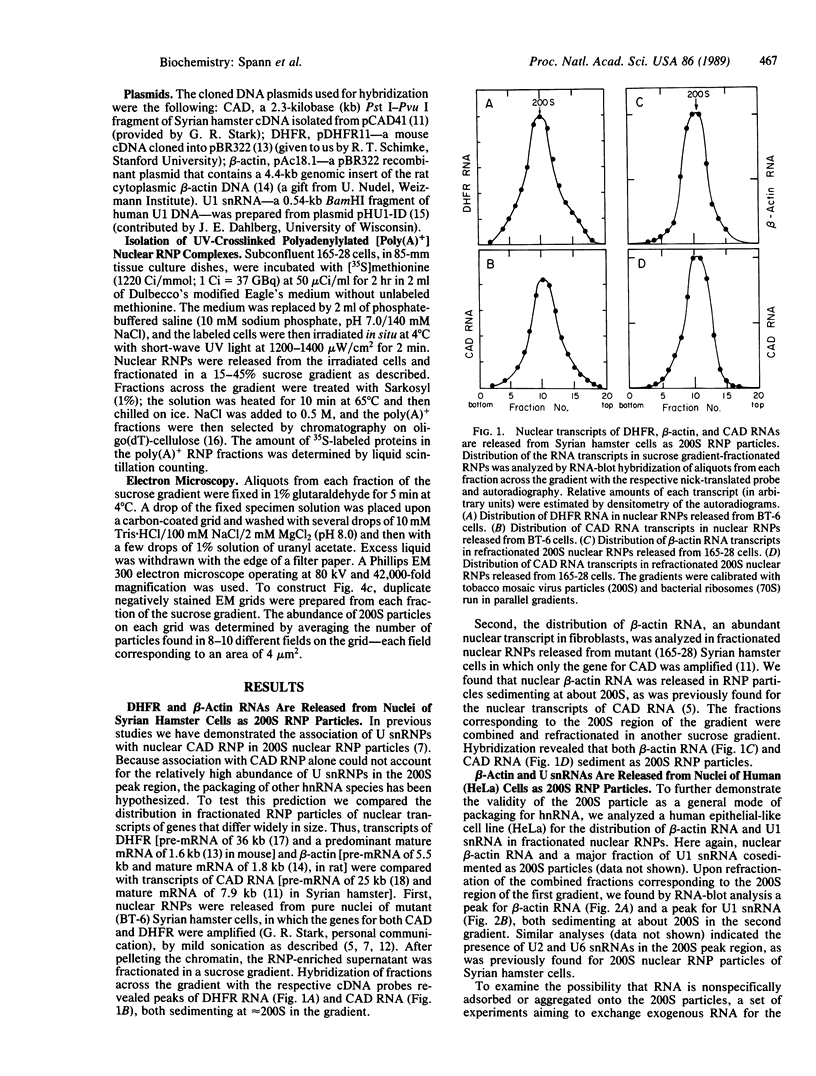
Abstract
We have previously shown that nuclear transcripts of the multifunctional enzyme, carbamoyl-phosphate synthetase, aspartate transcarbamylase, dihydroorotase RNA can be released from nuclei of Syrian hamster cells as compact ribonucleoprotein (RNP) particles that sediment at the 200S region in a sucrose gradient. The 200S nuclear RNP particles contain U1, U2, and U6 small nuclear RNPs, which are known to be required for splicing of pre-mRNA, as integral components of the particles. In this study we demonstrate that nuclear transcripts of dihydrofolate reductase in Syrian hamster cells and of beta-actin in both Syrian hamster and human cells are also released from the respective nuclei as 200S particles--despite the difference in length of these RNAs. Electron microscopy of the 200S particles revealed discrete compact composite structures with a cross section of approximately 50 nm. Finding that two more nuclear RNAs from two different cell types and two different species are released as 200S RNP particles suggests a general mode for packaging of heterogeneous nuclear RNA in large compact RNP particles the size of which is independent of the RNA length.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson K., Björkroth B., Daneholt B. The in situ structure of the active 75 S RNA genes in Balbiani rings of Chironomus tentans. Exp Cell Res. 1980 Dec;130(2):313–326. doi: 10.1016/0014-4827(80)90008-7. [DOI] [PubMed] [Google Scholar]
- Arad-Dann H., Isenberg D. A., Shoenfeld Y., Offen D., Sperling J., Sperling R. Autoantibodies against a specific nuclear RNP protein in sera of patients with autoimmune rheumatic diseases associated with myositis. J Immunol. 1987 Apr 15;138(8):2463–2468. [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Simonsen C. C., McEwan R. N., Schimke R. T. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982 Jul 10;257(13):7887–7897. [PubMed] [Google Scholar]
- Dreyfuss G., Choi Y. D., Adam S. A. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984 Jun;4(6):1104–1114. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu Rev Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- Economidis I. V., Pederson T. Structure of nuclear ribonucleoprotein: heterogeneous nuclear RNA is complexed with a major sextet of proteins in vivo. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1599–1602. doi: 10.1073/pnas.80.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Havron A., Sperling J. Specificity of photochemical cross-linking in protein-nucleic acid complexes: identification of the interacting residues in RNase- pyrimidine nucleotide complex. Biochemistry. 1977 Dec 13;16(25):5631–5635. doi: 10.1021/bi00644a038. [DOI] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. True genes for human U1 small nuclear RNA. Copy number, polymorphism, and methylation. J Biol Chem. 1984 Feb 10;259(3):2013–2021. [PubMed] [Google Scholar]
- Mayrand S., Pederson T. Nuclear ribonucleoprotein particles probed in living cells. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2208–2212. doi: 10.1073/pnas.78.4.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O. L., Jr, Bakken A. H. Morphological studies of transcription. Acta Endocrinol Suppl (Copenh) 1972;168:155–177. doi: 10.1530/acta.0.071s155. [DOI] [PubMed] [Google Scholar]
- Nudel U., Zakut R., Shani M., Neuman S., Levy Z., Yaffe D. The nucleotide sequence of the rat cytoplasmic beta-actin gene. Nucleic Acids Res. 1983 Mar 25;11(6):1759–1771. doi: 10.1093/nar/11.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Wahl G. M., Stark G. R. Structure of the gene for CAD, the multifunctional protein that initiates UMP synthesis in Syrian hamster cells. Mol Cell Biol. 1982 Mar;2(3):293–301. doi: 10.1128/mcb.2.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setyono B., Greenberg J. R. Proteins associated with poly(A) and other regions of mRNA and hnRNA molecules as investigated by crosslinking. Cell. 1981 Jun;24(3):775–783. doi: 10.1016/0092-8674(81)90103-3. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., McGrogan M., Nunberg J. H., Schimke R. T. Size heterogeneity in the 3' end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980 Nov;22(2 Pt 2):361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- Skoglund U., Andersson K., Björkroth B., Lamb M. M., Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983 Oct;34(3):847–855. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- Skoglund U., Andersson K., Strandberg B., Daneholt B. Three-dimensional structure of a specific pre-messenger RNP particle established by electron microscope tomography. Nature. 1986 Feb 13;319(6054):560–564. doi: 10.1038/319560a0. [DOI] [PubMed] [Google Scholar]
- Sperling J., Havron A. Photochemical cross-linking of neighboring residues in protein-nucleic acid complexes: rnase and pyrimidine nucleotide inhibitors. Biochemistry. 1976 Apr 6;15(7):1489–1495. doi: 10.1021/bi00652a020. [DOI] [PubMed] [Google Scholar]
- Sperling J., Sperling R. Photochemical cross-linking of histones to DNA nucleosomes. Nucleic Acids Res. 1978 Aug;5(8):2755–2773. doi: 10.1093/nar/5.8.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R., Spann P., Offen D., Sperling J. U1, U2, and U6 small nuclear ribonucleoproteins (snRNPs) are associated with large nuclear RNP particles containing transcripts of an amplified gene in vivo. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6721–6725. doi: 10.1073/pnas.83.18.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R., Sperling J., Levine A. D., Spann P., Stark G. R., Kornberg R. D. Abundant nuclear ribonucleoprotein form of CAD RNA. Mol Cell Biol. 1985 Mar;5(3):569–575. doi: 10.1128/mcb.5.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- van Eekelen C. A., van Venrooij W. J. hnRNA and its attachment to a nuclear protein matrix. J Cell Biol. 1981 Mar;88(3):554–563. doi: 10.1083/jcb.88.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]




