Abstract
Quiescent cells and tumor cells share the ability to evade irreversible cell fates. It was recently shown that the transcriptional regulator HES1 protects quiescent fibroblasts from differentiation or senescence. HES1 is highly expressed in rhabdomyosarcomas, and inhibition of HES1 restores differentiation in rhabdomyosarcoma cells. Pathways that lead to elevated HES1 levels, notch and hedgehog, are frequently upregulated in tumors. Compounds that inhibit these pathways induce differentiation and apoptosis in cancer cells and several are in clinical trials. HES1 might repress gene expression in part by recruiting histone deacetylases (HDACs). HDACs inhibit differentiation, whereas histone deacetylase inhibitors (HDACi’s) induce differentiation or apoptosis in tumors and are also showing promise as therapeutics. Small molecules that directly target HES1 itself were just identified. Thus, as we shall review in this article, therapies that target these pathways could be effective alone, in combination or in conjunction with traditional chemotherapeutics.
Introduction
From one perspective, quiescent cells, which include fibroblasts, lymphocytes, hepatocytes, stem cells and germ cells, are unarguably distinct from cancer cells. While quiescent cells respond to anti-proliferative signals from the environment by arresting their cell cycle, cancer cells fail to respond to such cues and continue to proliferate unabated [1]. From another perspective, however, cancer cells and quiescent cells actually share some similarities. Quiescent cells retain the capacity to re-enter the cell cycle upon receiving the appropriate cues, and therefore must ensure that they do not commit to typically irreversible pathways such as senescence, differentiation or apoptosis. Similarly, a subset of cells within a tumor can also remain in a non-dividing state of tumor dormancy. These cells, which may represent cancer stem cells, have been reported to exist in a quiescent state and thus to be mostly resistant to traditional chemotherapeutic agents, which are largely designed to kill proliferating cells [2, 3]. During dormancy, cancer cells endure low oxygen, acidic pH and nutrient deficiencies inside a tumor [4, 5]. Then, for reasons that remain unclear, these cells can become activated, proliferate and form a secondary tumor. For many tumor types, the presence of cells that might represent dormant tumor cells is closely associated with subsequent metastatic relapse [6]. Thus, an ability to survive in a reversible, out-of-cycle state is central to both quiescence and cancer.
Growing evidence has suggested that quiescence, instead of being a passive default state, is actively maintained by molecular mechanisms [7, 8]. Using DNA microarrays, researchers have identified molecular signatures of quiescence in hematopoietic stem cells [9], lymphocytes [8] and fibroblasts [10]. These studies have revealed that quiescence is associated with both downregulation and upregulation of a large number of transcripts. Gene expression changes have also been monitored in human diploid fibroblasts that enter quiescence in response to one of three independent signals – loss of adhesion, contact inhibition and mitogen withdrawal [11]. With each of these antiproliferative signals, there is a major reprogramming of gene expression. Among the gene expression changes that occur are some that are likely to enforce the nondividing state, for instance, regulation of the molecules involved in cell division itself. Other gene expression changes might ensure the reversibility of quiescence, for instance, by protecting the cells from damage induced by free radicals [11]. Yet other changes suggest pathways that quiescent cells utilize to protect themselves against senescence or differentiation. It has been hypothesized that these same pathways might be ‘co-opted’ by tumor cells to allow them to maintain their proliferative potential and avoid terminal cell fates [12].
The HES1 transcriptional repressor is one of the genes that might protect quiescent cells from a differentiated fate. Some tumor cells also rely on HES1 for protection against differentiation. We consider below several pathways that activate HES1 – the notch and hedgehog pathways – and an effector pathway of HES1 – histone deacetylases (HDACs). Small-molecule regulators of each of these pathways have shown promise as anti-cancer drugs and are being developed in clinical trials as summarized in Table 1. We will show how these compounds, individually and in combination, represent promising avenues for the treatment of multiple tumor types.
Table 1.
A selected subset of current clinical trials of inhibitors of the notch pathway, the hedgehog pathway and histone deacetylases
| Pathway | Target/Selectivity | Compound Name | Single agent/Combination | Disease | Trial Stage | NCT Number | Company/Institute |
|---|---|---|---|---|---|---|---|
| Notch | Gamma secretase | MK-0752 | Single agent | Pediatric Patients With Recurrent or Refractory CNS Malignancies | Phase I | 00923208 | Merck/NCI |
| Single agent | Metastatic or Locally Advanced Breast Cancer & Other Solid Tumors | Phase I | 00106145 | Merck | |||
| Followed by Docetaxel | Locally Advanced or Metastatic Breast Cancer | Phase I/II | 00645333 | Merck/U. Michigan Cancer Center | |||
| PF-03084014 | Single agent | Advanced Solid Tumor Malignancy And T-Cell Acute Lymphoblastic Leukemia/Lymphoblastic Lymphoma | Phase I | 00878189 | Pfizer | ||
| Hedgehog | Smoothened | IPI-926 | Single agent | Advanced and/or Metastatic Solid Tumor Malignancies | Phase I | 00761696 | Infinity Pharmaceuticals |
| Patched/Smoothened | GDC-0449 | Single agent | Patients With Ovarian Cancer in a Second or Third Complete Remission | Phase II | 00739661 | Genentech | |
| With concurrent chemotherapy & Bevacizumab | Metastatic Colorectal Cancer | Phase II | 00636610 | Genentech | |||
| Single agent | Recurrent or Refractory Medulloblastoma | Phase II | 00939484 | Genentech | |||
| Single agent | Recurrent Glioblastoma Multiforme | Phase II | 00980343 | Genentech/NCI | |||
| HDAC | class I and IIa HDAC | Valproic Acid(VPA) | With Fludarabine | Chronic Lymphocytic Leukemia | Phase II | 00524667 | CancerCare Manitoba/The Leukemia & Lymphoma Society |
| Pan HDAC | SAHA | With Tamoxifen | Advanced Breast Cancer Who Have Failed Prior Anti-hormonal Therapy | Phase II | 00365599 | Merck/H. Lee Moffitt Cancer Center& Research Institute | |
| With Paclitaxel& Carboplatin | Recurrent Ovarian Cancer | Phase II | 00772798 | Merck/Herlev Hospital/Odense University Hospital | |||
| With Bortezomib | Recurrent Glioblastoma Multiforme | Phase II | 00641706 | Merck/NCI | |||
| With Bortezomib | Diffuse Large B-Cell Lymphomas | Phase II | 00703664 | Merck/H. Lee Moffitt Cancer Center& Research Institute/NCI | |||
| Pan HDAC | PXD-101 (Belinostat) | Single agent | Resistant Epithelial Ovarian Tumors & Micropapillary/Borderline(LMP) Ovarian Tumors | Phase II | 00301756 | TopoTarget/Princess Margaret Hospital/Canada National Cancer Institute | |
| Single agent | Relapsed and Refractory Aggressive B-Cell Lymphomas | Phase II | 00303953 | TopoTarget/Southwest Oncology Group/National Cancer Institute |
HES1 protects quiescent cells and tumors from differentiation
To test whether quiescent cells actively resist a commitment to differentiation, the master regulator of muscle differentiation, the MyoD transcription factor, was introduced into fibroblasts and the induction of muscle genes was monitored [11]. Quiescent cells were less likely to induce muscle differentiation in response to MyoD in comparison with proliferating cells, indicating that quiescent cells have active mechanisms to resist a commitment to differentiation.
To further investigate the resistance to differentiation in quiescent cells, the basic helix-loop-helix transcriptional regulator HES1 was selected for detailed study [12, 13] because it was one of a few genes upregulated rapidly by each of three different quiescence signals. Furthermore, HES1 is expressed by almost all undifferentiated cells and is important for suppressing differentiation [14]. HES1-deficient mice show premature differentiation and severe defects in the brain, eye and pancreas [14, 15]. HES1 is expressed in neural precursor cells and expression disappears when cells initiate differentiation. However, in some situations, HES1 can also promote differentiation. For instance, in lymphocytes, HES1 is essential for expansion of T cell precursors, but also promotes differentiation in that it facilitates commitment to the CD8 lineage by repressing expression of the CD4 coreceptor. HES1 can also promote terminal neuronal and beta cell differentiation by silencing the repressor element silencer transcription factor REST [16]. For these reasons, HES1 was hypothesized to inhibit differentiation in quiescent fibroblasts.
Introduction of HES1 into proliferating fibroblasts made them resistant to a muscle differentiation program, whereas inhibition of HES1 in quiescent cells made them more responsive to differentiation-inducing transcription factors [12]. Thus, HES1 is important for preventing quiescent fibroblasts from undergoing differentiation [13]. Future experiments defining the genes regulated by HES1 in the context of fibroblast quiescence using chromatin immunoprecipitation followed by microarrays or next generation sequencing (ChIP-chip or ChIP-Seq) would further delineate HES1’s function in this context.
Many tumors are characterized by a relatively undifferentiated morphology, and induction of differentiation in these tumors might reduce their proliferative potential. The question was posed whether HES1 might be used by tumors to protect against differentiation. The levels of HES1 in rhabdomyosarcomas, tumors of skeletal muscle, were examined revealing that, strikingly, every cell line and patient sample exhibited elevated levels of HES1, with most between 5- and 50-fold higher than control skeletal muscle [12]. The elevated levels of HES1 were likely important for inhibiting differentiation, as expression of a dominant-negative form of HES1 forced rhabdomyosarcoma cell lines to cease proliferating and to undergo myogenic differentiation [12]. It was concluded that tumors and quiescent cells might share similar strategies for preventing differentiation. Furthermore, induction of HES1 in both quiescent cells and tumors is important for inhibiting their differentiation. High HES1 levels could reflect the stem-cell-like character of tumor cells, which may be derived from adult stem cells [17]. [17]. Mutations that result in elevated HES1 may provide a selective advantage to the particular cell, rendering it less responsive to signals for differentiation and hence predisposed to tumorigenesis.
Mechanisms that lead to HES1 induction and that allow HES1 to protect against differentiation could represent promising targets for tumor therapy (Figure 2). Targeting these pathways might coax tumors into a terminally differentiated state. Alternatively, because the tumors cells receive conflicting signals—on the one hand, to proliferate as a result of their genetic makeup, and on the other hand, to differentiate in response to the treatment—the tumors cells might apoptose. This would be similar to instances in which overexpression of an oncogene in the absence of growth factors results in apoptosis, which is interpreted as a response to conflicting proliferative signals [18]. We discuss two pathways that activate HES1, notch and hedgehog, below.
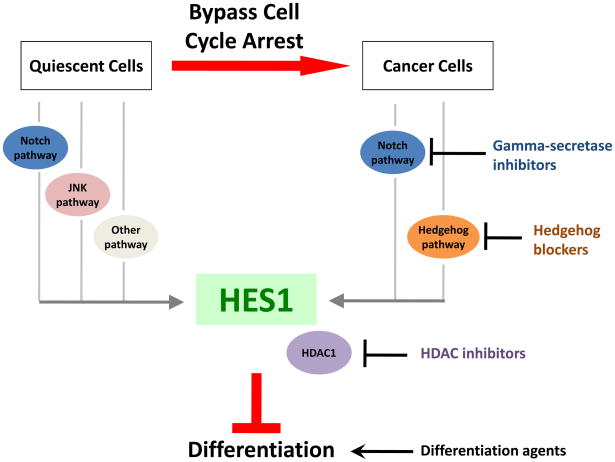
Figure 2. Role of HES1 in quiescence and cancer.
HES1 usually functions to inhibit differentiation in normal cells and tumors. HES1 is induced in quiescent cells, possibly through signaling from the notch pathway, the JNK pathway or other pathways. In cancer cells, HES1 upregulation likely occurs via the notch pathway or the hedgehog pathway. Small molecule inhibitors of the notch and hedgehog pathways have shown promise as anti-cancer agents possibly in part due to their effects on HES1. HES1 functions as a transcriptional repressor by interacting with histone deacetylases including HDAC1. HDACi’s have many cellular effects, including inhibition of HDAC1 activity and might thereby repress the effects of HES1. Agents that directly induce differentiation can also be effective as anti-cancer agents. Further, combinations of these agents can often be even more effective than the agents individually, possibly due to regulation of different sets of targets or a stronger regulation of key target genes.
The notch signaling pathway
The notch signaling pathway, the canonical pathway for the induction of HES1, facilitates the determination of cell fate during development and regulates differentiation in somatic stem cell populations [19]. Elimination of notch pathway components in Drosophila results in embryos with excess neuroblasts at the expense of epidermal precursors [20]. Mice with notch signaling defects exhibit a wide range of developmental abnormalities [21]. Aberrant upregulation of notch signaling has been observed in a variety of tumor types, including cervical, lung, colon, head and neck, renal and pancreatic cancer [22]. Thus, the notch pathway may be utilized by cancer cells to retain replicative potential rather than commit to differentiation.
In the canonical notch pathway, notch receptors on the cell surface receive signals from adjacent cells. Upon ligand binding, notch receptors are cleaved by α-secretase at the extracellular surface, and then by intracellularγ-secretase to release the intracellular portion of the Notch receptor (NICD). NICD translocates to the nucleus and associates with the RBP-Jκ transcription factor [23], which allows recruitment of coactivators that activate transcription of target genes, including members of the HES (Hairy/Enhancer of Split) family [24]. HES family members then repress transcription in conjunction with binding partners from the Transducin-like enhancer (TLE) family [25] (see Text Box 1).
Treating quiescent fibroblasts with a γ-secretase inhibitor, N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) resulted in increased expression of the muscle marker myosin heavy chain, indicating that the notch pathway plays a role in protecting quiescent fibroblasts from differentiation [12]. DAPT treatment of a rhabdomyosarcoma cell line suppressed HES1 expression and partially restored differentiation, demonstrating that notch signaling contributes to tumor initiation and/or progression in rhabdomyosarcomas and small molecules that target notch signaling might induce differentiation in cancer cells.
Notch signaling is involved in self-renewal in mammary stem cells [26], intestinal crypt progenitor cells [27], and hematopoietic stem cells [28]. Elevated levels of HES1 observed in rhabdomyosarcomas may reflect higher notch activity in the stem cells that give rise to those tumors. Indeed, HES1 expression has been detected in undifferentiated embryonic stem cells, fetal tissues, regenerating liver and tumors [29]. Higher HES1 levels might also reflect somatic mutations in the notch pathway. Activating mutations in notch signaling molecules are present in 40% of breast tumors [30], more than 50% of human T-cell acute lymphoblastic leukemias or lymphomas without specific chromosome translocations [31], and approximately 8% of diffuse large B-cell lymphomas [32]. Notch has also been associated with the stem cell-like population within tumors. Inhibition of notch signaling specifically reduced the ability of breast cancer cells to form mammospheres, an indication of pluripotent potential [33]. Finally, the tumorigenic potential of human adult neural stem cells has been associated with notch activation [34]. For these reasons, the role of the notch pathway might serve a similar function of inhibiting differentiation in quiescent cells and cancer stem cells.
Inhibition of the notch signaling pathway is a promising approach to treating tumors. In vitro, treatment with γ-secretase inhibitors results in apoptosis in breast cancer cells [35], slows the growth of osteosarcomas [36] and causes cell-cycle exit in T-cell acute lymphoblastic leukemia cells [37]. There is also evidence that inhibiting notch specifically targets a subset of cells with cancer stem cell-like properties. In medulloblastomas, suppression of notch signaling with γ-secretase inhibitors depleted a population of cells required for in vivo tumor formation, again supporting the notion that cells within a tumor with replicative capacity rely on the notch signaling pathway for survival [38]. Furthermore, neutralizing antibodies against a notch ligand resulted in reduced tumor growth and cancer stem cell frequency in human colon tumor xenografts [39].
A phase I clinical trial for a γ-secretase inhibitor, MK-0752 (Merck), has been launched for relapsed or refractory CNS malignancies [40], and two phase I/II clinical trials are under way to determine the safety and efficacy of MK-0752 in locally advanced or metastatic breast cancer patients [41]. These studies and others as summarized in Table 1 should provide insight into the benefits and side-effects of notch signaling inhibitors.
The hedgehog signaling pathway
HES1 can also be induced by signals besides the notch pathway (see Text Box 1). We focus here on regulation of HES1 by the hedgehog pathway [42, 43]. Hedgehog signaling is used to pattern organs during development, and is employed in adult tissue in the context of wound repair and tissue maintenance [44]. The hedgehog pathway often results in an inhibition of differentiation, although, in other conditions, hedgehog signaling can activate cells towards a particular fate [45]. Mutations that result in loss of hedgehog activity can lead to holoprosencephaly—a condition in which loss of ventral cell types in the forebrain causes a failure to separate the cerebral hemispheres into two lobes [46].
Hedgehog signaling is initiated when the morphogen hedgehog binds to the patched receptor, which relieves catalytic inhibition of the transmembrane protein smoothened [47]. Signaling through smoothened results in activation of transcription factors, including members of the Gli family. Gli activation results in gene expression changes that ultimately regulate differentiation, proliferation and tumorigenesis. Several studies have now shown that HES1 is transcriptionally activated by the hedgehog pathway [42, 43]. Microarray profiling of multipotent mesodermal cells revealed that overexpression of sonic hedgehog resulted in regulation of a large number of genes including HES1[42]. Signaling to HES1 did not reflect activation of the canonical notch pathway since HES1 was induced by hedgehog even in the presence of notch pathway inhibitor DAPT or a dominant negative form of RBP-Jκ. Subsequent analysis concluded that HES1 is a direct transcriptional target of the Gli transcription factor Gli2 based on chromatin immunoprecipitation [43].
As with notch signaling, hedgehog signaling is high in tissue stem cells, such as hematopoietic stem cells and neural stem cells [48]. Activation of the hedgehog pathway is also associated with tumorigenesis [49]. Inherited mutations in patched result in basal cell nevus syndrome. Mice heterozygous for patched develop medulloblastomas and skin lesions resembling basal cell carcinoma. In patients, almost all cases of sporadic basal cell carcinomas are due to hedgehog pathway activation, either through loss of heterozygosity at the patched locus and/or mutations in smoothened that diminish its ability to be inhibited by patched. Mutations causing increased hedgehog signaling have also been identified in medulloblastomas and prostate, pancreatic and colon cancer.
Hedgehog signaling is likely important for cancer stem cells. Elevated levels of hedgehog pathway components have been detected in putative pancreatic cancer stem cells [53] and breast cancer stem cells [54]. Hedgehog signaling might mediate resistance to chemotherapy and radiation therapy, as an increase in hedgehog activity was observed in esophageal cancers after chemotherapy and was associated with an increased proliferative fraction and tumor repopulation [55]. In pancreatic adenocarcinomas, a population of stem-cell-like, slowly cycling tumor cells exhibiting elevated levels of hedgehog pathway components survived chemotherapeutic treatment and repopulated the tumor rapidly [56]. Hedgehog might facilitate survival during chemotherapeutic treatment by increasing expression of cell cycle promoting proteins [55], by promoting DNA repair [55], or by increasing the expression of multi-drug resistance transporters [57].
In addition to activation of HES1 transcript levels by both notch and hedgehog, there may also be crosstalk between the pathways. Medulloblastomas from patched+/− mice [50, 51], smoothened overexpression mice [52], and human patients [52] express elevated levels of notch pathway components including the ligand Jagged1 and the Notch2 receptor. Inhibition of notch signaling in medulloblastomas from smoothened overexpression mice resulted in a reduction in viable cell numbers in cell lines, primary tumor cultures and xenografts [52]. Thus, hedgehog signaling and notch signaling may reinforce each other. Tumors, therefore, could express elevated levels of HES1 through notch signaling, hedgehog signaling, or a combination of both pathways.
The first known hedgehog pathway inhibitors, cyclopamine and jervine, were isolated from corn lilies as teratogens that caused cyclopia in mothers that ingested the plant [58]. They were subsequently shown to inhibit the hedgehog pathway by binding to smoothened. Treatment of medulloblastomas, hepatomas and gastrointestinal and lung cancer cell lines with cyclopamine in vitro results in reduced proliferation [49]. In a xenograft mouse model, cyclopamine treatment inhibited metastatic spread of pancreatic cancer cells [59]. Cyclopamine also inhibits the ability of gliomas to form gliomaspheres, indicative of pluripotent potential, and to regenerate tumors [60].
Because cyclopamine is low affinity and has poor oral bioavailability, alternatives have been syntheshesized, IPI-926 and GDC-0449. Clinical trials are being performed with these hedgehog antagonists [61]. GDC-0449 (Genentech) has a favorable toxicity profile and has shown initial efficacy in patients with basal cell carcinomas [58]. It is now being evaluated in phase II trials alone or in combination with other therapeutics for colorectal cancer, ovarian cancer, glioblastoma and medulloblastoma (www.Cancer.gov).
Histone deacetylases
In addition to the pathways that activate HES1, the mechanisms through which HES1 mediates its effects might also suggest promising strategies for anticancer therapy. HES1 can repress the transcription of its target genes through sequestration of other transcription factors or recruitment of cofactors from the TLE family [62]. Based on analogy with Drosophila, it has been suggested that TLE family members recruit histone deacetylases (HDACs) to aid in transcriptional repression [63]. By removing acetyl groups from histone tails, these enzymes modify chromatin configuration, often resulting in more tightly packed chromatin and repression of transcription [64]. Conversely, inhibition of HDAC activity results in the accumulation of acetylated core histones, which would be expected to cause a more open chromatin conformation and the transcriptional activation of target genes. As evidence that HDACs are involved in the effects of HES1, in some experiments, treatment with trichostatin A, a pan HDAC inhibitor, can at least partially reverse the repressive effects of HES1 [65, 66].
HDAC-mediated transcriptional repression has been implicated in quiescence. Mice with genetic disruption of mSin3b, which serves as a scaffold for HDACs, exhibit defective differentiation in multiple lineages and late embryonic lethality [67]. Cells derived from mSin3b-defective mice cycle normally under proliferative conditions but exhibit an impaired ability to exit the cell cycle in response to limiting growth factors. This may reflect the importance of downregulation of E2F target genes upon quiescence induction as mSin3b physically interacts with E2F promoters. Thus, HDAC activity might be associated with quiescence in the context of HES1 targets and more broadly, and the relevant targets might include genes associated with proliferation and differentiation.
Recruitment of HDACs to genes that would normally execute a differentiation program, resulting in repression of these same transcripts, is a clearly established tumorigenic pathway. As one example, the synthesis of a fusion protein (PML–RAR) between promyelocytic leukemia (PML) and the retinoic acid receptor alpha (RARα) results in a subtype of acute myeloid leukemia characterized by a block to differentiation at the promyelocytic stage. PML–RAR, in the absence of retinoic acid, binds to promoters of retinoic acid target genes, recruits an HDAC-containing complex and represses transcription [68]. Treatment of PML patients with high doses of retinoic acid releases HDAC activity from PML–RAR, permits transcription of target genes and induces terminal differentiation of leukemic blasts [69]. Thus, the recruitment of HDACs to inhibit gene expression is utilized not only by quiescent cells but also by tumors.
As would be expected if HDACs were involved in repressing expression of differentiation-promoting genes, treatment with HDAC inhibitors (HDACi’s) results in differentiation toward a variety of cell types including oligodendrocytes, myotubes, and adipocytes [70]. However, the mechanisms by which HDACi’s exert these effects are likely to be complex. Somewhat surprisingly, microarray studies have revealed that HDACi’s result in both repressive and activating effects [71–73]. Increased gene expression might reflect a direct effect of histone hyperacetylation, while decreased gene expression in response to treatment with HDACi’s could reflect indirect effects, for instance, downregulation of a transcriptional repressor. The effects of HDACi’s need not be limited to transcriptional effects mediated through HES1. HDACi’s can affect a large number of HDACs, any one of which can be recruited to chromatin through interactions with a wide array of cofactors, including, but clearly not limited to, HES1. In addition, many nonhistone proteins are also acetylated, including transcription factors, oncoproteins and tumor suppressors [74], and HDACs can remove the acetyl groups from these proteins as well. Experiments in which HDACi’s are applied while HES1 activity is inhibited could help to define the specific role of HES1 in HDACi-induced differentiation. In sum, HDAC-mediated repression of genes that activate differentiation pathways could represent another mechanism utilized by quiescent cells, stem cells and tumors to prevent the engagement of differentiation pathways and preserve pluripotency.
For a wide variety of tumors, treatment with HDACi’s induces differentiation or causes apoptosis [75]. Furthermore, numerous animal models have demonstrated the effectiveness of HDACi’s from various structural classes in antitumor activity [76]. For instance, in xenograft experiments in mice, treatment with the HDACi valproic acid decreased tumor growth and induced a more differentiated phenotype of neuroblastoma cells [77], the cyclic peptide prodrug depsipeptide (FK228) decreased the growth of leukemia and lymphoma cell lines [78], and the benzamide HDAC inhibitor MS-275 inhibited the growth of three different pediatric tumors [79].
One HDACi, vorinostat (SAHA, developed by Merck), has been approved by the FDA for the treatment of cutaneous T-cell lymphoma [80] and is also active against solid tumors. Other HDACi’s are in clinical development, with some promising results (reviewed in [81] and summarized in Table 1).
Combinations of therapies
Inhibitors of the notch pathway, the hedgehog pathway and histone deacetylases represent several therapeutic avenues available to reverse the anti-differentiation pathways activated in tumors, and thereby induce differentiation or apoptosis. Often, combinations of these compounds can be even more effective. Depending upon the tissue type or cellular context, multiple different pathways could contribute to an elevation of HES1 levels. For instance, both notch and hedgehog pathways might activate HES1, and inhibition of both of these pathways simultaneously might result in a better outcome than inhibition of either individually. Hedgehog-activated mice created by introduction of a constitutively active smoothened transgene develop medulloblastomas [52]. These medulloblastomas are characterized not only by high levels of hedgehog activity, but also high levels of HES1 and notch activity. Treatment with both anti-hedgehog and anti-notch compounds simultaneously resulted in essentially complete remission. In neural stem cells, HES1 activation can result from a combination of the notch pathway and the FGF signaling pathway [82]. FGF signaling results in beta-catenin stabilization and a complex of beta-catenin and the NICD promotes HES1 expression. In associated tumors, combination therapies that target both of these pathways might be more efficient than strategies aimed at one or the other pathway.
In other instances, targeting both HES1 levels and HES1 activity simultaneously might prove advantageous in treating cancer. As an example, combining a histone deacetylase inhibitor (SAHA) and an inhibitor of hedgehog signaling (SANT-1) resulted in a superadditive inhibition of cellular proliferation and colony formation in pancreatic cancer cells in vitro [83]. The combination of SAHA and SANT-1, but neither compound individually, resulted in cytodifferentiation as evidenced by expression of cytokeratin 7, a marker of pancreatic ductal differentiation. Treatment with a combination of the two compounds resulted in decreased proliferation and colony formation as a result of enhanced apoptosis, cell cycle arrest and differentiation.
Another strategy is to treat tumors with both a classical anti-chemotherapeutic agent designed to kill proliferating cells and a compound selected to induce differentiation or apoptosis in the quiescent stem cell compartment. For instance, inhibition of notch signaling in combination with conventional chemotherapeutic agents might be more effective than either treatment alone. Indeed, pretreatment of colon cancer cell lines with γ-secretase inhibitors enhances the effects of the DNA synthesis inhibitor oxaliplatin [84] and mitotic inhibitor taxanes [85]. An ongoing clinical trial of γ-secretase inhibitor MK-0752 includes an arm in which it will be given in combination with the mitotic inhibitor docetaxel to determine its efficacy in this combination approach [41].
The addition of HDACi’s to traditional cytotoxic drugs can also result in a stronger, more potent effect [86]. In breast cancer cell lines, the HDACi LAQ824 intensified the induction of apoptosis by the microtubule inhibitors docetaxel and epothilone B and the nucleoside analog gemcitabine [87]. Furthermore, HDACi’s improve the efficacy of radiation therapy too. For example, pretreatment with depsipeptide greatly increased radiation-induced apoptosis in squamous cell carcinoma cells [88]. HDACi’s also reduced cutaneous radiation toxicity following radiotherapy [89]. Thus, multiple strategies to eliminate quiescent, undifferentiated cells within a tumor, alone or in combination, represent promising avenues for development as therapeutic agents for a wide array of tumor types.
Concluding remarks
We discuss here one of the strategies used by quiescent cells to evade differentiation and how similar pathways are activated in tumors. There remain many avenues through which the insights described here can be translated into clinical treatment. Of particular interest would be cancer therapies that specifically target HES1 itself. Because HES1 lies at the crossroad of multiple signaling pathways, and is closely associated with tumor outcome [90], it represents an excellent target. While some tumors likely upregulate HES1 through the notch pathway, others through the hedgehog pathway, and others through distinct pathways, targeting HES1 directly could result in a higher response rate than for molecules that target only individual pathways. In addition, targeting HES1 itself might result in fewer side effects because the many other genes also regulated by the notch or hedgehog pathways would be unaffected. Targeting HES1 represents an opportunity for much greater specificity than treatment with HDACi’s, which have widespread effects on acetyl groups on histones and other non-histone proteins.
Because the possible benefits are substantial, it is especially exciting that the first HES1 dimer inhibitor isolated from natural products was just reported. Using an assay for HES1 dimerization, a natural products library was screened and two compounds that inhibit HES1-mediated downregulation of gene expression intracellularly were identified [91]. Of course, the sensitivity and efficacy of these molecules and related small molecules in vivo remains to be determined. Further, reducing HES1 activity systemically would be expected to affect the physiology of normal cells, especially stem cells, which could result in stem cell depletion, immune dysfunction and even aging phenotypes. Thus, design of a small molecule HES1 inhibitor represents a promising if challenging approach to therapy.
While we have focused here on inhibition of differentiation as a common characteristic between quiescent cells and tumors, there are likely many other important characteristics of quiescent cells that could also be engaged by tumor cells. For instance, expression of HES1 is central to the ability of quiescent cells to avoid senescence [12]. While activation of telomerase is commonly employed by tumors to evade senescence [92], they might also use mechanisms similar to those used by the quiescent cells to achieve this goal. In order to survive for long periods of time, quiescent cells likely also activate pathways that protect them from metabolic stress. Invocation of these same pathways may allow tumor cells to survive in the hypoxic and nutrient-deprived environment in the center of a tumor [4, 5].
Many cells within our bodies are quiescent, including fibroblasts, hepatocytes, lymphocytes, stem cells and germ cells. Pathologies of quiescence are likely to underlie a wide range of disorders, including autoimmune diseases, which are characterized by inappropriate proliferation of lymphocytes; fibrosis, in which there is excessive fibroblast activation after a wounding event; and even chronic wounds that might represent a failure of quiescent fibroblasts to reenter the cell cycle and coordinate a wound-healing response. Elucidating the molecular pathways invoked by quiescent cells could provide valuable information not only for treating cancer but also for tackling a wide range of other pathologies.
Supplementary Material
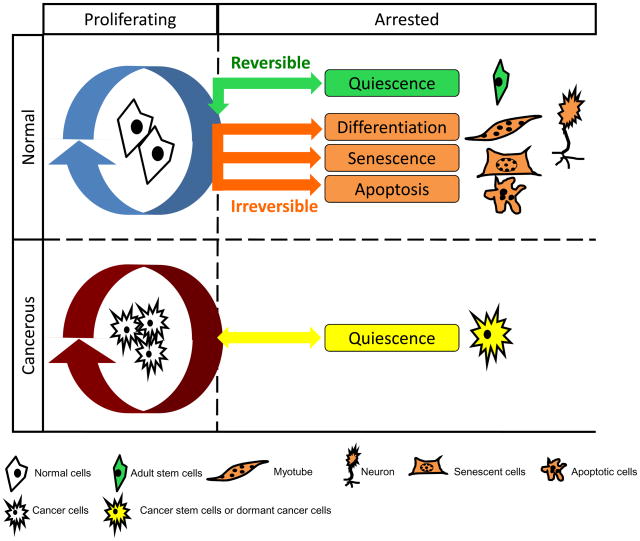
Figure 1. Comparison of different cellular fates.
Most cells within our body normally commit to one of five distinct cell fates: they can remain in the cell cycle and continue proliferation, or exit the cell cycle and become quiescent, senescent, differentiated or apoptotic. Quiescence is a reversible non-dividing state from which cells can be stimulated to proliferate in response to physiological signals. Senescence, differentiation, and apoptosis are all irreversible, terminal states. Senescence is a state of permanent cell-cycle arrest that can result from dysfunctional telomeres or stress. Differentiation represents the commitment to a lineage-specific cell type, such as muscle and neuron. Apoptosis is the process of programmed cell death. In contrast to normal cells, cancer cells are hyper-proliferative. They can escape senescence or apoptosis, and are poorly differentiated in many cases. A subpopulation of cells within a tumor, the cancer stem cells or dormant cancer cells, have been reported to exist in a quiescent state and thus to be responsible for the continued self-renewal capacity of the tumor.
References
- 1.Hanahan D, et al. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Holyoake T, et al. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94:2056–2064. [PubMed] [Google Scholar]
- 3.Jorgensen HG, et al. Lonafarnib reduces the resistance of primitive quiescent CML cells to imatinib mesylate in vitro. Leukemia. 2005;19:1184–1191. doi: 10.1038/sj.leu.2403785. [DOI] [PubMed] [Google Scholar]
- 4.Karantza-Wadsworth V, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White E, et al. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wikman H, et al. Cancer micrometastasis and tumour dormancy. APMIS. 2008;116:754–770. doi: 10.1111/j.1600-0463.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- 7.Gray JV, et al. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yusuf I, et al. Regulation of quiescence in lymphocytes. Trends Immunol. 2003;24:380–386. doi: 10.1016/s1471-4906(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 9.Venezia TA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer VR, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 11.Coller HA, et al. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sang L, et al. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sang L, et al. Fear of commitment: Hes1 protects quiescent fibroblasts from irreversible cellular fates. Cell Cycle. 2009;8:2161–2167. doi: 10.4161/cc.8.14.9104. [DOI] [PubMed] [Google Scholar]
- 14.Kageyama R, et al. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi M, et al. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 16.Abderrahmani A, et al. The hairy and enhancer of split 1 is a negative regulator of the repressor element silencer transcription factor. FEBS Lett. 2005;579:6199–6204. doi: 10.1016/j.febslet.2005.09.093. [DOI] [PubMed] [Google Scholar]
- 17.Klonisch T, et al. Cancer stem cell markers in common cancers - therapeutic implications. Trends Mol Med. 2008;14:450–460. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Lowe SW, et al. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 19.Hansson EM, et al. Notch signaling in development and disease. Semin Cancer Biol. 2004;14:320–328. doi: 10.1016/j.semcancer.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Campos-Ortega JA, et al. Molecular analysis of a cellular decision during embryonic development of Drosophila melanogaster: epidermogenesis or neurogenesis. Eur J Biochem. 1990;190:1–10. doi: 10.1111/j.1432-1033.1990.tb15538.x. [DOI] [PubMed] [Google Scholar]
- 21.Bolos V, et al. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, et al. Emerging role of Notch in stem cells and cancer. Cancer Lett. 2009;279:8–12. doi: 10.1016/j.canlet.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mumm JS, et al. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 24.Ohtsuka T, et al. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. Embo J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grbavec D, et al. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem Biophys Res Commun. 1996;223:701–705. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- 26.Dontu G, et al. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 28.Duncan AW, et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 29.Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol. 2007;31:461–466. [PubMed] [Google Scholar]
- 30.Bolos V, et al. Notch signalling in cancer stem cells. Clin Transl Oncol. 2009;11:11–19. doi: 10.1007/s12094-009-0305-2. [DOI] [PubMed] [Google Scholar]
- 31.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 32.Lee SY, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 2009;100:920–926. doi: 10.1111/j.1349-7006.2009.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farnie G, et al. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99:616–627. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- 34.Casalbore P, et al. Tumorigenic potential of olfactory bulb-derived human adult neural stem cells associates with activation of TERT and NOTCH1. PLoS One. 2009;4:e4434. doi: 10.1371/journal.pone.0004434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasul S, et al. Inhibition of gamma-secretase induces G2/M arrest and triggers apoptosis in breast cancer cells. Br J Cancer. 2009;100:1879–1888. doi: 10.1038/sj.bjc.6605034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka M, et al. Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100:1957–1965. doi: 10.1038/sj.bjc.6605060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao SS, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69:3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- 38.Fan X, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 39.Hoey T, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009:5. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Deangelo DJ, et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. ASCO Annual Meeting; 2006. Abstract No: 6585. [Google Scholar]
- 41.Krop IE, et al. Phase I pharockinetic (PK), and pharmacodynamic (PD) trial of the novel oral Notch inhibitor MK-0752 in patients (pts) with advanced breast cancer (BC) and other solid tumors. ASCO Annual Meeting; 2006. Abstract No: 10574. [Google Scholar]
- 42.Ingram WJ, et al. Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene. 2008;27:1489–1500. doi: 10.1038/sj.onc.1210767. [DOI] [PubMed] [Google Scholar]
- 43.Wall DS, et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adolphe C, et al. An in vivo comparative study of sonic, desert and Indian hedgehog reveals that hedgehog pathway activity regulates epidermal stem cell homeostasis. Development. 2004;131:5009–5019. doi: 10.1242/dev.01367. [DOI] [PubMed] [Google Scholar]
- 45.Briscoe J, et al. A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol Cell. 2001;7:1279–1291. doi: 10.1016/s1097-2765(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 46.Roessler E, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 47.Ingham PW, et al. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 48.Jiang J, et al. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hovhannisyan A, et al. From teratogens to potential therapeutics: natural inhibitors of the Hedgehog signaling network come of age. Planta Med. 2009;75:1371–1380. doi: 10.1055/s-0029-1185979. [DOI] [PubMed] [Google Scholar]
- 50.Dakubo GD, et al. Expression of Notch and Wnt pathway components and activation of Notch signaling in medulloblastomas from heterozygous patched mice. J Neurooncol. 2006;79:221–227. doi: 10.1007/s11060-006-9132-2. [DOI] [PubMed] [Google Scholar]
- 51.Yokota N, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 52.Hallahan AR, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 53.Li C, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka H, et al. The Hedgehog signaling pathway plays an essential role in maintaining the CD44+CD24−/low subpopulation and the side population of breast cancer cells. Anticancer Res. 2009;29:2147–2157. [PubMed] [Google Scholar]
- 55.Sims-Mourtada J, et al. Hedgehog: an attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin Cancer Res. 2006;12:6565–6572. doi: 10.1158/1078-0432.CCR-06-0176. [DOI] [PubMed] [Google Scholar]
- 56.Dembinski JL, et al. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis. 2009 doi: 10.1007/s10585-009-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sims-Mourtada J, et al. Sonic Hedgehog promotes multiple drug resistance by regulation of drug transport. Oncogene. 2007;26:5674–5679. doi: 10.1038/sj.onc.1210356. [DOI] [PubMed] [Google Scholar]
- 58.Garber K. Hedgehog drugs begin to show results. J Natl Cancer Inst. 2008;100:692–697. doi: 10.1093/jnci/djn169. [DOI] [PubMed] [Google Scholar]
- 59.Feldmann G, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bar EE, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borzillo GV, et al. The Hedgehog signaling pathway as a target for anticancer drug discovery. Curr Top Med Chem. 2005;5:147–157. doi: 10.2174/1568026053507732. [DOI] [PubMed] [Google Scholar]
- 62.Paroush Z, et al. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 63.Chen G, et al. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang GG, et al. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Fischer A, et al. Delta-Notch--and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takata T, et al. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- 67.David G, et al. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc Natl Acad Sci U S A. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grignani F, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 69.Lo-Coco F, et al. Acute promyelocytic leukemia: recent advances in diagnosis and management. Semin Oncol. 2008;35:401–409. doi: 10.1053/j.seminoncol.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 70.Haumaitre C, et al. Directing cell differentiation with small-molecule histone deacetylase inhibitors: the example of promoting pancreatic endocrine cells. Cell Cycle. 2009;8:536–544. doi: 10.4161/cc.8.4.7610. [DOI] [PubMed] [Google Scholar]
- 71.Mitsiades CS, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci U S A. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mariadason JM, et al. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–4572. [PubMed] [Google Scholar]
- 73.Glaser KB, et al. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2:151–163. [PubMed] [Google Scholar]
- 74.Drummond DC, et al. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 75.Botrugno OA, et al. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280:134–144. doi: 10.1016/j.canlet.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 76.Lane AA, et al. Histone Deacetylase Inhibitors in Cancer Therapy. J Clin Oncol. 2009 doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 77.Cinatl J, Jr, et al. Induction of differentiation and suppression of malignant phenotype of human neuroblastoma BE(2)-C cells by valproic acid: enhancement by combination with interferon-alpha. Int J Oncol. 2002;20:97–106. [PubMed] [Google Scholar]
- 78.Kosugi H, et al. In vivo effects of a histone deacetylase inhibitor, FK228, on human acute promyelocytic leukemia in NOD/Shi-scid/scid mice. Jpn J Cancer Res. 2001;92:529–536. doi: 10.1111/j.1349-7006.2001.tb01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jaboin J, et al. MS-27–275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res. 2002;62:6108–6115. [PubMed] [Google Scholar]
- 80.Duvic M, et al. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16:1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 81.Cang S, et al. New clinical developments in histone deacetylase inhibitors for epigenetic therapy of cancer. J Hematol Oncol. 2009;2:22. doi: 10.1186/1756-8722-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shimizu T, et al. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol Cell Biol. 2008;28:7427–7441. doi: 10.1128/MCB.01962-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chun SG, et al. Combined targeting of histone deacetylases and hedgehog signaling enhances cytoxicity in pancreatic cancer. Cancer Biol Ther. 2009:8. doi: 10.4161/cbt.8.14.8633. [DOI] [PubMed] [Google Scholar]
- 84.Meng RD, et al. Use of the Notch signaling pathway to predict disease progression and distant recurrence-free surival in early stage colon cancer. ASCO Annual Meeting; 2008. Abstraqct No: 14500. [Google Scholar]
- 85.Akiyoshi T, et al. Gamma-secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology. 2008;134:131–144. doi: 10.1053/j.gastro.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 86.Carew JS, et al. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008;269:7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 87.Fuino L, et al. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol Cancer Ther. 2003;2:971–984. [PubMed] [Google Scholar]
- 88.Zhang Y, et al. Histone deacetylase inhibitors FK228, N-(2-aminophenyl)-4-[N-(pyridin-3-yl-methoxycarbonyl)amino- methyl]benzamide and m-carboxycinnamic acid bis-hydroxamide augment radiation-induced cell death in gastrointestinal adenocarcinoma cells. Int J Cancer. 2004;110:301–308. doi: 10.1002/ijc.20117. [DOI] [PubMed] [Google Scholar]
- 89.Chung YL, et al. Antitumor histone deacetylase inhibitors suppress cutaneous radiation syndrome: Implications for increasing therapeutic gain in cancer radiotherapy. Mol Cancer Ther. 2004;3:317–325. [PubMed] [Google Scholar]
- 90.Fan X, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 91.Arai MA, et al. The first Hes1 dimer inhibitors from natural products. Bioorg Med Chem Lett. 2009;19:5778–5781. doi: 10.1016/j.bmcl.2009.07.146. [DOI] [PubMed] [Google Scholar]
- 92.Beatty GL, et al. Telomerase as a universal tumor antigen for cancer vaccines. Expert Rev Vaccines. 2008;7:881–887. doi: 10.1586/14760584.7.7.881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.