Abstract
Aphid species within the genus Tuberculatus exhibit a variety of interactions with ants, ranging from close associations to non-attendance. An ant-attended species, Tuberculatus quercicola, and two non-attended species, Tuberculatus japonicus and Tuberculatus paiki, are sympatric and hosted by the tree species Quercus dentata (Fagaceae). An undescribed ant-attended species of Tuberculatus (sp. A) and several non-attended Tuberculatus species are found on Quercus crispula trees. The population genetic structure was examined for the species sympatric on 11 Q. dentata trees and on 11 Q. crispula trees using five microsatellite loci. To determine the extent to which ant-attended or non-attended species migrate between subpopulations, flight intercept traps were placed in the study sites. Ant-attended species exhibited lower allelic richness and showed increased genetic differentiation between subpopulations compared with those of non-attended species. The number of non-attended species caught in traps increased with seasonal abundance; however, few ant-attended species were trapped, despite their abundance. These results suggest that populations of ant-attended aphids are composed of fragmented local subpopulations that are connected by low dispersal rates, leading to considerable population differentiation.
Keywords: mutualism, flight traps, Quercus
1. Introduction
Some aphid species exhibit mutualistic interactions with ants, where aphids receive protection against predators as a reward for providing honeydew for attending ants. Despite their beneficial services, attending ants often have negative impacts on aphids, including a decrease in aphid body size or embryo number owing to the costs of increased honeydew production (Stadler & Dixon 1998; Yao et al. 2000; Yao & Akimoto 2002) and suppression of colony development (Katayama & Suzuki 2002). Moreover, attending ants act as inhibitor agents for the dispersal of aphids. Ant mandibular secretions can inhibit alate development (Kleinjan & Mittler 1975) and ant semiochemicals can reduce the walking activity of apterous aphids (Oliver et al. 2007).
The aphid genus Tuberculatus feeds on Fagaceae and exhibits various mutualistic interactions with ants, ranging from non-attendance to intermediate or close associations (figure 1). Tuberculatus aphids do not alternate host plants throughout their life history. During the summer, regardless of colony density or ant attendance, all nymphs develop into alate viviparous females that produce parthenogenetic offspring. In preliminary experiments conducted on ant-attended species Tuberculatus quercicola, the removal of attending ants always resulted in the extinction of T. quercicola colonies (Yao et al. 2000).
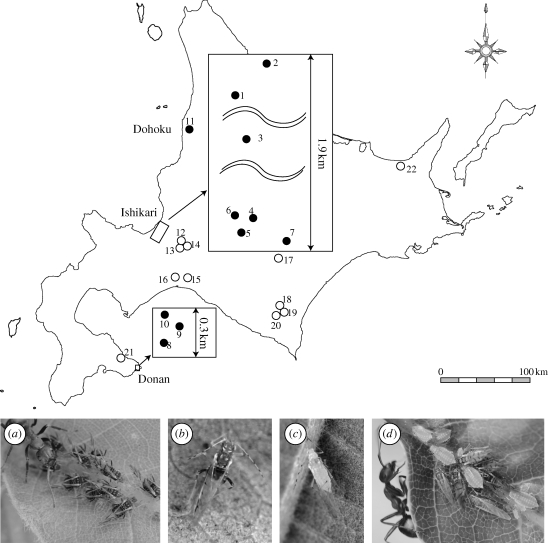
Figure 1.
The distribution of Quercus trees from where aphid samples were collected. Closed circles indicate Q. dentata on which ant-attended species (a) T. quercicola and non-attended species, (b) T. japonicus and (c) T. paiki, were found sympatrically. Open circles indicate Q. crispula on which ant-attended species (d) T. sp. A were colonized. The number on circle corresponds to tree number shown in table 1.
This study examined the population genetic structure of ant-attended and non-attended aphid species, focusing on the two ant-attended species T. quercicola on Quercus dentata and Tuberculatus sp. A on Quercus crispula and the two non-attended species Tuberculatus japonicus and Tuberculatus paiki sympatric on Q. dentata. Furthermore, a field study was conducted to assess the number of aphids in flight and their population dynamics, using flight intercept traps and weekly observations.
2. Material and methods
(a). Sample collection
During 2006, either all three or two (T. quercicola and T. japonicus) of these species were collected from 11 Q. dentata trees distributed throughout Hokkaido, in northern Japan. An ant-attended undescribed species T. sp. A and several non-attended species were collected from 11 Q. crispula trees between 2005 and 2008 (table 1 and figure 1). Both ant-attended species were associated with the ant Formica yessensis. To avoid multiple collections of an aphid clone, a single aphid was collected from each sampled leaf.
Table 1.
Number of individuals used for genotyping (N) and indices of genetic diversity (AP, G/n and FIS) for Tuberculatus aphids. Lower two rows indicate total number and mean ± s.d. A Wilcoxon signed-rank test was conducted between species on Q. dentata. A Mann–Whitney U test was conducted between T. sp. A and non-attended species on Q. dentata. Different letters on mean values indicate a significant difference between species. p < 0.05. NA indicates ‘not analysed’ because all individuals in the population were homozygous.
|
T. quercicola |
T. japonicus |
T. paiki |
T. sp. A |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | AP | G/n | FIS | N | AP | G/n | FIS | N | AP | G/n | FIS | N | AP | G/n | FIS | ||
| tree 1 | 16 | 3.34 | 0.94 | 0.46 | 16 | 7.59 | 0.94 | 0.19 | 11 | 8.69 | 1 | 0.17 | tree 12 | 16 | 1.59 | 0.25 | 0.40 |
| tree 2 | 16 | 2.10 | 0.63 | 0.05 | 16 | 7.51 | 1 | 0.21 | 10 | 7.60 | 1 | 0.18 | tree 13 | 16 | 1.20 | 0.13 | 1 |
| tree 3 | 16 | 1 | 0.06 | NA | 16 | 8.27 | 1 | 0.16 | 11 | 5.21 | 1 | 0.13 | tree 14 | 16 | 2.43 | 0.63 | 0.70 |
| tree 4 | 16 | 3.33 | 0.94 | 0.47 | 16 | 7.05 | 0.94 | 0.23 | 16 | 7.41 | 1 | 0.10 | tree 15 | 16 | 1.74 | 0.50 | 0.47 |
| tree 5 | 16 | 2.87 | 0.88 | 0.20 | 16 | 7.37 | 0.94 | 0.12 | 16 | 6.99 | 1 | 0.21 | tree 16 | 16 | 1.80 | 0.81 | 0.52 |
| tree 6 | 16 | 3.58 | 1 | 0.18 | 16 | 7.86 | 1 | 0.09 | 16 | 8.16 | 1 | 0.18 | tree 17 | 19 | 2.66 | 0.89 | 0.15 |
| tree 7 | 16 | 3.55 | 0.88 | 0.27 | 16 | 7.61 | 1 | 0.24 | 16 | 7.22 | 1 | 0.06 | tree 18 | 20 | 2.37 | 0.95 | 0.39 |
| tree 8 | 17 | 2.12 | 0.47 | 0.66 | 24 | 7.70 | 1 | 0.25 | 32 | 6.84 | 1 | 0.21 | tree 19 | 8 | 2.20 | 0.88 | 0.13 |
| tree 9 | 8 | 1.80 | 0.75 | 0.39 | 16 | 7.01 | 1 | 0.09 | 16 | 6.90 | 1 | 0.09 | tree 20 | 8 | 2.40 | 0.88 | 0.21 |
| tree 10 | 8 | 1.40 | 0.50 | 0.15 | 8 | 7.20 | 1 | 0.19 | 16 | 7.86 | 1 | 0.28 | tree 21 | 8 | 1 | 0.13 | NA |
| tree 11 | 20 | 4.18 | 0.95 | 0.35 | 16 | 7 | 1 | 0.24 | — | — | — | — | tree 22 | 16 | 1.79 | 0.56 | 0.23 |
| 165 | 2.66ac | 0.73ac | 0.32ac | 176 | 7.47b | 0.98b | 0.18b | 160 | 7.29b | 1b | 0.16ab | 159 | 1.93c | 0.60c | 0.42c | ||
| 1.03 | 0.29 | 0.18 | 0.40 | 0.03 | 0.06 | 0.94 | 0 | 0.07 | 0.53 | 0.31 | 0.27 | ||||||
(b). Microsatellite DNA analysis
Genomic DNA was extracted from an entire aphid following the Chelex procedure (Walsh et al. 1991). The genotypes of individual aphids were examined using five microsatellite DNA primers (Tq-15, -17, -21, -23 and -26) (Yao et al. 2003). The polymerase chain reaction amplifications and determination of allele sizes were the same as described in Yao et al. (2003).
(c). Genetic diversity
Linkage disequilibrium between loci was tested using Fisher's exact probability test in Genepop (Raymond & Rousset 1995). Mean number of alleles per polymorphic locus (AP) corrected for the small sample size was calculated using Microsatellite Analyzer (Dieringer & Schlötterer 2003). To examine whether the distribution of clonal aphids was restricted within a tree, G/n (Llewellyn et al. 2003) was calculated in each species, where G is the number of different genotypes (clones) in a tree and n was the number of individuals collected. The extent of genetic structuring within trees was investigated by calculating the inbreeding coefficient FIS in FSTAT (Goudet 1995). The differences in genetic diversity indices between species were tested using a Wilcoxon signed-rank test with a sequential Bonferroni correction for multiple comparisons on the pairs of aphid populations collected on Q. dentata. Because an insufficient number of individuals per species was available for the non-attended Tuberculatus aphids on Q. crispula, genetic diversity indices of T. sp. A were compared with those of the three species inhabiting Q. dentata using a Mann–Whitney U test.
(d). Genetic differentiation among subpopulations
Genetic intraspecific differences among subpopulations were examined using an analysis of molecular variance (AMOVA) in Arlequin (Schneider et al. 2000). Quercus dentata trees applied to the AMOVA were divided into the following regions: the Ishikari region, which included trees 1–7; the Donan region, which included trees 8–10; and the Dohoku region, which contained tree 11. AMOVA for Q. crispula was conducted only on T. sp. A populations. Quercus crispula was limited to one tree per region, therefore hierarchical groups in AMOVA were simply divided into two groups: ‘among trees’ and ‘within trees’.
(e). Trap survey
Winged aphids were trapped at two sites, Ishikari (area corresponding to Q. dentata, trees 1–7, approx. 0.14 km2) and Tomakomai (area corresponding to Q. crispula, tree 16, approx. 2.08 × 10−4 km2), from June to October 2008. A flight intercept trap was constructed of a cylindrical tomato cage (120 cm tall and 30 cm in diameter) and transparent packaging tape. Six tape clippings (6 cm width and 80 cm long) were vertically attached to the cages. Ten and five traps were located at an average distance of 12 and 4 m away from adjacent trees at the Ishikari and Tomakomai sites, respectively. To investigate the demographic patterns of winged Tuberculatus aphids that colonized host trees, five Q. dentata and three Q. crispula host trees located adjacent to the traps were chosen, and 20 and 10 shoots were randomly selected from each tree. The numbers of winged Tuberculatus aphids caught in the traps and observed in the trees were counted weekly and accumulated through the seasons. The Fisher exact test with a sequential Bonferroni correction for multiple comparisons was conducted on the comparison between species. Because it was difficult to identify non-attended Tuberculatus species inhabiting Q. crispula without a microscope, all non-attended species on the Tomakomai site were pooled for statistical analyses.
3. Results
(a). Genetic diversity
Linkage disequilibrium was not detected for any pair of loci across all populations of the three species on Q. dentata; however, it was detected for one pair of loci (Tq-15 and Tq-26) in T. sp. A. The AP and G/n ratio were significantly lower in T. quercicola than in the two non-attended species. FIS values were significantly higher in T. quercicola than in T. japonicus, but no significant difference was detected between T. paiki and either other species. The AP and G/n ratio were significantly lower in T. sp. A than in the two non-attended species. Significantly higher FIS values were found in T. sp. A than in the two non-attended species (table 1).
(b). Genetic differentiation among subpopulations
The AMOVA revealed that variation among regions and variation among trees within regions were higher in T. quercicola than in non-attended species. In T. sp. A, variation among and within trees was 58 and 42 per cent, respectively (table 2).
Table 2.
AMOVA for T. quercicola, T. japonicus, T. paiki and T. sp. A. (ss indicates sum of squared deviations in each hierarchical group. Variance components and fixation indices were tested using a non-parametric permutation approach (1023 permutations).)
| source of variation | d.f. | ss | variance components | percentage of variation | fixation indices | p |
|---|---|---|---|---|---|---|
| T. quercicola | ||||||
| among regions | 2 | 102.3 | 0.453 | 24.83 | FCT = 0.248 | <0.001 |
| among trees within regions | 8 | 106.6 | 0.426 | 23.32 | FSC = 0.310 | <0.001 |
| within trees | 319 | 301.8 | 0.946 | 51.85 | FST = 0.482 | <0.001 |
| total | 329 | 510.7 | 1.825 | |||
| T. japonicus | ||||||
| among regions | 2 | 19.4 | 0.059 | 2.70 | FCT = 0.027 | <0.001 |
| among trees within regions | 8 | 33.7 | 0.069 | 3.13 | FSC = 0.032 | <0.001 |
| within trees | 341 | 703.3 | 2.063 | 94.17 | FST = 0.058 | <0.001 |
| total | 351 | 756.5 | 2.190 | |||
| T. paiki | ||||||
| among regions | 1 | 25.9 | 0.097 | 5.28 | FCT = 0.053 | <0.05 |
| among trees within regions | 8 | 69.1 | 0.234 | 12.72 | FSC = 0.134 | <0.001 |
| within trees | 310 | 468.2 | 1.510 | 82.00 | FST = 0.180 | <0.001 |
| total | 319 | 563.2 | 1.842 | |||
| T. sp. A | ||||||
| among trees | 10 | 287.9 | 0.979 | 57.71 | ||
| within trees | 307 | 220.3 | 0.718 | 42.29 | FST = 0.577 | <0.001 |
| total | 317 | 508.3 | 1.697 | |||
(c). Trap survey
On both sites, only a few ant-attended aphids were trapped. The Fisher exact test showed a significant difference in the ratio of aphids caught in traps and observed in trees between ant-attended and non-attended species at both sites (table 3).
Table 3.
Total numbers of winged aphids across trees and across traps on Q. dentata at the Ishikari site and Q. crispula at the Tomakomai site. The Fisher exact test, p < 0.001 for both pairs in the Ishikari site and p < 0.01 for between T. sp. A and combined non-attended Tuberculatus species. Different letters on the numbers of trapped individuals indicate a significant difference between species.
| species | no. trapped | |
|---|---|---|
| Q. dentata | ||
| T. quercicola | 1342 | 8a |
| T. japonicus | 200 | 52b |
| T. paiki | 1315 | 137c |
| Q. crispula | ||
| T. sp. A | 194 | 2a |
| non-attended Tuberculatus species | 218 | 20b |
4. Discussion
The present study clearly demonstrated that mutualistic interactions with ants resulted in low dispersal, low genetic diversity and high genetic differentiation of mutualistic aphids. Many factors influence genetic variation at polymorphic loci (Lowe et al. 2004); however, population size has the greatest impact because it can alter the probability of loss or fixation of alleles through random changes in allele frequency. The lower G/n ratio per tree and the small fraction of trapped individuals indicated that the distribution of ant-attended clonal aphids was more spatially restricted within a single tree than that of non-attended clonal aphids. Furthermore, higher FIS values in the two ant-attended species may be attributed to local inbreeding, suggesting that the effective population size of the ant-attended species is limited to relatively small ranges compared with that of non-attended species. Besides limited dispersal, the loss of genetic diversity can be partly accounted by the relative phylogenetic position of the ant-attended species.
The AMOVA showed high levels of genetic differentiation in ant-attended species. The percentage of variation ‘among trees within regions’ was higher in T. quercicola than in non-attended species, indicating that limited gene flow and associated genetic differentiation occurred even at a microgeographic scale of 1.9 (the Ishikari region) or 0.3 km (the Donan region). Genetic differentiation of subdivided populations has been addressed in light of the effects of extinction and recolonization (Pannell & Charlesworth 1999). Many factors involving the formation and maintenance of aphid–ant mutualisms are labile, such that most aphid species forego the potential benefits of associating with ants (Bristow 1991). Of these factors, the difference in susceptibility of host plants to aphids and the availability of suitable ants, in particular, play vital roles in mutualisms. The longevity of aphid colonies is influenced by their attractiveness to ants during the season. Even after the acquisition of ant attendance, aphid colonies occasionally become less attractive owing to employment of alternative resources by the ants (Offenberg 2001) or competition from neighbouring aphids for the services of ant mutualists (Cushman & Addicott 1989). Taken together, aphids with low rates of dispersal and unstable relationships with ants, patterns of recurrent local extinction and recolonization events would be expected in ant-attended aphid colonies.
Given that ant-attended colonies lasted for more than 30 days compared with the experimentally ant-excluded colonies (Yao et al. 2000), the reduction in dispersal in ant-attended aphids would be a selective advantage in protection against predators. However, it is unclear why ant-attended Tuberculatus aphids have fully developed wings. No wing dimorphism has been reported in Tuberculatus species. To determine whether possession of wings arise from phylogenetic constraints or hidden adaptive significance, anatomical and physiological studies on the formation and maintenance of wings are further needed.
Acknowledgements
This study was supported in part by a Grant-in-Aid for Scientific Research (no. 15570009 and 19570010).
References
- Bristow C. M.1991Why are so few aphids ant-tended? In Ant–plant interactions (eds Huxley C. R., Cutler D. F.), pp. 104–119 Oxford, UK: Oxford University Press [Google Scholar]
- Cushman J. H., Addicott J. F.1989Intra- and interspecific competition for mutualists: ants as a limited and limiting resource for aphids. Oecologia 79, 315–321 (doi:10.1007/BF00384310) [DOI] [PubMed] [Google Scholar]
- Dieringer D., Schlötterer C.2003Microsatellite Analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 3, 167–169 (doi:10.1046/j.1471-8286.2003.00351.x) [Google Scholar]
- Goudet J.1995Fstat. A computer program to calculate F-statistics, Version 1.2. J. Hered. 86, 485–486 [Google Scholar]
- Katayama N., Suzuki N.2002Cost and benefit of ant attendance for Aphis craccivora (Hemiptera: Aphididae) with reference to aphid colony size. Can. Entomol. 134, 241–249 [Google Scholar]
- Kleinjan J. E., Mittler T. E.1975A chemical influence of ants in wing development in aphids. Entomol. Exp. Appl. 18, 384–388 (doi:10.1007/BF00628368) [Google Scholar]
- Llewellyn K. S., Loxdale H. D., Harrington R., Brookes C. P., Clark S. J., Sunnucks P.2003Migration and genetic structure of the grain aphid (Sitobion avenae) in Britain related to climate and clonal fluctuation as revealed using microsatellites. Mol. Ecol. 12, 21–34 (doi:10.1046/j.1365-294X.2003.01703.x) [DOI] [PubMed] [Google Scholar]
- Lowe A., Harris S., Ashton P.2004Ecological genetics Oxford, UK: Blackwell Publishing [Google Scholar]
- Offenberg J.2001Balancing between mutualism and exploitation: the symbiotic interaction between Lasius ants and aphids. Behav. Ecol. Sociobiol. 49, 304–310 (doi:10.1007/s002650000303) [Google Scholar]
- Oliver T. H., Mashanova A., Leather S. R., Cook J. M., Jansen V. A. A.2007Ant semiochemicals limit apterous aphid dispersal. Proc. R. Soc. B 274, 3127–3131 (doi:10.1098/rspb.2007.1251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannell J. R., Charlesworth B.1999Neutral genetic diversity in a metapopulation with recurrent local extinction and recolonization. Evolution 53, 664–676 (doi:10.2307/2640708) [DOI] [PubMed] [Google Scholar]
- Raymond M., Rousset F.1995Genepop (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249 [Google Scholar]
- Schneider S., Roessli D., Excoffier L.2000Arlequin ver. 2.000: a software for population genetics data analysis. Geneva, Switzerland: Genetics and Biometry Laboratory, University of Geneva [Google Scholar]
- Stadler B., Dixon A. F. G.1998Costs of ant attendance for aphids. J. Anim. Ecol. 67, 454–459 (doi:10.1046/j.1365-2656.1998.00209.x) [Google Scholar]
- Walsh P. S., Metzger D. A., Higuchi R.1991Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513 [PubMed] [Google Scholar]
- Yao I., Akimoto S.2002Flexibility in the composition and concentration of amino acids in honeydew of the drepanosiphid aphid Tuberculatus quercicola. Ecol. Entomol. 27, 745–752 (doi:10.1046/j.1365-2311.2002.00455.x) [Google Scholar]
- Yao I., Shibao H., Akimoto S.2000Costs and benefits of ant attendance to the drepanosiphid aphid Tuberculatus quercicola. Oikos 89, 3–10 (doi:10.1034/j.1600-0706.2000.890101.x) [Google Scholar]
- Yao I., Akimoto S., Hasegawa E.2003Isolation of microsatellite markers from the drepanosiphid aphid Tuberculatus quercicola (Homoptera, Aphididae). Mol. Ecol. Notes 3, 542–543 (doi:10.1046/j.1471-8286.2003.00504.x) [Google Scholar]