Abstract
Study Objectives:
To screen the PER3 promoter for polymorphisms and investigate the phenotypic associations of these polymorphisms with diurnal preference, delayed sleep phase disorder/syndrome (DSPD/DSPS), and their effects on reporter gene expression.
Design:
Interspecific comparison was used to define the approximate extent of the PER3 promoter as the region between the transcriptional start site and nucleotide position −874. This region was screened in DNA pools using PCR and direct sequencing, which was also used to screen DNA from individual participants. The different promoter alleles were cloned into a luciferase expression vector and a deletion library created. Promoter activation was measured by chemiluminescence.
Setting:
N/A
Patients or Participants:
DNA samples were obtained from volunteers with defined diurnal preference (3 × 80, selected from a pool of 1,590), and DSPD patients (n = 23).
Interventions:
N/A
Measurements and Results:
We verified three single nucleotide polymorphisms (G −320T, C −319A, G −294A), and found a novel variable number tandem repeat (VNTR) polymorphism (−318 1/2 VNTR). The −320T and −319A alleles occurred more frequently in DSPD compared to morning (P = 0.042 for each) or evening types (P = 0.006 and 0.033). The allele combination TA2G was more prevalent in DSPD compared to morning (P = 0.033) or evening types (P = 0.002). Luciferase expression driven by the TA2G combination was greater than for the more common GC2A (P < 0.05) and the rarer TA1G (P < 0.001) combinations. Deletion reporter constructs identified two enhancer regions (−703 to −605, and −283 to −80).
Conclusions:
Polymorphisms in the PER3 promoter could affect its expression, leading to potential differences in the observed functions of PER3.
Citation:
Archer SN; Carpen JD; Gibson M; Lim GH; Johnston JD; Skene DJ; von Schantz M. Polymorphism in the PER3 promoter associates with diurnal preference and delayed sleep phase disorder. SLEEP 2010;33(5):695-701.
Keywords: Circadian rhythms, clock genes, genetic polymorphisms, sleep disorders
TWO DISTINCTIVE BIOLOGICAL TIMING PROCESSES INTERACT IN THE CREATION OF A REST-ACTIVITY CYCLE - THE CIRCADIAN OSCILLATOR AND THE SLEEP homeostat.1 In mammals, the circadian oscillation is driven by a negative feedback loop involving the Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes, whose transcription is initiated early in the morning. The translated proteins form heteropolymers with each other and with casein kinase Iδ (CK1δ and ε (CK1ε),2 which provide the phosphorylation of specific sites that influence protein stability and nuclear translocation.3 In the nucleus, these heteropolymers inhibit the positive transcription factors (CLOCK/NPAS2 and BMAL1/2) that stimulate Per/Cry transcription, thereby closing the cycle. The genetic basis of the sleep homeostat is currently less well understood.4 However, many reports of genetic factors involved in sleep pertain to clock genes involved in the circadian oscillator, which may affect sleep either via its interaction with the sleep homeostat, or via pleiotropic action of clock genes. Thus, altered sleep homeostasis has been reported in knockout mice lacking both Cry1 and Cry25 or Npas2.6
In a few human pedigrees, altered free-running period is seen in mutations that cause the circadian rhythm sleep disorder ASPD/ASPS (advanced sleep phase disorder or syndrome). In affected individuals, the preferred sleep/wake cycle is advanced in relation to the external 24-h cycle by several hours.7 Mutations causing familial ASPD have been found in the PER28 and CSNK1D (CK1δ9) genes. Associations with extreme diurnal preference, which is a much more easily obtained correlate of intrinsic period and sleep timing,10,11 have been reported for polymorphisms in CLOCK,12 PER1,13 PER2,14 and PER3.15–17 In contrast to ASPD, delayed sleep phase disorder/syndrome (DSPD/DSPS) is not generally believed to necessarily reflect a longer intrinsic period (although this has been reported in one case18). Rather, it may be associated with an altered phase relationship between the oscillator and the sleep-wake cycle and/or abnormal entrainment, and possibly altered sleep homeostasis.19 Genetic studies have indicated polymorphisms in PER315,17,20 as risk factors for DSPD.
The human PER3 gene is substantially more polymorphic than the other PER paralogs21 and does not display signs of either positive or balancing selection.22 A series of experiments with double knockout mouse models in different combinations of the three Per paralogs showed that, unlike Per1 and Per2, Per3 is neither necessary nor sufficient to drive circadian rhythmicity in the absence of the other two.23 Four haplotypes composed of different combinations of five polymorphisms affecting the encoded protein sequence were first identified in a Japanese population.20 One of these haplotypes was defined by the missense polymorphism V647G, and was reported to associate with DSPD. Another polymorphism described but not further analyzed in this study was a coding-region variable number tandem repeat (VNTR) polymorphism, consisting of an 18-amino-acid motif repeated 4 or 5 times (PER34 and PER35). We have previously reported an association between the PER35 allele and extreme morning preference, and between the PER34 allele and extreme evening preference as well as DSPD.15 In a subsequent prospective study, PER35/5 homozygotes were found to have reduced sleep latency, and spend a substantially higher proportion of their sleep time in slow wave sleep (SWS).24 When kept awake during the subsequent night, they displayed a higher amount of theta activity, and a dramatic reduction in cognitive test performance.25 A more recent study has confirmed similar effects of the same polymorphism on the sleep deprived EEG.26 Thus, the previously reported association of the PER3 VNTR with diurnal preference may in fact be a reflection not so much of differences in circadian parameters, as of the increased amount of sleep pressure experienced by PER35/5 homozygotes.27 In other words, as supported by the double knockout mouse study,23 Per3 may have a more important role in sleep homeostatic mechanisms than in the circadian oscillator.
Given the importance of the coding-region polymorphisms in the human PER3 gene in influencing diurnal preference and as a risk factor for DSPD, we extended our studies into the promoter region, for which polymorphisms had not previously been reported. We discovered a novel cluster of polymorphisms in the PER3 promoter region, provided an improved definition of the promoter region itself, and investigated the effect of the polymorphism cluster on gene expression.
MATERIALS AND METHODS
Subjects
Horne-ÖOstberg (HÖO)28 diurnal preference scores and buccal DNA samples were obtained from a population of 1,590 volunteers aged between 18 and 81 years, as described previously.29,16 Genetic analyses were conducted on the 5% of subjects with the highest (extreme morning preference) and the lowest (extreme evening preference) HÖO scores, which were corrected for age but not for gender by regression analysis (n = 80 in each group). In addition, an equal number of individuals with an intermediate HÖO score close to the regression line were selected. A separate group of patients diagnosed with DSPD (n = 23)13 according to the version of the International Classification of Sleep Disorders that was current at the time of diagnosis30 were also investigated. HÖO scores and demographic data for all four groups are shown in Table 1. DNA from each subject was extracted as described previously.29,16 All studies were performed in accordance with the Declaration of Helsinki, and had received a favorable opinion from the University of Surrey Ethics Committee.
Table 1.
Circadian phenotype and demographic data for study participants
| Group | HÖ score (mean ± SD) | Gender ratio (female/male) | Age (mean ± SD) |
|---|---|---|---|
| Morning | 72 ± 7 | 44/36 | 37.1 ± 10.6 |
| Evening | 30 ± 6 | 44/36 | 41.6 ± 16.3 |
| Intermediate | 53 ± 4 | 38/42 | 41.6 ± 16.3 |
| DSPD | N/A | 11/12 | 27.9 ± 14.9 |
Polymorphism Detection
An approximate definition of the PER3 promoter region was obtained by identifying an upstream segment highly conserved between the human, rat and mouse genomes, according to the Ensembl genome browser. This suggested a region spanning between the predicted transcriptional start site and nucleotide position −874. Screening of the promoter region was performed in pools of DNA samples prepared from subjects with extreme diurnal preference, as well as from panels of pooled DNA samples from specific ethnic groups (Coriell Institute, Camden, NY), as previously described.14 The polymorphism cluster identified through this process was contained within the amplicon obtained using the primers CGCGGCGCCGGCTGCTGACC and TCACCCCTGCCCCAACTTTC and the following amplification parameters: 95°C for 4 min, followed by 35 cycles of 95°C for 1 min, 63°C for 1 min, and 72°C for 1 min. The PER3 promoter region is highly GC-rich and difficult to amplify by PCR. For amplification of this region, the Failsafe system (Epicentre Biotechnologies, Madison, WI) was used, identifying buffer premix E as optimal. Genotyping of all amplicons was conducted by direct sequencing (CEQ2000, Beckman Coulter, Fullerton, CA). The same conditions were used for genotyping of participant DNA samples. Some amplicons were cloned using the TA Cloning Kit (Promega, Madison, WI) prior to sequencing.
Promoter Expression Constructs
Fragments representing the four different observed promoter allelic combinations (PH1–PH4 in Figure 1) (–874 to +130 in the canonical sequence) were inserted into the pGL3-Enhancer vector (Promega). The existence of these four combinations on single chromosomes was confirmed by sequencing of TA-cloned PCR products amplified from individual participants. Vector constructs, including a control vector with no insert, were purified using the Pureyield Midiprep system (Promega), quantified (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, DE), and sequenced using vector-specific primers. Prior to transfection and expression, human embryonic kidney cells (HEK293) were seeded into 96-well plates (1 × 104 cells/well) and grown for 48 h in minimum essential medium (Eagle) containing 2 mM L-glutamine and Earle's BSS (basic sodium salts), 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acids, supplemented with 10% heat-inactivated horse serum, and 1 mM sodium pyruvate (all cell culture reagents from Sigma, St. Louis, MO). Then 48 ng of the expression constructs were transiently transfected into HEK293 cells using the FuGene 6 Transfection Reagent (Roche Applied Science, Basel, Switzerland). To normalize the data for transfection efficiency, 2 ng of a Renilla luciferase vector (pRL-SV40, Promega) was co-transfected into the same cells. All transfections were completed in triplicate and repeated in two or three separate experiments. The effects of the putative promoter inserts on the expression of the downstream luciferase gene were then quantified by measuring luciferase activity after 24 h using the Dual-Glo Luciferase Assay System (Promega) and a Lumicount Luminator (VICTOR3 Multilabel counter, PerkinElmer, Boston, MA).
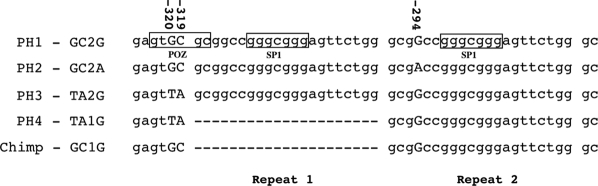
Figure 1.
Location of the three promoter SNPs genotyped in this study relative to the novel 1 or 2 repeat VNTR. The promoter allele combinations PH1 – PH4 are shown, together with the aligned sequence from the chimpanzee (release 2.1).
To further define the PER3 promoter region and identify potential enhancer or suppressor elements within it, the most frequently occurring allelic combination (PH1) was subjected to nested deletion mutagenesis (Erase-a-Base, Promega). The following nested deletions were generated with respect to the full-length fragment size: 833 (−703 to +130), 735 (−605 to +130), 413 (−283 to +130), and 210 (−80 to +130) (numbering in brackets indicates start and end positions with respect to the transcriptional start site). These deleted fragments were created within the pGL3-Enhancer vector, which was re-ligated, purified, and sequenced. These constructs were then used, together with the full-length promoter fragment and a control vector with no insert, in the same transfection reporter experiment described above. The full-length 1,004-bp approximate PER3 promoter region sequence was also analyzed for the existence of potential transcription factor binding sites (TFBS) (MatInspector, Genomatix, Munich, Germany). Only predicted TFBS with core and matrix sequence similarity values of at least 1.00 and 0.89, respectively, were considered (maximum = 1.00 for each).
A two-sided Fisher exact test (Prism 5.0a, GraphPad, La Jolla, CA) was used to determine statistical significance of differences in allele frequencies between subject groups. Bonferroni correction for multiple comparisons has been used but because there are significant levels of linkage disequilibrium across the PER3 gene,31 this is likely to be overly conservative for multiple polymorphism analysis.32 The two-sided Fisher exact test was also used to test for differences between groups in predicted haplotype frequencies. The χ2 statistic was used to test for deviation from Hardy-Weinberg equilibrium. One-way ANOVA (Bonferroni corrected) was used to determine significant differences between transfection results.
RESULTS
Promoter Region Polymorphisms
The presence of three promoter region single nucleotide plymorphisms (SNPs) G−320T (rs2797687), C−319A (rs2794664), and G−294A (rs228730) was verified in the DNA pools and subsequently in individual samples. During the sequencing of the amplicons, four individuals were identified whose DNA sequence could not be resolved beyond position −297, suggesting the heterozygous presence of an insertion or deletion polymorphism (indel). To resolve this, the amplicons from these individuals were cloned, and individual clones were sequenced. This revealed that the promoter region of PER3 contains a VNTR polymorphism (−318 1/2 VNTR), with one or two tandem copies of a 21-bp motif (see Figure, 1). These four individuals were heterozygous with alleles containing both one and two tandem units, while the remaining participants (n = 259) were all homozygous for two units. The genotype and allele frequencies for all the promoter polymorphisms in the different subject groups are shown in Table 2. Allele frequencies for the G−294A and the −318 1/2 VNTR polymorphisms did not differ significantly between any of the subject groups studied. The frequencies of the −320T and −319A alleles, however, were significantly higher in DSPD (T = 0.17, A = 0.17) than in extreme morning types (T = 0.07, A = 0.07) (P = 0.042) and extreme evening types (T = 0.04, A = 0.06) (P = 0.006 and 0.033, respectively). Only the difference between DSPD patients and evening types with respect to the −320T allele remained statistically significant after Bonferroni correction (corrected P = 0.024). The −320T and −319A allele frequencies were also significantly higher in the intermediate group (T = 0.12, A = 0.19) compared to the evening group (T = 0.04, A = 0.06; P = 0.023 and 0.001, respectively), the latter significant also after Bonferroni correction with P = 0.004. Intermediates also showed a higher −319A allele frequency (A = 0.19) compared to the morning group (A = 0.07; P = 0.001, P = 0.004 after Bonferroni correction). The higher −320T and −319A allele frequencies within the intermediate group were due to a larger number of heterozygotes for these two SNPs, compared to the other subject groups. When the diurnal preference groups were combined, the −320T allele (but not the −319A) remained significantly higher in the DSPD group (P = 0.046, OR = 2.521, 95% CI = 1.096 – 5.799). The −318 VNTR was in Hardy-Weinberg equilibrium within the subject groups, as was the G−294A within the intermediate, evening, and DSPD groups.
Table 2.
Genotype and allele frequencies in the extreme morning, intermediate, and extreme evening preference groups (n = 80 in each group), and DSPD patients (n = 23) for the four PER3 promoter region polymorphisms.
| Genotype frequencies |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G-320T |
C-319A |
-318 VNTR |
G-294A |
|||||||||
| GG | GT | TT | CC | CA | AA | 1/1 | 1/2 | 2/2 | GG | GA | AA | |
| Morning | 0.93 | 0.01 | 0.06 | 0.93 | 0.01 | 0.06 | 0 | 0.03 | 0.97 | 0.86 | 0.11 | 0.03 |
| Intermediate | 0.85 | 0.06 | 0.09 | 0.70 | 0.21 | 0.09 | 0 | 0 | 1.0 | 0.86 | 0.14 | 0 |
| Evening | 0.95 | 0.01 | 0.04 | 0.91 | 0.05 | 0.04 | 0 | 0.03 | 0.97 | 0.74 | 0.25 | 0.01 |
| DSPD | 0.83 | 0 | 0.17 | 0.83 | 0 | 0.17 | 0 | 0 | 1.0 | 0.83 | 0.17 | 0 |
| Allele frequencies | ||||||||
|---|---|---|---|---|---|---|---|---|
| G-320T |
C-319A |
-318 VNTR |
G-294A |
|||||
| G | T | C | A | 1 | 2 | G | A | |
| Morning | 0.93 | 0.07 | 0.93 | 0.07 | 0.01 | 0.99 | 0.92 | 0.08 |
| Intermediate | 0.88 | 0.12 | 0.81 | 0.19 | 0 | 1.0 | 0.93 | 0.07 |
| Evening | 0.96 | 0.04 | 0.94 | 0.06 | 0.01 | 0.99 | 0.86 | 0.14 |
| DSPD | 0.83 | 0.17 | 0.83 | 0.17 | 0 | 1.0 | 0.91 | 0.09 |
| P values, odds ratio, 95% confidence intervals |
||||
|---|---|---|---|---|
| M vs I | 0.001, 3.26, 1.57-6.74 | |||
| I vs E | 0.023, 2.95, 1.20-7.22 | 0.001, 3.61, 1.70-7.64 | ||
| M vs D | 0.042, 2.85, 1.07-7.58 | 0.042, 2.85, 1.07-7.58 | ||
| E vs D | 0.006, 4.60, 1.57-13.48 | 0.033, 3.16, 1.17-8.55 | ||
P values are given for statistically significant comparisons of allele frequencies between morning (M), intermediate (I), evening (E), and DSPD (D) groups, followed by odds ratios and 95% confidence intervals.
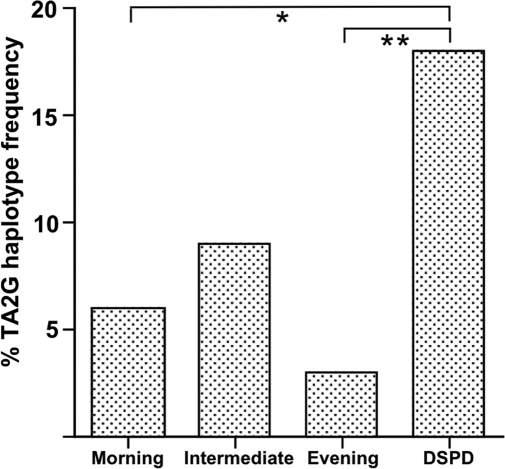
When the genotype frequencies were analyzed to predict allele combination frequencies (PHASE v2.0.233), four commonly occurring haplotypes (predicted frequency 1% or higher) were identified, all of which were observed directly through cloning and sequencing of amplified genomic DNA. These were: GC2G, GC2A, TA2G, and the rare TA1G haplotype, which were predicted to occur in the combined diurnal preference groups with frequencies of 79%, 10%, 6%, and 1%, respectively. The TA2G combination was predicted to occur more in the DSPD group (18%) compared to both morning (6%) and evening (3%) groups (P = 0.03, OR = 0.32, 95% CI = 0.12 – 0.86; P = 0.002, OR = 0.29, 95% CI = 0.10 – 0.81, respectively) (Figure 2). The latter remained significant after Bonferroni correction (P = 0.008).
Figure 2.
Predicted frequencies for the TA2G haplotype in the extreme morning, intermediate, and extreme evening preference groups, and DSPD patients. P = 0.033 (*), P = 0.002 (**).
Analysis of Promoter Activity
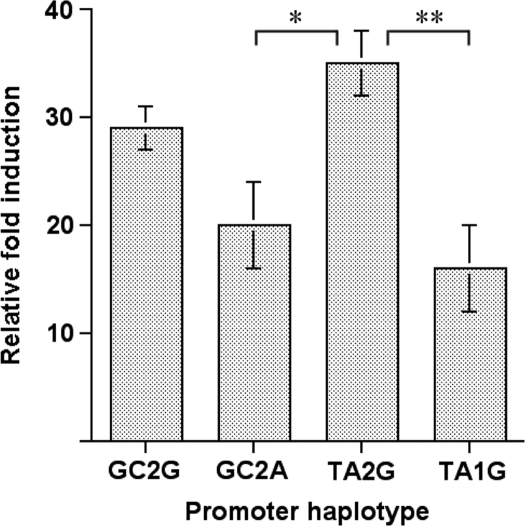
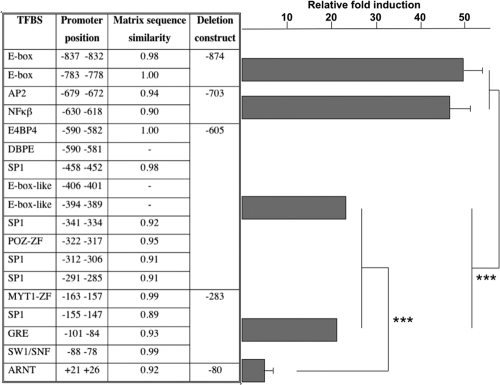
The relative induction levels of luciferase activity for the four allele combinations in the promoter region, compared to a control vector with no insert, are shown in Figure 3. The largest induction of gene expression was found with the TA2G combination, which was significantly greater than the GC2A (P < 0.05) and the TA1G (P < 0.01) combinations. With the serial deletion promoter constructs, significant differences were observed in the relative fold induction between the −874/−703 and the −605/−283 constructs (P < 0.0001), and also between the −605/−283 constructs and the −80 construct (P < 0.0001) (Figure 4).
Figure 3.
Relative induction of luciferase gene expression, compared to vector control with no insert, for the 4 different allelic combination constructs. N = 9 for each construct. P < 0.05 (*), P < 0.01 (**).
Figure 4.
The table part of the figure shows predicted transcription factor binding sites (TFBS), promoter position relative to the transcriptional start site, matrix sequence similarity scores (maximum = 1.00) in the different deletion constructs. To the right is shown relative induction of luciferase gene expression, compared to the vector control with no insert, for the 5 promoter serial deletion constructs. N = 6 for each construct. P < 0.0001 (***).
Transcription Factor Binding Sites
Analysis of the PER3 promoter region sequence predicted with a high degree of probability the presence of the following TFBS, in addition to others with lower probability values; Two E-box sites, a DBPE (D-element binding protein, or D-box) site, as well as binding sites for: ARNT (aryl hydrocarbon receptor nuclear translocator) heterodimer, NF-κβ (nuclear factor-κβ), E4BP4 (adenovirus E4 promoter ATF-site binding protein), PPAR (peroxisome proliferators-activated receptor), AP-2 (activator protein 2), POZ (poxvirus and zinc finger) domain, MYT1 (myelin transcription factor 1) zinc finger domain, Sp1 (specificity protein 1), GRE (glucocorticoid response element), and SW1/SNF complex (Figure 4).
DISCUSSION
We have discovered a polymorphic cluster in the promoter region of PER3, and identified an allelic combination that both associates with DSPD, and increases the efficiency of the PER3 promoter in a reporter gene assay.
The two SNPs in the promoter that associate with DSPD are immediately adjacent, and form part of a polymorphism cluster within the promoter region (Figure 1). Immediately following these two SNPs is a VNTR polymorphism with one or two copies of a 21-bp unit. The two-unit allele is by far the most common one, and contains within it the fourth polymorphism of the cluster (G−294A). This form is found in allelic combinations both with GC and with TA in the preceding SNPs, whereas we only found the much rarer one-unit VNTR allele combined with TA. Interestingly, the chimpanzee sequence in Genbank carries the other major human SNP combination, GC, followed by one unit of the VNTR (which one would a priori hypothesize to be the ancestral form). This may indicate either that the SNPs go back to the common ancestor of the two species, in which case the chimpanzee would be predicted to be polymorphic as well, or that the region is intrinsically unstable, so that the same SNPs have emerged at least twice.
Analysis of the genotype frequencies predicted four main allelic haplotype combinations, one of which, TA2G, was predicted to occur at a significantly higher frequency in DSPD patients compared to extreme morning and evening types. If one assumes that DSPD is a more extreme form of evening preference, then it is unclear why this haplotype was not predicted to occur more frequently within the extreme evening preference group. However, as discussed above, DSPD cannot necessarily be considered as one extreme of a diurnal preference spectrum. No significant differences were observed for TA2G haplotype frequency between the different diurnal preference groups. Specific information about ethnicity was neither available for the normal volunteers, nor for the DSPD patients. However, sampling of both took place either in the UK or in the Netherlands, and it is therefore unlikely that ethnic differences constitute a significant confounding factor.
We employed a reporter gene system both to investigate the effects of the four allelic combinations of the promoter polymorphisms found in a significant number of individuals, and to dissect the function of the different regions of the most common promoter sequence. A significant association with the T and A SNP alleles and DSPD was found only when combined with the 2-unit VNTR. This parallels the finding that the TA2G combination produced a level of promoter activation that was significantly higher than that of TA1G (Figure 3). The other two allelic combinations, GC2G and GC2A, gave intermediate levels of reporter gene expression.
The 1,004-bp PER3 upstream region was predicted to contain multiple cis-acting elements for transcription factor binding (Figure 4). Sp1 binding sites tend to occur in clusters in GC-rich regions, which is consistent with the finding of a cluster of five potential sites near the middle of the PER3 promoter fragment. These sites were in close proximity to the promoter polymorphisms described above. Each VNTR repeat unit contained a putative binding site for Sp1, which is implicated in the repression of mouse Per1 transcription34 and may explain the reduced level of expression driven by the TA1G construct. Although it does not affect the predicted Sp1 sites, the G−320T and C−319A SNP duplex is positioned in the middle of the predicted POZ domain, which is disrupted in the TA alleles. Transcriptional repression by binding to the POZ domain is known to be mediated by interfering with the DNA binding activity of Sp1 in promoters,35 a process that is also methylation-state sensitive.36
In the nested deletions of the approximate PER3 promoter, the two largest fragments (−874 and −703) produced the highest level of expression, but with little difference in expression between them (Figure 4). The only cis-acting elements predicted to be missing in the shorter fragment were two canonical E-box sites. This is an interesting observation, as E-boxes (although an almost universal feature of clock gene promoters) are missing from the mouse Per3 promoter,37 although their presence in the human promoter did not appear to influence the amplitude of basal expression significantly.
The removal of 98 bp between the −703 and the −605 fragment had a significant effect on levels of reporter expression. This sequence contains an NF-κβ binding site. NF-κβ is a gene expression enhancer that has been shown to be active in the hamster suprachiasmatic nucleus (SCN), where it is involved with circadian regulation and entrainment.38 It also shows increased activity in the hypothalamus and cortex of sleep-deprived mice,39,40 and its accumulation is associated with sleep onset and the activation of sleep-promoting substances.41 This is an interesting observation given the link between PER3 and sleep homeostasis in humans.24
The reduction from the −605 to the −283 fragment removed a large section of sequence that contains the promoter polymorphisms, the POZ domain, the E4BP4 site, and four Sp1 sites. The E4BP4 site also overlaps a DBPE site, which is conserved between humans and mice and has previously been proposed to drive mPer3 expression.37 E4BP4 acts as a negative regulator of mouse Per2 and is required, together with a non-canonical E-box, for robust circadian expression of Per2.42,43 The presence of two closely spaced non-canonical E-box elements within the promoters of human PER1-3 has been reported to be necessary for sustaining circadian rhythms in culture.44 The absence of a significant difference in expression between the −605 and −283 fragments may be explained by the simultaneous deletion of multiple enhancer and repressor elements, or because they control rhythmic, as opposed to basal, promoter activity. It is also possible that a polymorphism in this region has a stronger effect on promoter activation than its absence.
The smallest fragment, with the lowest level of transactivation, additionally lacks the potential glucocorticoid response element (GRE), a binding site for glucocorticoid receptors, which appear to be expressed in most tissues except the SCN.45 Glucocorticoids activate these receptors, which then bind promoter GREs and can either induce or repress gene expression.46 The synthetic glucocorticoid dexamethasone has been shown to induce robust expression of mouse Per gene expression in culture, including Per3.47 Thus, it seems likely that the presence of a GRE in the PER3 promoter may serve to entrain its expression in peripheral oscillators.
Thus, we have discovered a novel polymorphic hotspot in the promoter region, and provided evidence suggesting that it may be important both for gene expression levels and for determining phenotype within the normal and pathological range of diurnal preference. Predicted transcription factor binding sites are putative and will need to be verified experimentally. Future work will also be required to determine the haplotypic combinations between this polymorphic promoter cluster and the previously described polymorphisms affecting PER3 protein structure, and how these combine to affect phenotype. This will be important in understanding more fully the phenotypic associations that we have previously described for the PER3 VNTR polymorphism.48
DISCLOSURE STATEMENT
This was not an industry supported study. Dr. Skene has received research support from Philips Lighting. The other authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
This work was funded by an MRC quota studentship (to JDC), and grants from the Wellcome Trust (074293), and the EU 6th Framework project (EUCLOCK, 018471). We thank Marcel Smits and Adrian Williams for providing DSPD patient DNA samples.
REFERENCES
Note: gene names should be in italics, but may not appear so in original title
- 1.Borbely AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1:195–204. [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Walton KM, Fisher K, Rubitski D, et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330:430–9. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 4.Franken P, Dijk DJ. Circadian clock genes and sleep homeostasis. Eur J Neurosci. 2009;29:1820–9. doi: 10.1111/j.1460-9568.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- 5.Wisor JP, O'Hara BF, Terao A, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franken P, Dudley CA, Estill SJ, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006;103:7118–23. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones CR, Campbell SS, Zone SE, et al. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat Med. 1999;5:1062–5. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- 8.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Padiath QS, Shapiro RE, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–4. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 10.Duffy JF, Dijk DJ, Hall EF, Czeisler CA. Relationship of endogenous circadian melatonin and temperature rhythms to self-reported preference for morning or evening activity in young and older people. J Investig Med. 1999;47:141–50. [PMC free article] [PubMed] [Google Scholar]
- 11.Duffy JF, Rimmer DW, Czeisler CA. Association of intrinsic circadian period with morningness-eveningness, usual wake time, and circadian phase. Behav Neurosci. 2001;115:895–9. doi: 10.1037//0735-7044.115.4.895. [DOI] [PubMed] [Google Scholar]
- 12.Mishima K, Tozawa T, Satoh K, Saitoh H, Mishima Y. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med Genet B Neuropsychiatr Genet. 2005;133:101–4. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- 13.Carpen JD, von Schantz M, Smits M, Skene DJ, Archer SN. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet. 2006;51:1122–5. doi: 10.1007/s10038-006-0060-y. [DOI] [PubMed] [Google Scholar]
- 14.Carpen JD, Archer SN, Skene DJ, Smits M, von Schantz M. A single-nucleotide polymorphism in the 5'-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–7. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 15.Archer SN, Robilliard D, Skene DJ, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–5. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 16.Jones KH, Ellis J, von Schantz M, Skene DJ, Dijk DJ, Archer SN. Age-related change in the association between a polymorphism in the PER3 gene and preferred timing of sleep and waking activities. J Sleep Res. 2007;16:12–6. doi: 10.1111/j.1365-2869.2007.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira DS, Tufik S, Louzada FM, et al. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- 18.Campbell SS, Murphy PJ. Delayed sleep phase disorder in temporal isolation. Sleep. 2007;30:1225–8. doi: 10.1093/sleep/30.9.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okawa M, Uchiyama M. Circadian rhythm sleep disorders: characteristics and entrainment pathology in delayed sleep phase and non-24-h sleep-wake syndrome. Sleep Med Rev. 2007;11:485–96. doi: 10.1016/j.smrv.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Ebisawa T, Uchiyama M, Kajimura N, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–6. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Schantz M. Phenotypic effects of genetic variability in human clock genes on circadian and sleep parameters. J Genet. 2008;87:513–9. doi: 10.1007/s12041-008-0074-7. [DOI] [PubMed] [Google Scholar]
- 22.Nadkarni NA, Weale ME, von Schantz M, Thomas MG. Evolution of a length polymorphism in the human PER3 gene, a component of the circadian system. J Biol Rhythms. 2005;20:490–9. doi: 10.1177/0748730405281332. [DOI] [PubMed] [Google Scholar]
- 23.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–36. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 24.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–8. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 25.Groeger JA, Viola AU, Lo JC, von Schantz M, Archer SN, Dijk DJ. Early morning executive functioning during sleep deprivation is compromised by a PERIOD3 polymorphism. Sleep. 2008;31:1159–67. [PMC free article] [PubMed] [Google Scholar]
- 26.Goel N, Banks S, Mignot E, Dinges DF. PER3 polymorphism predicts cumulative sleep homeostatic but not neurobehavioral changes to chronic partial sleep deprivation. PLoS One. 2009;4:e5874. doi: 10.1371/journal.pone.0005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis J, von Schantz M, Jones KH, Archer SN. Association between specific diurnal preference questionnaire items and PER3 VNTR genotype. Chronobiol Int. 2009;26:464–73. doi: 10.1080/07420520902820970. [DOI] [PubMed] [Google Scholar]
- 28.Horne JA, ÖOstberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- 29.Robilliard D, Archer SN, Arendt J, et al. The 3111Clock gene polymorphism is not associated with sleep and circadian rhythmicity in phenotypically characterized human subjects. J Sleep Res. 2002;11:305–12. doi: 10.1046/j.1365-2869.2002.00320.x. [DOI] [PubMed] [Google Scholar]
- 30.The international classification of sleep disorders: diagnostic and coding manual. 2nd ed. Westchester, IL: American Academy of Sleep Medicine; 2005. [Google Scholar]
- 31.Nievergelt CM, Kripke DF, Barrett TB, et al. Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141:234–41. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–9. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 33.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Song X, Ma Y, Liu J, Yang D, Yan B. DNA binding, but not interaction with Bmal1, is responsible for DEC1-mediated transcription regulation of the circadian gene mPer1. Biochem J. 2004;382:895–904. doi: 10.1042/BJ20040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DK, Suh D, Edenberg HJ, Hur MW. POZ domain transcription factor, FBI-1, represses transcription of ADH5/FDH by interacting with the zinc finger and interfering with DNA binding activity of Sp1. J Biol Chem. 2002;277:26761–8. doi: 10.1074/jbc.M202078200. [DOI] [PubMed] [Google Scholar]
- 36.Inoue S, Oishi M. Effects of methylation of non-CpG sequence in the promoter region on the expression of human synaptotagmin XI (syt11) Gene. 2005;348:123–34. doi: 10.1016/j.gene.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marpegan L, Bekinschtein TA, Freudenthal R, et al. Participation of transcription factors from the Rel/NF-kappa B family in the circadian system in hamsters. Neurosci Lett. 2004;358:9–12. doi: 10.1016/j.neulet.2003.12.112. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Gardi J, Kushikata T, Fang J, Krueger JM. Nuclear factor-kappaB-like activity increases in murine cerebral cortex after sleep deprivation. Am J Physiol. 1999;276:R1812–8. doi: 10.1152/ajpregu.1999.276.6.R1812. [DOI] [PubMed] [Google Scholar]
- 40.Brandt JA, Churchill L, Rehman A, et al. Sleep deprivation increases the activation of nuclear factor kappa B in lateral hypothalamic cells. Brain Res. 2004;1004:91–7. doi: 10.1016/j.brainres.2003.11.079. [DOI] [PubMed] [Google Scholar]
- 41.Schulze G. Sleep protects excitatory cortical circuits against oxidative damage. Med Hypotheses. 2004;63:203–7. doi: 10.1016/j.mehy.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 42.Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007;35:648–55. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohno T, Onishi Y, Ishida N. The negative transcription factor E4BP4 is associated with circadian clock protein PERIOD2. Biochem Biophys Res Commun. 2007;354:1010–5. doi: 10.1016/j.bbrc.2007.01.084. [DOI] [PubMed] [Google Scholar]
- 44.Nakahata Y, Yoshida M, Takano A, et al. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol Biol. 2008;9:1. doi: 10.1186/1471-2199-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balsalobre A, Brown SA, Marcacci L, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 46.So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci U S A. 2008;105:5745–9. doi: 10.1073/pnas.0801551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–36. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dijk DJ, Archer SN. PERIOD3, circadian phenotypes, and sleep homeostasis. Sleep Med Rev. 2009 doi: 10.1016/j.smrv.2009.07.002. epub PMID 19716732. [DOI] [PubMed] [Google Scholar]