-0.99, P = 0.04) in random-effect model, and the G allele decreased colon cancer risk. No significant association was observed between PPAR-γ 34 C>G and rectal cancer.
CONCLUSION: PPAR-γ 34 C>G is associated with colon cancer risk, but not associated with CRC and rectal cancer risk.
Keywords: Peroxisome proliferator-activated receptor-γ, Colorectal cancer, Polymorphism, Meta-analysis
INTRODUCTION
Colorectal cancer (CRC) is one of the major causes of cancer death in developed countries, with over 9500 new cases in Netherlands in 2002 alone, for instance[1]. In 2003, it was estimated that about 147 500 new cancer cases and 57 100 deaths were caused by CRC in the USA[2]. With more than 71 000 new occurrences per year, the incidence and mortality rate of CRC in Germany is almost the highest all over the world[3]. Epidemiological and experimental evidences attributed CRC to both genetic and experimental factors were involved. Accumulating evidences suggested that the peroxisome proliferator-activated receptor-γ (PPAR-γ) gene is related to CRC, which has been implicated in the pathogenesis of CRC in animal models and clinical studies. PPAR-γ is a member of the nuclear hormone receptor super-family, and plays a pivotal role in regulating adipocyte differentiation, glucose and lipid metabolism, insulin sensitivity, atherogenesis and immune[4]. A proline to alanine substitution has been detected in the PPAR-γ gene which is a common structural polymorphism (34 C>G, rs1801282) located at codon 12 (Pro12Ala) of PPAR-γ2-specific exon B. Although many studies were performed to investigate the relationship between the polymorphism 34 C>G of PPAR-γ gene and CRC, results were contradictory. Our study aims to confirm the former data of the association between PPAR-γ gene 34 C>G and CRC.
MATERIALS AND METHODS
Identification and eligibility of relevant studies
A systematic literature search in PubMed and Google was carried out in January 2010 using ‘PPAR gene’, ‘association’ and ‘CRC’ with restriction to ‘human’ or ‘homo sapiens’. Additional articles were identified through references cited in retrieved articles. Publications containing the same or overlapping data from the same authors were excluded. Studies were considered as eligible for the meta-analysis if the frequency of relevant genotypes was reported in both CRC cases and CRC-free controls, or in both colon cancer cases and CRC-free controls, as well as in both rectal cancer cases and CRC-free controls. Moreover, all of them were case-control studies or cohort studies. Nine articles reported on the analysis of the association between PPAR-γ Pro12Ala and CRC[1,2,4-10], four of which focused on the association between the polymorphism PPAR-γ Pro12Ala and colon cancer risk or rectal cancer risk[4-7].
Data extraction and statistical analysis
For each study, information was gathered about the first author, year of publication, country where the study was conducted and the distribution of each PPAR-γ 34 C>G genotype in cases and controls (Table 1). Some calculated data collected from the original data of the articles were applied in the subsequent meta-analysis.
Table 1.
Study characteristics
| No. | Ref. | Yr | Country |
Number of cases |
Number of controls |
34G allele fre-quency in control | P1 | ||||||||
| CC | CG | GG | Total | CG + GG | CC | CG | GG | Total | CG + GG | ||||||
| 1 | Landi et al[5] | 2003 | Spain (W, E) | 311 | 46 | 3 | 360 | 49 | 243 | 61 | 5 | 309 | 66 | 0.11 | 0.773 |
| 2 | Gong et al[2] | 2005 | USA (W) | 129 | 30 | 4 | 163 | 34 | 153 | 52 | 7 | 212 | 59 | 0.16 | 0.743 |
| 3 | Murtaugh et al[6] | 2005 | USA (W) | 1840 | - | - | 2371 | 531 | 2283 | - | - | 2972 | 689 | - | - |
| 4 | Jiang et al[4] | 2005 | India (A) | 240 | 57 | 4 | 301 | 61 | 230 | 57 | 4 | 291 | 61 | 0.11 | 1.000 |
| 5 | Siezen et al[1] | 2006 | Netherlands (W, E) | 160 | 40 | 1 | 201 | 41 | 325 | 71 | 2 | 398 | 73 | 0.09 | 0.783 |
| 6 | Koh et al[7] | 2006 | Singapore (A) | 345 | - | - | 362 | 17 | 1075 | - | - | 1164 | 89 | - | - |
| 7 | Gunter et al[8] | 2006 | USA (W) | 153 | 41 | 4 | 198 | 45 | 146 | 37 | 1 | 184 | 38 | 0.11 | 0.838 |
| 8 | Theodoropoulos et al[9] | 2006 | Greek (W, E) | 164 | 48 | 10 | 222 | 58 | 118 | 70 | 12 | 200 | 82 | 0.24 | 0.950 |
| 9 | Vogel et al[10] | 2007 | Denmark (W, E) | 252 | 96 | 7 | 355 | 103 | 550 | 190 | 13 | 753 | 203 | 0.14 | 0.816 |
P value for Hardy-Weinberg equilibrium (HWE) for peroxisome proliferator-activated receptor-γ (PPAR-γ) 34C>G polymorphism among controls. W: Western country; E: European country; A: Asian country; -: No data was shown in reference.
Percentage of GG genotype in controls of each study was calculated, followed by Hardy-Weinberg Equilibrium (HWE) test in controls to determine the reliability of data, using a Chi-squared Goodness-of-fit Test by SPSS 13.0. Analysis was also conducted on inter-ethnicity difference in minor allele frequency. One-way ANOVA was used to compare more than two independent groups, while two-tailed t test was used to compare two independent groups by SPSS 13.0 software.
To investigate the effect of each allele, the ORs of G allele were calculated, referenced by C. Subsequently, pairwised combinations of genotypes were used to determine the hereditary models, including GG vs CC, CG vs CC, and GG vs CG, CG vs CC + GG, GG vs CC + CG and CG vs GG + GC, and the later genotype was used as a reference in each pair.
We also conducted meta-analyses for a combination of CG and GG genotypes vs CC genotype in each sub-groups (European and USA population). In addition, stratified analyses were performed based on the case collection, including meta-analyses on the association between PPAR-γ 34 C>G and colon cancer risk and rectal cancer risk.
Heterogeneity among studies was tested to estimate which effect model, the fixed-effect one or the random-effect one, should be used. With a P > 0.05, the included studies were considered homogeneous and the fixed-effect model should be selected, otherwise, random-effect model should be used.
All of the meta-analyses above were conducted using Review Manager 4.2 software. The two-sided P < 0.05 was considered statistically significant.
RESULTS
Nine studies published from 2003-2007 were about the analysis on the relationship between PPAR-γ 34 C>G polymorphism and CRC risk, with a total number of 4533 cases and 6483 controls (Table 1). Seven studies (3870 cases/5028 controls) were conducted in Western countries, including 4 in Europe[1,5,9,10] and 3 in the USA[2,6,8]. Another two studies were performed in Asian countries[4,7]. There were four studies concerning colon cancer or rectal cancer with a total number of 2073 cases/3735 controls and 1321 cases/2765 controls, respectively[4-7].
The genotype distribution in the control groups in each study did not depart from the HWE with P > 0.05, except for two studies, in which HWE test could not be performed because of the incomplete data (Table 1).
The G allele frequency of PPAR-γ 34 C>G was 0.13 in the control group (626 cases/4694 controls) and 0.12 in the case group (424 cases/3600 controls), respectively. No statistical significance was found between case group and control group (P = 0.381). The G allele frequency of PPAR-γ 34 C>G in the Western controls (seven studies) was 0.14, the same as in the European controls (four studies). And in USA controls of three studies, G allele frequency of PPAR-γ 34 C>G was 0.13. In conclusion, there is no inter-ethnicity difference in minor allele frequency (P = 0.968).
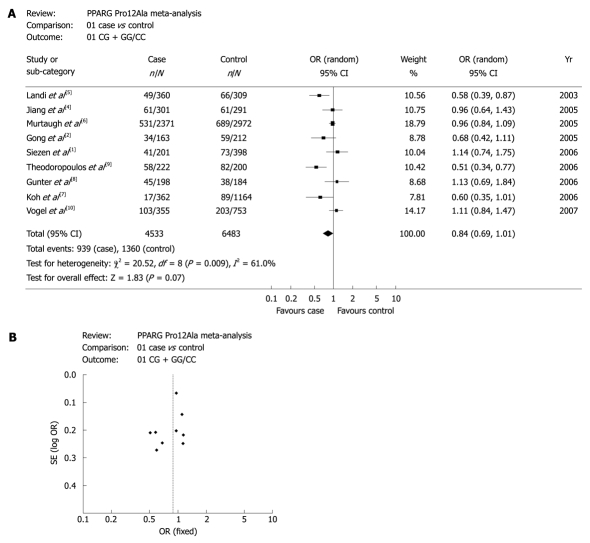
Overall and subgroup-specific summary ORs and 95% CIs for the relationship between PPAR-γ 34 C>G and CRC risk are summarized in Table 2. For G vs C allele, GG vs CC, CG vs CC, GG vs CG, CG vs CC + GG, GG vs CC + CG and CG + GG vs CC genotypes of the overall study population, the fixed-effect and the random-effect ORs (95% CIs) were listed, respectively. Test for heterogeneity indicated that studies in the analyses for G vs C allele, CG vs CC, CG vs CC + GG, CG + GG vs CC genotypes, respectively were heterogeneous with P < 0.05. Hence, random-effect models were selected. And for the rest ones with P > 0.05, fixed-effect model was used. As a result, no statistical significance was observed among the above analyses. However, the data of CG + GG vs CC genotypes was marginally significant with OR = 0.84 (95% CI: 0.69-1.01, P = 0.07).
Table 2.
Overall and group-specific summary statistics for PPAR-γ 34 C>G and colorectal, colon and rectal cancers
| No. of studies | Polymorphisms | P1 | Fixed-effect OR (95% CI), P2 | Random-effect OR (95% CI), P3 | |
| Colorectal cancer | |||||
| Total | 7 | G vs C | 0.010 | 0.88 (0.77, 1.00), 0.060 | 0.86 (0.69, 1.08), 0.210 |
| 7 | GG vs CC | 0.720 | 0.85 (0.53, 1.35), 0.480 | 0.83 (0.52, 1.33), 0.440 | |
| 7 | CG vs CC | 0.020 | 0.86 (0.74, 1.00), 0.050 | 0.83 (0.65, 1.07), 0.160 | |
| 7 | GG vs CG | 0.990 | 1.10 (0.68, 1.79), 0.690 | 1.09 (0.67, 1.78), 0.720 | |
| 7 | CG vs CC + GG | 0.020 | 0.86 (0.74, 1.00), 0.060 | 0.84 (0.66, 1.07), 0.150 | |
| 7 | GG vs CC + CG | 0.830 | 0.91 (0.57, 1.44), 0.680 | 0.89 (0.56, 1.43), 0.630 | |
| 9 | CG + GG vs CC | 0.009 | 0.90 (0.82, 0.99), 0.030 | 0.84 (0.69, 1.01), 0.070 | |
| Western | 6 | G vs C | 0.006 | 0.87 (0.75, 1.00), 0.050 | 0.85 (0.65, 1.11), 0.230 |
| 6 | GG vs CC | 0.600 | 0.83 (0.51, 1.36), 0.470 | 0.81 (0.49, 1.34), 0.420 | |
| 6 | CG vs CC | 0.009 | 0.84 (0.72, 1.00), 0.040 | 0.81 (0.60, 1.09), 0.170 | |
| 6 | GG vs CG | 0.970 | 1.12 (0.67, 1.87), 0.670 | 1.11 (0.66, 1.86), 0.700 | |
| 6 | CG vs CC + GG | 0.010 | 0.85 (0.72, 1.00), 0.050 | 0.82 (0.61, 1.09), 0.160 | |
| 6 | GG vs CC + CG | 0.730 | 0.90 (0.55, 1.47), 0.680 | 0.88 (0.54, 1.45), 0.630 | |
| 7 | CG + GG vs CC | 0.006 | 0.91 (0.82, 1.01), 0.070 | 0.85 (0.68, 1.06), 0.140 | |
| USA | 3 | CG + GG vs CC | 0.320 | 0.95 (0.84, 1.07), 0.360 | 0.94 (0.80, 1.11), 0.450 |
| Europe | 4 | CG + GG vs CC | 0.002 | 0.84 (0.70, 1.00), 0.060 | 0.79 (0.52, 1.19), 0.260 |
| Colon cancer | 4 | CG + GG vs CC | 0.030 | 0.83 (0.72, 0.96), 0.010 | 0.80 (0.65, 0.99), 0.040 |
| Rectal cancer | 4 | CG + GG vs CC | 0.050 | 0.98 (0.82, 1.18), 0.860 | 0.84 (0.58, 1.22), 0.370 |
P value for test for heterogeneity;
P value for fixed-effect model;
P value for random-effect mode.
Limited results in studies from Western countries, the fixed-effect and random-effect ORs (95% CIs ) for G vs C allele, GG vs CC, CG vs CC, GG vs CG, CG vs CC + GG, GG vs CC + CG and CG + GG vs CC genotypes are listed in Table 2, respectively. Same to the former analyses of the total study population, there was no evidence for the association between PPAR-γ 34 C>G and CRC risk.
Further subgroup-specific analyses were performed in the European and USA studies. In studies from European countries, summary ORs (95% CIs) for CG + GG vs CC genotypes in fixed-effect model and random-effect model were 0.84 (0.70, 1.00) and 0.94 (0.80, 1.11), respectively. In studies from the USA, the corresponding ORs (95% CIs) were 0.79 (0.52, 1.19) and 0.95 (0.84, 1.07), respectively. No evidence was found for the association between PPAR-γ 34 C>G and CRC risk in each of the two study populations.
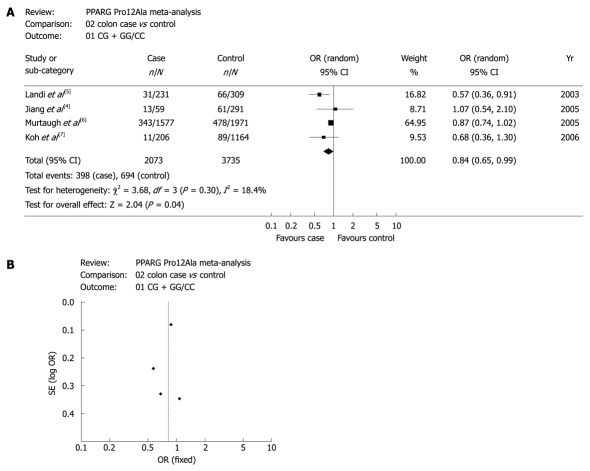
However, when stratified analyses were performed, the results were different. As shown in Table 2, four studies were involved in the meta-analyses for the association of PPAR-γ 34 C>G with colon cancer risk and rectal cancer risk. In the four studies, only the data for CG + GG vs CC genotypes was sufficient enough for the analyses. As P values of the heterogeneity test in the two meta-analyses were less than 0.05, random-effect model was used. Summary ORs (95% CIs) for CG + GG vs CC genotypes in colon cancer studies and rectal cancer studies were 0.80 (0.65, 0.99) and 0.84 (0.58, 1.22), respectively. Statistical significance was observed in the association meta-analysis between PPAR-γ 34 C>G and colon cancer risk (P = 0.04), indicating that PPAR-γ 34 C>G was associated with colon cancer risk, and G allele decreased the colon cancer risk. However, there was no evidence for the association between PPAR-γ 34 C>G and rectal cancer risk (P = 0.37).
Figure 1A shows the forest plots of meta-analysis for CG + GG vs CC genotypes to confirm the association between PPAR-γ 34 C>G and CRC risk in the overall study population. The association between PPAR-γ 34 C>G and CRC risk had a marginally statistical significance. Figure 1B is the funnel plot, suggesting that there was no publication bias in the studies.
Figure 1.
Association between peroxisome proliferator-activated receptor-γ (PPAR-γ) 34 C>G and colorectal cancer risk (CG + GG vs CC). A: Forest plot-studies are sorted in order of publication year; B: Funnel plots for the associations.
Figure 2A displays the forest plots of the study on the association between PPAR-γ 34 C>G and colon cancer risk. Obviously, there was a significant association of PPAR-γ 34 C>G with colon cancer risk. Publication bias was not found in this study as shown in Figure 2B. Other forest plots and funnel plots of meta-analyses on the association between PPAR-γ 34 C>G and CRC risk, colon cancer risk and rectal cancer risk were not shown. However, the results are presented in Table 2.
Figure 2.
Association between PPAR-γ 34 C>G and colon cancer risk (CG + GG vs CC). A: Forest plot-studies are sorted in order of publication year; B: Funnel plots for the associations.
DISCUSSION
CRC is one of the leading causes of cancer death in the developed countries[11,12]. Both sporadic and hereditary CRC is caused by a set of molecular events[13]. Accumulated evidences indicate that lipid metabolism, especially the one involved in the arachidonic acid (AA)-pathway, appears to play a critical role in the development of colorectal tumor[14]. PPAR-γ gene, one of the most important components of the AA-pathway, has been verified to express in a variety of tumor cells. And it will lead to either inhibition of cell proliferation or induction of apoptosis after bonding with ligands[15,16]. Many studies reported that PPAR-γ was also expressed in colon tumors, normal colon mucosa and colon cancer cell lines[17-19]. Genomics research showed that there was a polymorphism in the coding region 34 C>G in PPAR-γ which resulted in the amino acid change of Pro12 Ala[20]. So far, many studies have been performed on the association between PPAR-γ 34 C>G polymorphism and CRC risk, but produced controversial results.
According to our search of references, no systematic review has been published on the analysis of the association between PPAR-γ 34 C>G and CRC. In order to confirm the data on the associations between PPAR-γ gene polymorphism and CRC, we did a meta-analysis based on nine studies from Europe, Asia and the USA.
Our results showed that there was no evidence for the association between PPAR-γ 34C>G and CRC if all of the subjects in the nine studies were included. Subgroup-specific meta-analyses also indicated that there was no association between PPAR-γ 34C>G and CRC in European, Asian and the USA studies. As shown in our analysis, G allele might decrease CRC risk, although statistical difference was not significant in these meta-analyses.
Many studies have indicated that both genetic and environmental factors are involved in the development of colorectal tumor[21]. Environmental factors include ethnicity, gender, diet, age, NSAIDS use, BMI, smoking, drinking, family history of disease and so on. It was inferred that there must be interaction between the environmental factors and PPAR-γ gene, which had been proved in many researches, including the nine studies. Therefore, interaction is one of the factors of the meta-analyses. Further analysis should be conducted to confirm the influence of PPAR-γ 34 C>G on CRC risk.
Stratified meta-analysis indicated that PPAR-γ 34 C>G was associated with colon cancer, and the G allele decreased the colon cancer risk. No evidence was observed for the association between PPAR-γ 34 C>G and rectal cancer.
In conclusion, no evidence was observed for the association between PPAR-γ 34 C>G and CRC risk and rectal cancer risk. However, PPAR-γ 34 C>G is associated with colon cancer risk, which is meaningful to early diagnosis, prevention and individual-based treatment of colon cancer. Furthermore, 34 C>G of PPAR-γ gene might be a potential therapeutic target for colon cancer.
COMMENTS
Background
Colorectal cancer (CRC) is one of the major causes of cancer death in the developed countries. Accumulated evidences suggest that the peroxisome proliferator-activated receptor-γ (PPAR-γ) gene is related to CRC.
Research frontiers
Accumulated evidences indicate that lipid metabolism, especially the one in the arachidonic acid (AA)-pathway, appears to play a critical role in the development of CRC. PPAR-γ gene, one of the most important components of the AA-pathway, has been verified to express in a variety of tumor cells. A proline to alanine substitution in the PPAR-γ gene was detected, which might be associated with CRC.
Innovations and breakthroughs
Many studies have been performed about the association between the polymorphism 34 C>G of PPAR-γ gene and CRC, but got conflicting results. In order to confirm the data, meta-analyses, as a better statistical analysis technique, were performed in this report.
Applications
In this report, the association between PPAR-γ gene polymorphism 34 C>G and colon cancer risk was observed, and the G allele decreased the colon cancer risk, which is meaningful to early diagnosis, prevention and individual-based treatment of colon cancer. Furthermore, 34 C>G of PPAR-γ gene might be a potential therapeutic target for colon cancer.
Terminology
Susceptibility to colon cancer differs according to genotype of PPAR-γ 34 C>G, and people with G allele might have a lower colon cancer risk.
Peer review
This is an interesting meta-analysis on the association between PPAR-γ 34C>G polymorphism and CRC risk. The main finding of the paper was that PPAR-γ 34 C>G was weakly associated with colon cancer risk, but was not associated with CRC and rectal cancer risk. The methods are properly used; authors report a high quality meta-analysis.
Footnotes
Supported by The National Natural Science Foundation of China, No. 30772534
Peer reviewers: Peter L Lakatos, MD, PhD, Assistant Professor, 1st Department of Medicine, Semmelweis University, Koranyi S 2A, Budapest H1083, Hungary; Ulrike S Stein, PhD, Assistant Professor, Max-Delbrück-Center for Molecular Medicine, Robert-Rössle-Straße 10, 13125 Berlin, Germany
S- Editor Tian L L- Editor Ma JY E- Editor Zheng XM
References
- 1.Siezen CL, Bueno-de-Mesquita HB, Peeters PH, Kram NR, van Doeselaar M, van Kranen HJ. Polymorphisms in the genes involved in the arachidonic acid-pathway, fish consumption and the risk of colorectal cancer. Int J Cancer. 2006;119:297–303. doi: 10.1002/ijc.21858. [DOI] [PubMed] [Google Scholar]
- 2.Gong Z, Xie D, Deng Z, Bostick RM, Muga SJ, Hurley TG, Hebert JR. The PPAR{gamma} Pro12Ala polymorphism and risk for incident sporadic colorectal adenomas. Carcinogenesis. 2005;26:579–585. doi: 10.1093/carcin/bgh343. [DOI] [PubMed] [Google Scholar]
- 3.Sieg A, Friedrich K. Perspectives of colorectal cancer screening in Germany 2009. World J Gastrointest Endosc. 2009;1:12–16. doi: 10.4253/wjge.v1.i1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J, Gajalakshmi V, Wang J, Kuriki K, Suzuki S, Nakamura S, Akasaka S, Ishikawa H, Tokudome S. Influence of the C161T but not Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-gamma on colorectal cancer in an Indian population. Cancer Sci. 2005;96:507–512. doi: 10.1111/j.1349-7006.2005.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, de Oca J, Capella G, Canzian F. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63:3560–3566. [PubMed] [Google Scholar]
- 6.Murtaugh MA, Ma KN, Caan BJ, Sweeney C, Wolff R, Samowitz WS, Potter JD, Slattery ML. Interactions of peroxisome proliferator-activated receptor {gamma} and diet in etiology of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1224–1229. doi: 10.1158/1055-9965.EPI-04-0681. [DOI] [PubMed] [Google Scholar]
- 7.Koh WP, Yuan JM, Van Den Berg D, Ingles SA, Yu MC. Peroxisome proliferator-activated receptor (PPAR) gamma gene polymorphisms and colorectal cancer risk among Chinese in Singapore. Carcinogenesis. 2006;27:1797–1802. doi: 10.1093/carcin/bgl001. [DOI] [PubMed] [Google Scholar]
- 8.Gunter MJ, Canzian F, Landi S, Chanock SJ, Sinha R, Rothman N. Inflammation-related gene polymorphisms and colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2006;15:1126–1131. doi: 10.1158/1055-9965.EPI-06-0042. [DOI] [PubMed] [Google Scholar]
- 9.Theodoropoulos G, Papaconstantinou I, Felekouras E, Nikiteas N, Karakitsos P, Panoussopoulos D, Lazaris ACh, Patsouris E, Bramis J, Gazouli M. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol. 2006;12:5037–5043. doi: 10.3748/wjg.v12.i31.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel U, Christensen J, Dybdahl M, Friis S, Hansen RD, Wallin H, Nexø BA, Raaschou-Nielsen O, Andersen PS, Overvad K, et al. Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutat Res. 2007;624:88–100. doi: 10.1016/j.mrfmmm.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 11.O’Morain C, Qasim A. Concept of chemoprevention in colorectal cancer. World J Gastrointest Oncol. 2009;1:21–25. doi: 10.4251/wjgo.v1.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Guan YS, Pan WM, Luo ZM, Wei JH, Zhao L, Wu H. Clinical value of F-FDG PET/CT in assessing suspicious relapse after rectal cancer resection. World J Gastrointest Oncol. 2009;1:55–61. doi: 10.4251/wjgo.v1.i1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tejpar S, Van Cutsem E. Molecular and genetic defects in colorectal tumorigenesis. Best Pract Res Clin Gastroenterol. 2002;16:171–185. doi: 10.1053/bega.2001.0279. [DOI] [PubMed] [Google Scholar]
- 14.Jones R, Adel-Alvarez LA, Alvarez OR, Broaddus R, Das S. Arachidonic acid and colorectal carcinogenesis. Mol Cell Biochem. 2003;253:141–149. doi: 10.1023/a:1026060426569. [DOI] [PubMed] [Google Scholar]
- 15.Grommes C, Landreth GE, Heneka MT. Antineoplastic effects of peroxisome proliferator-activated receptor gamma agonists. Lancet Oncol. 2004;5:419–429. doi: 10.1016/S1470-2045(04)01509-8. [DOI] [PubMed] [Google Scholar]
- 16.Theocharis S, Margeli A, Vielh P, Kouraklis G. Peroxisome proliferator-activated receptor-gamma ligands as cell-cycle modulators. Cancer Treat Rev. 2004;30:545–554. doi: 10.1016/j.ctrv.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Osawa E, Nakajima A, Wada K, Ishimine S, Fujisawa N, Kawamori T, Matsuhashi N, Kadowaki T, Ochiai M, Sekihara H, et al. Peroxisome proliferator-activated receptor gamma ligands suppress colon carcinogenesis induced by azoxymethane in mice. Gastroenterology. 2003;124:361–367. doi: 10.1053/gast.2003.50067. [DOI] [PubMed] [Google Scholar]
- 18.Shimada T, Kojima K, Yoshiura K, Hiraishi H, Terano A. Characteristics of the peroxisome proliferator activated receptor gamma (PPARgamma) ligand induced apoptosis in colon cancer cells. Gut. 2002;50:658–664. doi: 10.1136/gut.50.5.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen GG, Lee JF, Wang SH, Chan UP, Ip PC, Lau WY. Apoptosis induced by activation of peroxisome-proliferator activated receptor-gamma is associated with Bcl-2 and NF-kappaB in human colon cancer. Life Sci. 2002;70:2631–2646. doi: 10.1016/s0024-3205(02)01510-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhou XP, Smith WM, Gimm O, Mueller E, Gao X, Sarraf P, Prior TW, Plass C, von Deimling A, Black PM, et al. Over-representation of PPARgamma sequence variants in sporadic cases of glioblastoma multiforme: preliminary evidence for common low penetrance modifiers for brain tumour risk in the general population. J Med Genet. 2000;37:410–414. doi: 10.1136/jmg.37.6.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zambirinis CP, Theodoropoulos G, Gazouli M. Undefined familial colorectal cancer. World J Gastrointest Oncol. 2009;1:12–20. doi: 10.4251/wjgo.v1.i1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]