Abstract
Rationale:
Studies of the genetics of obstructive sleep apnea may be facilitated by identifying intermediate traits with high heritability that quantify etiological pathways, such as those related to respiratory control. Electrocardiogram (ECG)-based sleep spectrograms, measuring the coupling between respiratory modulation of ECG QRS-wave amplitude and heart rate variability, may provide measures of sleep state and ventilatory dynamics during sleep. We evaluated the familial aggregation of distinctive spectrographic biomarkers of unstable sleep, related to elevated-low frequency cardiopulmonary coupling (e-LFC), to assess their utility in genetic studies.
Methods:
622 participants from 137 families from the Cleveland Family Study underwent standardized polysomnography (PSG). From the ECG signal on the PSG, the interbeat interval time series and the corresponding ECG-derived respiratory signal were extracted, and the low frequency (0.01-0.1 Hz) component of their coupling was computed using a fully automated method. Narrow sense heritability of e-LFC was calculated using variance component methods.
Results:
A spectral marker of abnormal low frequency cardiopulmonary coupling (e-LFC) demonstrated moderate correlation with apnea hypopnea index (AHI; r = 0.35, P < 0.0001). The heritability estimate for e-LFC, after adjusting for age and sex was 0.32 (P < 10-5) and remained unchanged after additionally adjusting for body mass index or AHI. In biological relatives of those with sleep apnea, a related marker of e-LFC was more prevalent than in controls (P = 0.05).
Conclusions:
Approximately 30% of the variability of e-LFC, measured from a continuous ECG during sleep, is explained by familial factors other than BMI. ECG-based spectrographic measures of cardiopulmonary coupling may provide novel phenotypes for characterizing subgroups of individuals with different propensities and genetic etiologies for sleep apnea or for other conditions associated with sleep fragmentation.
Citation:
Ibrahim LH; Jacono FJ; Patel SR; Thomas RJ; Larkin EK; Mietus JE; Peng CK; Goldberger AL; Redline S. Heritability of abnormalities in cardiopulmonary coupling in sleep apnea: use of an electrocardiogram-based technique. SLEEP 2010;33(5):643-646.
Keywords: Obstructive sleep apnea, heritability, cardiopulmonary coupling, ECG spectrogram
OBSTRUCTIVE SLEEP APNEA (OSA) IS A COMPLEX DISEASE WITH A STRONG GENETIC COMPONENT1. A NUMBER OF GENETIC ETIOLOGICAL PATHWAYS, such as those that influence adiposity, craniofacial structure, and control of breathing, may increase disease susceptibility.2,3 Measuring and quantifying variation in such intermediate phenotypes may be particularly useful in identifying subgroups of individuals who share specific genetic variants that increase risk of sleep apnea. Of particular interest are intermediate traits associated with control of ventilation, a phenotype that could be a target for pharmacological or other interventions. However, these traits have not been well investigated primarily because of inadequacies in measurement techniques. Therefore, new approaches are needed to measure these traits in large populations.
Periodic oscillations of respiration with a near-identical period length (i.e., Cheyne-Stokes breathing) are considered a manifestation of ventilatory control irregularities.4,5 Thomas et al.6 recently developed an electrocardiogram (ECG)-based technique that measures cardiopulmonary coupling using heart rate variability and an ECG-derived respiration signal. ECG spectrographic measures were found to correlate strongly with electroencephalographic (EEG) measures of sleep stability, but not conventional sleep stage distributions. The resultant sleep spectrogram is a map of coupled sleep oscillations, demonstrating spontaneously transitioning periods of high frequency coupling (HFC, “stable” sleep), low frequency coupling (LFC, “unstable” sleep), and very low frequency coupling (wake or REM sleep). A spectrographic subset of LFC, denoted as elevated LFC (e-LFC), is increased in severely fragmented sleep states including sleep apnea. It has been suggested that narrow band e-LFC (e-LFCNB) may be a marker of strong chemoreflex modulation of sleep respiration, suggested by its association with central apneas and an increased risk of positive airway pressure treatment-emergent central sleep apnea.7 We hypothesized that a measure that characterizes pathologic oscillations in breathing will be heritable, and that heritability of this trait may be independent of body mass index (BMI), and thus be potentially useful as an intermediate phenotype to understand the genetic basis of OSA.
METHODS
Study Population
Subjects were participants in the Cleveland Family Study, a longitudinal community-based study of OSA probands and their family members along with neighbor control families, established to evaluate the genetic aspects of OSA. Families informative for genetic studies of sleep apnea were recruited for extensive in-laboratory phenotyping. These informative families included individuals from sibships at the extremes of the AHI distribution or containing ≥ 3 generations of family members. Additional details about the methods of recruitment and data collection have been previously described.8 The analytic sample consisted of 622 participants from 137 families.
Data Collection
Each participant underwent in-laboratory overnight polysomnography (PSG) using the Compumedics E-Series System (Abbotsford, Australia) conducted in the General Clinical Research Center at University Hospitals Case Medical Center (Cleveland, OH). Respiration was measured using abdominal and thoracic inductance plethysmography sampled at 32 Hz. Institutional review board approval was obtained; written informed consent was obtained from each participant.
Apneas and hypopneas were defined using Sleep Heart Health Study criteria 9 modified to include the nasal pressure signal.
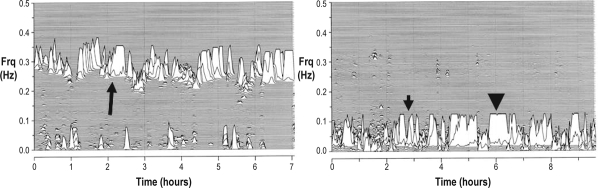
Sleep Spectrogram Analysis (Figure 1)
Figure 1.
The ECG-derived sleep spectrogram. The left part of the figure shows a healthy pattern dominated by high frequency cardiopulmonary coupling (long arrow). The right part of the figure shows a potentially pathologic (“unstable”) pattern with absence of high frequency cardiopulmonary coupling, which is replaced by low frequency coupling. Note the clear separation of high frequency and low frequency coupling bands. Further, the latter has two important spectral subcomponents: broad and narrow bands that may be abnormally elevated. The short arrow (right side) identifies broad-band elevated-low frequency coupling and the arrow-head identifies narrow band elevated-low frequency coupling. Both examples are from the Cleveland Family study participants. Frq, frequency of coupled respiration and heart rate variability.
Details of the method have been published and are available in the online Appendix (www.journalsleep.org). In brief, using a continuous single lead ECG, we combined information from heart rate variability and ECG-derived respiration (EDR). The latter reflects amplitude variations in the QRS complex that result from shifts in the cardiac electrical axis relative to the electrodes during respiration, and changes in thoracic impedance as the lungs fill and empty. After filtering for outliers and cubic spline re-sampling at 2 Hz, the cross-spectral power and coherence of these 2 signals were calculated over a 1024-sample (8.5-min) window using the fast Fourier transform applied to 3 overlapping 512-sample sub-windows within the 1024-sample coherence window. The window was then advanced by 256 samples (2.1 min) and the computations repeated until the entire NN interval/EDR series was analyzed. For each 1024-sample window, the product of the coherence and cross-spectral power was used to calculate the ratio of coherent cross power in the low frequency (0.01-0.1 Hz) band to that in the high frequency (0.1-0.4 Hz) band. This ratio was used to classify each successive sampling window as high frequency coupling (“stable” state) or low frequency coupling (“unstable” state). Very low frequency coupling (wake or REM sleep) was calculated using the ratio of coherent cross power in the 0-0.01 Hz band to the power in the 0.01-0.4 Hz band.
We further identified a subset of low frequency coupling, called elevated-Low Frequency Coupling (e-LFC). The frequency band defining e-LFC had been previously determined using data from the open access PhysioNet Sleep Apnea Database (http://www.physionet.org/physiobank/database/apnea-ecg/), identifying the frequency range that provided the maximum combined sensitivity and specificity for apnea/ hypopnea detection. To further identify periods of breathing with relatively constant cycle length (e.g., as may occur with periodic breathing patterns), we also quantified the temporal dispersion of consecutive spectral peaks of apnea detection, using an algorithm that identified the pattern of narrow spectral band e-LFC (e-LFCNB); i.e., requiring a minimum power in this band of 0.3 normalized units and a coupling frequency of each pair of consecutive measurements remaining within 0.0059 Hz of each other over 5 consecutive overlapping coherence windows (16.9 continuous min). To detect subtle effects of the respiratory chemoreflex on sleep-breathing for this analysis, we relaxed the criteria to 3 consecutive overlapping coherence windows, or 12.7 continuous min with a narrow spectral pattern. (See appendix at www.journalsleep.org for further details).
It is important to note that the low frequency component of the sleep spectrogram is not equivalent to the low frequency component of the HRV spectrum, but more specifically represents relatively low frequency respiratory-coupled oscillations in heart rate Thus the frequency bands of the ECG-derived sleep spectrogram are distinct from the standard HRV bands, although an overlap exists.
Calculation of Heritability
Narrow sense heritabilities (defined as the proportion of total variance due to additive genetic effects) were estimated using a variance component approach as implemented in the software package SOLAR.10 A square root transformation was applied to meet normality assumptions. Heritability of e-LFCNBwas unable to be measured because of excessive kurtosis. Instead, e-LFCNB was expressed as a dichotomous variable (present/ absent) and its prevalence in biologic relatives (parents, siblings, offspring,) of OSA cases was compared to non-biologic relatives in all families.
RESULTS
The sample consisted of 42% (n = 260) males and 58% females, and included a slight predominance of African Americans (56%). Overall, the mean BMI was 31.2 kg/m2 and mean age was 40.1 years. The AHI varied from 0 to 119, with approximately 25% of the cohort meeting criteria for OSA (AHI ≥ 15). As expected, compared to subjects without OSA, those with OSA were heavier (mean BMI 37 kg/m2), older (mean age 52 years), and consisted of a greater proportion of men (57%). Of note, the frequency of central apneas was very low (0.40 central apneas per hour of sleep).
Spearman correlation coefficients demonstrated that e-LFC and e-LFCNB were modestly associated with age (r = 0.29 and r = 0.15, respectively: P ≤ 0.0001), BMI (r = 0.13 and r = 0.11, respectively: P < 0.001), and central apnea index (r = 0.13 for both: P < 0.002). The e-LFC and e-LFCNB were somewhat more strongly associated with AHI (r = 0.36 and r = 0.26, respectively: P < 0.0001).
The variation of indices of e-LFC and e-LFCNB with levels of AHI was also analyzed. There was an increase in the magnitude of e-LFC (as a proportion of total sleep time) by AHI levels (AHI < 5 = 0.062, AHI 5-15 = 0.071, AHI 15-30 = 0.078, AHI > 30 = 0.154; P < 0.0001), after adjusting for age, sex, and BMI. A similar finding was seen with e-LFCNB (AHI < 5 = 0.006, AHI 5-15 = 0.006, AHI 15-30 = 0.007, AHI > 30 = 0.015; P < 0.0001) after adjusting for age, sex, and BMI.
The age and sex adjusted heritability estimate (h2) for AHI was similar to that observed for e-LFC (AHI h2 = 0.31, e-LFC h2 = 0.32, P < 10-5 for all estimates; Table 1) After adjusting for age, sex, and BMI, the h2for AHI was attenuated, while e-LFC increased slightly (AHI h2 = 0.24, e-LFC h2 = 0.35, P < 10-5 for all estimates). When further adjusting e-LFC for AHI, the h2 for e-LFC increased (h2 = 0.41, P < 10-5).
Table 1.
Heritability estimates for elevated low frequency coupling and apnea hypopnea index
| Covariate | Age and Sex Adjusted | Age, Sex, and BMI Adjusted | Age, Ses, BMI and AHI Adjusted |
|---|---|---|---|
| e-LFC | h2= 0.32 | h2= 0.35 | h2= 0.41 |
| SE = 0.08 | SE = 0.08 | SE = 0.09 | |
| AHI | h2= 0.31 | h2= 0.24 | xxx |
| SE = 0.08 | SE = 0.08 | ||
e-LFC, elevated low frequency coupling; AHI, apnea hypopnea index; h2, heritability estimate; SE, standard error; P-value <10-5 for all estimates
The prevalence of e-LFCNBin biologic relatives of affected probands was 17%, which was higher than in comparable biologic relatives of control probands (7%: P = 0.05). In contrast, among non-biologic relatives of case and control families, which mainly consisted of spouses, no difference in the prevalence of e-LFCNBwas observed (10% vs. 8% respectively, P > 0.99).
DISCUSSION
The key findings from our analysis are that: (1) Low frequency cardiopulmonary oscillations, which are a marker of unstable sleep state, have a strong heritable component; and (2) An ECG-based spectrographic biomarker of putative chemoreflex modulation of sleep-respiration is increased in biological relatives of sleep apnea patients.
OSA is a disease with a strong genetic component.11–15 An enhanced understanding of the genetics of OSA may be achieved by identifying intermediate traits with high heritability. Such intermediate phenotypes may be more powerful than conventional polysomnographic metrics in dissecting the underlying specific genetic etiology. Etiological pathways that may be involved with increasing OSA susceptibility include those that influence central obesity, craniofacial morphology, and control of ventilation.16 Obesity is easily measured and has been studied extensively. Because of the complexities in techniques for measuring other phenotypes in large samples, they have not been as well investigated. The propensity for unstable sleep or sleep fragmentation has not been previously considered as an endophenotype for sleep apnea genetic risk, although there is evidence that arousals may amplify the effects of sleep apnea.17,18
A new approach for identifying subgroups of patients with OSA is by mapping cardiopulmonary coupling during sleep, and the technique described here provides an example of a practical method for this assessment. Since such measurements are readily obtainable from routine PSG or even from portable monitors or Holter recordings, we examined their potential utility for genetic epidemiologic studies by quantifying their heritability, their association with AHI and BMI, and their distributions among biological relatives of probands compared to control family members. Our analyses demonstrated that even after considering demographic factors including BMI, e-LFC and e-LFCNB were significantly associated with AHI. In age- and sex-adjusted analyses, the heritability for the AHI and the continuous measure from the spectrogram, the e-LFC, were each approximately 0.30. However, after further adjusting for BMI, the heritability of the AHI decreased and the e-LFC was modestly increased. Furthermore, after adjusting for AHI, the heritability of e-LFC further increased to 0.41. These findings suggest that e-LFC, although associated with OSA, is determined by familial factors other than those associated with obesity, such as ventilatory control mechanisms or sleep stability. As such, the measurement of e-LFC provides unique information beyond that provided by the AHI.
Although the heritability of e-LFCNB,a putative marker for chemoreflex modulation of sleep-respiration and OSA, could not be calculated because of kurtosis of this measure, when dichotomized it was more prevalent in biological relatives of affected families than in members of control families. In contrast, there was no difference in the prevalence of this marker between the unrelated family members of cases and controls. Individuals with a genetic predisposition for OSA may possibly be identified by quantifying respiratory-heart rate variations that result in precisely timed oscillations that are suggestive of chemoreflex-driven sleep-respiratory pathology, which, in their more extreme forms, may present as central sleep apnea or periodic breathing.
A strength of our study is the moderately large sample enriched with family members with OSA, allowing calculation of heritability for standard and novel measures. However, a larger sample of control families would have enabled a more detailed examination in how the spectral measurements varied in those with and without a presumed genetic predisposition to OSA.
As proposed intermediate phenotypes, e-LFC or e-LFCNBmay provide information on one or more physiological traits, such as breathing variability, sleep state stability/arousability, and perhaps other cardiopulmonary functions. It is likely that these indices reflect traits that predispose to conditions characterized by variations in breathing pattern and sleep stability, such as, but not limited to, OSA. For example, in exploratory analyses, we found that e-LFCNB was not significantly associated with periodic leg movements, but was associated with a subset of PLMs that had concurrent arousals (r = 0.26). Given that the heritability of this trait is greater after adjusting for BMI, it may prove to be particularly valuable for identifying subgroups of individuals with common physiological predispositions to OSA and other disorders associated with sleep fragmentation.
In summary, the findings provide evidence for the potential utility of temporally derived measures of heart rate and breathing variation as intermediate phenotypes in genetic epidemiological studies. Further work defining the relationship of these measurements to those of traditional ventilatory control measurements and sleep state dynamics would assist with further understanding physiological differences among individuals with OSA. However, these preliminary data point to the feasibility of using relatively simple measurements from the PSG to obtain highly heritable phenotypes that provide information different from the AHI or BMI.
DISCLOSURE STATEMENT
This was not an industry supported study. Dr. Peng is a co-founder of DynaDx Corporation which specializes in signal analysis software. Dr. Redline directed a research study supported by Dymedic Corp. She received no direct income from Dymedic. Dr. Redline has received the use of equipment from Respironics. Dr. Goldberger is a co-inventor of the Cardiopulmonary sleep spectrogram software that received a US patent and is licensed to Embla, Inc., via the Beth Israel Medical Center, Boston, MA. Dr. Goldberger has also consulted for DynaDx Corp. Dr. Thomas and Mr. Mietus own a sleep spectrogram patent licensed to Embla, Inc. The other authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
We are indebted to the dedicated staff of the Cleveland Family Study, including Joan Aylor, Kathryn Clark, Jennifer Frame, Heather Rogers, Rawan Salem, and Xiaobei Zhu, as well as the nurses working in the University Hospitals Case Medical Center General Clinical Research Center. We are particularly grateful for the participation of the members of the Cleveland Family Study whose continuing enthusiasm has made this study possible.
This study had the following grant support: HL46380, HL81385, UL1-RR024989, VA Research Service, KL2-RR024990, G. Harold and Leila Y. Mathers Foundation, James S. McDonnell Foundation, and the NIH-sponsored Research Resource for Complex Physiologic Signals (UO1EB008577).
ABBREVIATIONS
- ECG
Electrocardiogram
- e-LFC
elevated-low frequency cardiopulmonary coupling
- AHI
apnea hypopnea index
- PSG
polysomnography
- OSA
obstructive sleep apnea
- EEG
electroencephalographic
- e-LFCNB
narrow band e-LFC
- HFC
high frequency cardiopulmonary coupling
- LFC
low frequency cardiopulmonary coupling
- EDR
ECG derived respiratory signal
- h2
heritability
APPENDIX
ECG-Derived Cardiopulmonary Coupling Assessment
Cardiopulmonary coupling analysis and generation of sleep spectrograms
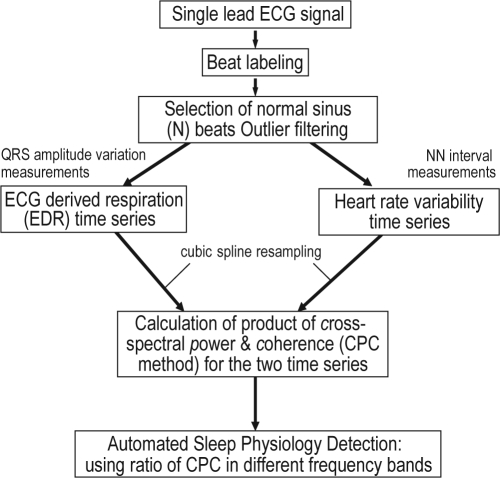
The cardiopulmonary coupling technique is based on a continuous electrocardiogram (ECG) signal and employs Fourier-based techniques to analyze 2 features of the signal: (1) the variability of the cardiac interbeat (RR) interval series and (2) the fluctuations in QRS amplitude induced by respiration--the ECG-derived respiration (EDR) signal. These signals have 2 basic patterns: a high frequency component due to physiological sinus arrhythmia that reflects breath-to-breath fluctuations, and a low frequency component that reflects cyclic variation across multiple breaths. Using the Fourier transform, the R-R interval time series and the associated EDR signals are first decomposed into a set of sinusoidal oscillations with specific amplitudes and phases at each frequency. Two factors are considered in evaluating the strength of the coupling between these 2 signals: (1) If, at a given frequency, both signals have relatively large oscillation amplitudes, then it is likely that these 2 signals are coupled with each other. This can be measured by computing the cross-spectral power, i.e., the product of the powers of the two individual signals at a given frequency. (2) If 2 oscillations at a given frequency are synchronized with each other (i.e., they maintain a constant phase relationship), this can be measured by computing the coherence of these signals. We use the product of the coherence and the cross-spectral power to weight these 2 effects in order to quantify the degree of the cardiopulmonary coupling.
The sequential steps in the derivation of the cardiopulmonary coupling measures are diagrammed in Supplementary Figures 1 and 2. Using a single lead ECG, an automated beat detection algorithm1,2 is used to detect beats and classify them as either normal or ectopic based on their morphology and timing. In addition, amplitude variations in the QRS complex due to shifts in the cardiac electrical axis relative to the electrodes during respiration and changes in thoracic impedance are determined. These fluctuations in the mean cardiac electrical axis (typically between 1 degree and 12 degrees peak-to-peak) correlate with phasic changes in the respiratory cycle. From these amplitude variations, a surrogate ECG derived respiratory signal (EDR) is obtained as previously described.3,4 A time series of normal-to-normal sinus (N-N) intervals and the time series of the EDR associated with these N-N intervals are then extracted from the original R-R interval time series. Outliers due to false or missed R-wave detections are removed using a sliding window average filter with a length of 41 data points, where central points lying outside 20% of the window average are rejected. Since Fourier analysis requires evenly sampled data, the resulting N-N interval series and its associated EDR signal are resampled at 2 Hz using cubic spline interpolation. At this sampling rate the Nyquist frequency allows detection of coupling frequencies up to 1 Hz. The cross-spectral power and coherence of these 2 signals are calculated over a 1024 sample (8.5 min) window using the fast Fourier transform applied to the 3 overlapping 512 sample subwindows within the 1024 coherence window. In each sub-window, the DC components and linear trends are removed and the data windowed using the Hanning (cosine) function before calculation of the Fourier transform. The 1024 coherence window is then advanced by 256 samples (2.1 min) and the calculation repeated until the entire N-N interval/EDR series are analyzed.
Supplementary Figure 1.
Sequential steps in the derivation of cardiopulmonary coupling measures.
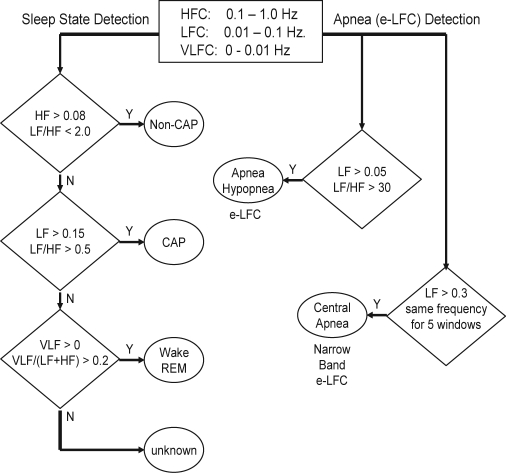
Supplementary Figure 2.
Sequential steps in the detection of sleep physiology from cardiopulmonary coupling measures.
HFC, High Frequency Coupling; LFC, Low Frequency Coupling; VLFC, Very Low Frequency Coupling; e-LFC, elevated-Low Frequency Coupling
For each 1024 window, the product of the coherence and cross-spectral power is used to generate a spectrogram of coupling powers at each frequency vs. time. This technique thus generates a moving average of the oscillatory frequencies of the coupling between heart rate and respiration. During sleep, a predominance of power in the low-frequency band is associated with periodic sleep behaviors and periodic respiration during SDB, while a predominance of power in the high-frequency band is associated with physiologic respiratory sinus arrhythmia and deep sleep with stable respiration. To quantify the low and high frequency coupling power distributions, in each 1024 window the coherence and cross power product is used in calculating the ratio of the sum of the 2 maximal coherent cross power peaks in the low-frequency (0.01-0.1 Hz) band to the sum of the 2 maximal peaks in the high-frequency (0.1-0.4 Hz) band.
Prior analysis of polysomnographic data using cardiopulmonary coupling spectrograms indicated that the low and high-frequency coupling regimes has only weak correlation with standard sleep staging but did follow cyclic alternating pattern (CAP) scoring, where low-frequency coupling is associated with CAP and high-frequency coupling with non-CAP. It was also determined that the ratio of the sum of the 2 maximal peaks in the very low frequency (0-0.01Hz) to the combined power of the 2 maximal peaks in each of the low- and high-frequency bands could be used to estimate wake/REM periods where a predominance of power in the very low-frequency band is associated with wake/REM periods. For each of the 3 sleep states of non-CAP, CAP, and combined wake/REM, separate receiver-operator curves were calculated over a range of power thresholds, and the thresholds giving the maximum combined sensitivities and specificities for that state were selected as optimal for the detection of that state. Using these thresholds, sleep demonstrating predominantly non-CAP, CAP, and wake/REM states could be identified.
Cardiopulmonary coupling analysis and estimation of elevated low frequency coupling (e-LFC) subtypes
Analysis of the PhysioNet Sleep Apnea Database5 using the cardiopulmonary coupling technique indicated that elevated power in the low frequency coupling region coincided with periods of scored apnea/hypopnea. Sensitivities and specificities for minute-by-minute apnea/hypopnea detection were calculated for a range of low frequency coupling powers and low/high coupling ratios. Receiver operator curves were then calculated and the thresholds giving the maximum combined sensitivity and specificity for apnea/hypopnea detection was selected as optimal. These detection thresholds required that the minimum low frequency power be greater than 0.05 normalized units and that the low to high frequency ratio be >30 to define periods of probable apnea/hypopnea, which we term elevated LFC (e-LFC). Thus, e-LFC is defined here as a subset of low frequency coupled cardiopulmonary oscillations, periods of which correlated significantly with periods of manually scored apneas and hypopneas in the PhysioNet Sleep Apnea Database.
Some spectrograms from the PhysioNet Sleep Apnea Database demonstrated periods of near-constant frequency spectral peaks in the e-LFC region that was reminiscent of the oscillations of heart rate variability seen in Cheyne-Stokes respiration in heart failure patients, which has a relatively constant cycle length. To explore this phenomenon further, we applied the algorithm to the PhysioNet Congestive Heart Failure Database,6 with the expectation that the database would provide more prolonged episodes with central periodic oscillations. Since the period of central apnea can be as slow as 120 seconds or longer we used the frequency band between 0.006 and 0.1 Hz to define narrow spectral band e-LFC (putative central sleep apnea, periodic breathing, or complex sleep apnea). We required (1) a minimum power in this band of 0.3 normalized units and (2) that the coupling frequency of each pair of consecutive measurements remains within 0.0059 Hz of each other over 5 consecutive sampling windows (totaling 16.9 continuous minutes). Periods of e-LFC not meeting these criteria were defined as broad spectral band e-LFC (putative obstructive sleep apnea). The amounts of broad and narrow spectral band coupling in e-LFC bands are again expressed as the percentage of windows detected in relation to the total sleep period. Thus, the narrow spectral band e-LFC identified periods with oscillations that have a single dominant coupling frequency, suggesting central sleep apnea or periodic breathing. The broad spectral band e-LFC identified periods with oscillations that have variable coupling frequencies, suggesting an alternate process, which we posited was dominance of anatomic upper airway obstructive processes. As it takes 16.9 min of continuous narrow-band cardiopulmonary coupling to reach the detection threshold, we estimated that this would be approximately equal to an averaged central apnea index of 5/h of sleep, assuming 6 h of sleep and a periodic breathing cycle length of approximately 35 sec. Thus the cardiopulmonary coupling technique can be used to detect apnea/hypopnea and differentiate these into obstructive vs. central.
Parametric variations of number of sampling windows required to designate as narrow spectral bands can be varied, the minimum being 3 consecutive measurements that remain within 0.00059 Hz of each other. This requires 12.7 min of continuous nearly identical in timing low frequency coupled HRV-respiratory oscillations. Central apneas and classic periodic breathing are rare in subjects selected for epidemiological studies: to better detect subtle chemoreflex influences, we relaxed the detection threshold to 3 consecutive measurements with the above characteristics.
REFERENCES
- 1.Moody GB, Mark RG. Comput Cardiol. 1982. Development and evaluation of a 2-lead ECG analysis program; pp. 39–44. [Google Scholar]
- 2.Mark RG, Moody GB. Arrhythmia analysis, automated. In: Webster JG, editor. Encyclopedia of medical devices and instrumentation. New York: Wiley,; 1988. pp. 120–30. [Google Scholar]
- 3.Moody GB, Mark RG, Zoccola A, Mantero S. Derivation of respiratory signals from multi-lead ECGs. Comput Cardiol. 1985;12:113–6. [Google Scholar]
- 4.Moody GB, Mark RG, Bump MA, et al. Clinical validation of the ECG-derived respiration (EDR) technique. Comput Cardiol. 1986;13:507–10. [Google Scholar]
- 5. http://www.physionet.org/physiobank/database/apnea-ecg.
- 6. http://www.physionet.org/physiobank/database/chfdb.
REFERENCES
- 1.Redline S, Tosteson T, Tishler PV, Carskadon MA, Millman RP. Studies in the genetics of obstructive sleep apnea. Familial aggregation of symptoms associated with sleep-related breathing disturbances. Am Rev Respir Dis. 1992;145:440–4. doi: 10.1164/ajrccm/145.2_Pt_1.440. [DOI] [PubMed] [Google Scholar]
- 2.Palmer LJ, Redline S. Genomic approaches to understanding obstructive sleep apnea. Respir Physiol Neurobiol. 2003;135:187–205. doi: 10.1016/s1569-9048(03)00044-2. [DOI] [PubMed] [Google Scholar]
- 3.Strohl KP, Saunders NA, Feldman NT, Hallett M. Obstructive sleep apnea in family members. N Engl J Med. 1978;299:969–73. doi: 10.1056/NEJM197811022991801. [DOI] [PubMed] [Google Scholar]
- 4.Cherniack NS, Longobardo GS. Cheyne-Stokes breathing. An instability in physiologic control. N Engl J Med. 1973;288:952–7. doi: 10.1056/NEJM197305032881810. [DOI] [PubMed] [Google Scholar]
- 5.Cherniack NS. Respiratory dysrhythmias during sleep. N Engl J Med. 1981;305:325–30. doi: 10.1056/NEJM198108063050606. [DOI] [PubMed] [Google Scholar]
- 6.Thomas RJ, Mietus JE, Peng CK, Goldberger AL. An electrocardiogram-based technique to assess cardiopulmonary coupling during sleep. Sleep. 2005;28:1151–61. doi: 10.1093/sleep/28.9.1151. [DOI] [PubMed] [Google Scholar]
- 7.Thomas RJ, Mietus JE, Peng CK, et al. Differentiating obstructive from central and complex sleep apnea using an automated electrocardiogram-based method. Sleep. 2007;30:1756–69. doi: 10.1093/sleep/30.12.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redline S, Tishler PV, Tosteson TD, et al. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med. 1995;151:682–7. doi: 10.1164/ajrccm/151.3_Pt_1.682. [DOI] [PubMed] [Google Scholar]
- 9.Quan SF, Howard BV, Iber C, et al. The Sleep Heart Health Study: design, rationale, and methods. Sleep. 1997;20:1077–85. [PubMed] [Google Scholar]
- 10.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer LJ, Buxbaum SG, Larkin EK, et al. Whole genome scan for obstructive sleep apnea and obesity in African-American families. Am J Respir Crit Care Med. 2004;169:1314–21. doi: 10.1164/rccm.200304-493OC. [DOI] [PubMed] [Google Scholar]
- 12.Guilleminault C, Partinen M, Hollman K, Powell N, Stoohs R. Familial aggregates in obstructive sleep apnea syndrome. Chest. 1995;107:1545–51. doi: 10.1378/chest.107.6.1545. [DOI] [PubMed] [Google Scholar]
- 13.Patel SR, Blackwell T, Redline S, et al. Osteoporotic Fractures in Men Research Group; Study of Osteoporotic Fractures Research Group. Shared genetic basis for obstructive sleep apnea and adiposity measures. Int J Obes (Lond) 2008;32:1825–34. doi: 10.1038/sj.ijo.0803803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redline S, Tosteson T, Tishler PV, Carskadon MA, Millman RP. Studies in the genetics of obstructive sleep apnea. Familial aggregation of symptoms associated with sleep-related breathing disturbances. Am Rev Respir Dis. 1992;145(2Pt 1):440–4. doi: 10.1164/ajrccm/145.2_Pt_1.440. [DOI] [PubMed] [Google Scholar]
- 15.Sundquist J, Li X, Friberg D, Hemminki K, Sundquist K. Obstructive sleep apnea syndrome in siblings: an 8-year Swedish follow-up study. Sleep. 2008;31:817–23. doi: 10.1093/sleep/31.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redline S, Young T. Epidemiology and natural history of obstructive sleep apnea. Ear Nose Throat J. 1993;72(20-1):24–6. [PubMed] [Google Scholar]
- 17.Terzano MG, Parrino L, Boselli M, Spaggiari MC, Di Giovanni G. Polysomnographic analysis of arousal responses in obstructive sleep apneas syndrome by means of the cyclic alternating pattern. J Clin Neurophysiol. 1996;13:145–55. doi: 10.1097/00004691-199603000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Younes M. Role of arousals in the pathogenesis of obstructive sleep apnea. Am J Respir Crit Care Med. 2004;169:623–33. doi: 10.1164/rccm.200307-1023OC. [DOI] [PubMed] [Google Scholar]