Abstract
The emergence and spread of H5N1 avian influenza viruses from Asia through to Europe and Africa pose a significant animal disease problem and have raised concerns that the virus may pose a pandemic threat to humans. The epizootological factors that have influenced the wide distribution of the virus are complex, and the variety of viruses currently circulating reflects these factors. Sequence analysis of the virus genes sheds light on the H5N1 virus evolution during its emergence and spread, but the degree of virus variation at the level of an individual infected bird has been described in only a few studies. Here, we describe some results of a study in which turkeys, ducks and chickens were infected with either one of two H5N1 or one of three H7N1 viruses, and the degree of sequence variation within an individual infected avian host was examined. We developed ‘deep amplicon’ sequence analysis for this work, and the methods and results provide a background framework for application to disease outbreaks in the field.
Keywords: chickens, turkeys, ducks, sequence variation, avian influenza viruses
1. Introduction
The emergence of H5N1 highly pathogenic avian influenza (HPAI) virus since it appeared in Hong Kong in 1997 (Claas et al. 1998) has resulted in the exceptional spread of the virus to now cover over 60 countries spreading from China and the eastern regions of Russia in the east, through to Burkina Faso in West Africa and Scotland in northwest Europe. The extent of the spread of the H5N1 virus seems unprecedented and has not been seen previously in the second half of the twentieth century: in all other outbreaks of HPAI since the 1950s, control by culling and sanitary procedures usually resulted in the rapid containment and elimination of the virus. Culling, sanitary control measures and vaccination have been insufficient to contain H5N1 infection, although, for example, in Japan, South Korea and several European Union member states (OIE 2009), early detection and culling of infected poultry proved effective in controlling HPAI H5N1 influenza.
The widespread dissemination of H5N1 viruses in poultry has resulted in a number of human infections and has been recognized to pose a threat of a human influenza pandemic. At the end of March 2009, over 410 human cases of infection had been confirmed, with a case-fatality rate of over 60 per cent. Most of the human cases have been associated with exposure to sick poultry, and there have been only few clusters of infection where human-to-human transmission cannot be ruled out (WHO 2009). Human cases have been reported in 15 countries, with Indonesia, Vietnam, Egypt and China bearing the largest burden of human disease, accounting for 75 per cent of the cases. With the exception of Indonesia, human H5N1 influenza cases in each of these countries have been confirmed in the first three months of 2009. At the time of writing, the pandemic threat was defined by WHO as Phase 3, in which human infection is recognized from a new subtype but human-to-human transmission is rare. Should the rate of clusters of human-to-human transmission increase, then the pandemic risk phase will be increased (WHO 2005).
It is striking that control of disease in animals shows an impact in the reduction of human infections. This is illustrated by the events observed in Vietnam between 2004 and 2007, where high numbers of human cases were seen in both 2004 and 2005 but dropped to zero in 2006, following the initiation of a vaccination campaign of poultry in 2005 when 160 million of an estimated population of 250 million poultry were vaccinated. The numbers of human cases rose somewhat in the following 2 years, but remain at a much lower level than that reported in 2004 and 2005 in the presence of continued poultry vaccination.
The reasons for the failure to eliminate H5N1 virus in domesticated poultry are complex but are likely to be associated with the widespread distribution of the virus before its presence is recognized and sanitary measures become enforced. A major factor that hampers control measures is likely to be the maintenance of highly pathogenic H5N1 viruses in different avian host species, a factor previously thought to be uncommon with other HPAI viruses. H5N1 infections have been observed in a diverse range of avian species: an estimate of the range of species infected with H5N1 viruses can be made from a survey of the genetically characterized viruses whose sequences have been made publicly available (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). As of March 2009, with over 2300 H5N1 haemagglutinin (HA) genes of avian origin analysed, approximately 1000 were from chickens, 650 from ducks, 200 from geese and around 50 each from swans and turkeys. These numbers can only be used as a very rough estimate of prevalence of the virus since domestic species have been under closer surveillance than wild birds in which infections are more difficult to detect. It is notable that species not normally associated with avian influenza virus infection have been found to be infected, and examples include sparrows, crows, magpies and storks, and birds of prey that may have fed on infected poultry, e.g. falcons (Monne et al. 2008), kestrels (Smith et al. 2009), vultures (Ducatez et al. 2007a,b) and buzzards (Hars et al. 2008). The presence of H5N1 viruses in the bar-headed goose population around Qinghai Lake in late 2005 (Chen et al. 2005) provides a striking example of wild-bird infection, and it is postulated that virus was carried from Qinghai Lake westwards through bird migration. The observations that, following experimental infection, a small number of ducks of several species showed severe disease signs lend support to the notion that spread through wild birds is feasible (Hulse-Post et al. 2005; Sturm-Ramirez et al. 2005; Keawcharoen et al. 2008; Kim et al. 2008; Londt et al. 2008), yet the relative importance of wild birds in transmission of the H5N1 virus remains an open question.
The prevalence of H5N1 in wild birds has been studied intensively in Europe since 2006, and the results of a 2006 survey have been reported recently (Hesterberg et al. 2009). The survey showed that H5N1 viruses were found only rarely in dabbling ducks, but swans, diving ducks, mergansers and grebes showed higher rates of infection and usually associated with disease. The survey, of over 120 000 birds sampled in 2006 with only 591 detected as positive for H5N1, concluded that this virus was not able to be sustained in the wild-bird population within the European Union. The continuing spread of infection seems to be very complex, and Yasué et al. (2006) highlighted several important points in a review of the evidence for transmission of virus by wild birds over large distances. For example, in Qinghai Lake, bar-headed geese were not the only wild-bird species affected and there were subsequent reports of domesticated geese in the area. Also, which host infected the bar-headed goose population? And when were the geese first affected by H5N1 influenza? Yasué et al. (2006) pointed out that bar-headed geese overwinter in India and arrive following their trans-Himalayan migration at the lake in March. The signs of H5N1 infection were first recorded around Qinghai Lake in May to July. It may be pertinent to note that Vijaykrishna et al. (2008) estimate that the H5N1 virus was introduced into Indonesia and Vietnam three to six months prior to the first recognition of H5N1 infection in each country, so there may be a considerable delay in some areas between the index infection and the recognition of infection. Wild-bird migration has been proposed to be associated with the trans-African spread of H5N1 to Western Africa. Ducatez et al. (2006) adduced that, in the emergence of virus in Nigeria, three separate introductions of the H5N1 virus coincided with the flight paths of migratory birds; nevertheless, commercial links are also known to exist between the poultry industries of Nigeria and the Far East, and the possibility of infected poultry importation or associated contaminated products cannot be excluded. The relative importance of the role of migratory birds in spreading H5N1 virus compared with the spread through human agricultural and food production activities needs careful consideration. It seems prudent to focus on the domesticated duck and poultry sector for the detection of H5N1 infection and for the control of the infection once found.
The widespread geographical distribution of H5N1 viruses, possibly combined with circulation in wild-bird species, has led to a wide diversity of H5N1 viruses. Phylogenetic analyses of the HA genes of H5N1 viruses have led to the construction of 10 clades defined according to phylogenetic criteria. However, as the number of genes sequenced has increased and as the virus continues to evolve, the initial phylogenetic classification of 10 clades has become more elaborate with the division of some clades into subclades and third-order clades (WHO/OIE/FAO Working Group 2008, 2009).
The rapid evolution of the virus into the 10 recognized clades is likely to be associated with the dependence of the virus on an RNA-dependent RNA polymerase for replication. RNA viruses are considered subject to more sequence variation than viruses with less error-prone DNA-dependent DNA polymerases. There has been a long debate about whether RNA viruses exist as a virus quasi-species (e.g. Smith et al. 1997; Holmes & Moya 2002; Moya et al. 2004) or whether they behave in a manner of standard population genetics, with selection at the individual virus rather than selection at the level of the virus population, a feature of a quasi-species. It has been pointed out many times (reviewed in the aforementioned references) that sequence variability does not imply quasi-species behaviour per se. RNA viruses are thought of as having large populations, but they may undergo severe bottlenecks in transmission between hosts of the same, or different, species and result in a small effective population size (Moya et al. 2004); it is well established in classical population biology that bottlenecks increase the fixation of neutral mutation (Maynard Smith 1989) and hence virus population dynamics will influence the frequency of the fixation of mutations.
In the work we describe below, as part of a more detailed study into host selection of variant viruses, we investigated whether different avian influenza viruses showed any variation in the degree of diversity from the consensus sequence of the virus as they replicated in different hosts. We examined viruses of different natural history: a clade 2.2 H5N1 virus isolated from turkeys during the early phase of the introduction of this clade into Europe; an H5N1 virus, also of Eurasian origin, that represented a virus present prior to the explosive spread of the current H5N1 viruses and three H7N1 subtype viruses from an outbreak of avian influenza in Italy in 1999–2000. These viruses were used to infect groups of chickens, turkeys and ducks, and the variation in a region of approximately 1000 nucleotides in the virus HA gene was examined. In comparison, we also examined a smaller number of clones in a second RNA segment encoding the NS1 and NS2 (NEP) polypeptides of the virus; this gene was chosen as it underwent adaptive changes as the H7N1 epidemic emerged in Italy (Dundon et al. 2006) and showed considerable reassortment and change as the H5N1 panzootic emerged (Li et al. 2004; Duan et al. 2008).
2. Material and methods
(a). Virus stocks
The following virus strains were propagated in 9-day-old specified pathogen free embryonated fowls' eggs (Charles River, USA), and viral titres were calculated by 50 per cent egg infectious dose (EID50): A/chicken/Italy/1279/99 (H7N1), abbreviated Italy/1279; A/ostrich/Italy/984/00 (H7N1), abbreviated Italy/984; A/turkey/Italy/3466/99 (H7N1), Italy/3466; A/turkey/England/50-92/91 (H5N1), England/50-92; and A/turkey/Turkey/1/2005 (H5N1), Turkey/05. Italy/984, England/50-92 and Turkey/05 were HPAI viruses; Italy/1279 and Italy/3466 were low pathogenicity avian influenza (LPAI) viruses.
(b). Infection of turkeys, chickens and ducks
Pre-inoculation blood tests, together with buccal and cloacal swabs, were taken to ensure that all birds were free of influenza infection prior to the start of the experiment. Birds, in groups of 10 turkeys, chickens or ducks, were inoculated with 102, 104 or 106 EID50 of virus delivered intranasally in a volume of 0.1 ml. The birds were monitored twice daily for clinical signs, and daily buccal and cloacal swabs were taken. Any bird deemed unable to reach food or water, or unduly ill, was humanely euthanized and recorded as mortality for that day. Each experiment lasted a maximum of three weeks. Any birds surviving at that time were humanely euthanized.
(c). Viral RNA isolation and determination of viral titres in buccal and cloacal swabs
Buccal and cloacal swabs were taken, and viral titres were determined by an influenza matrix gene one-step quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) as described previously (European Union 2006; Londt et al. 2008).
(d). Deep amplicon sequencing
cDNA amplicons (bases 9–1152 bp for H7 and 9–1206 bp for H5) of the HA gene or bases 8–850 for RNA segment 8 encoding the NS1 and NS2 (NEP) polypeptides were prepared from virus inocula, buccal and cloacal swabs by a two-step RT-PCR. The RT reactions were performed using an influenza virus universal oligonucleotide primer, 5′-AGCAAAAGCAGG-3′ with the Verso cDNA Kit (Thermo Scientific) according to the suppliers' instructions. For PCR amplification, each RT reaction (5 µl) was supplemented with 10× buffer, 250 µM each dNTP, 0.2 µM forward and reverse primers in a 50 µl final reaction volume and 1 µl pfu Ultra II fusion HS DNA polymerase was added (Stratagene, enzyme units were not defined by the manufacturer). The amplification was performed using an initial denaturation step (95°C for 1 min), followed by 35 cycles of amplification (95°C for 20 s, 55°C for 20 s and 72°C for 45 s) and a final extension (72°C for 3 min). In most cases, PCR products were gel purified using a gel extraction kit (QIA quick, Qiagen), and the gene products were cloned into the pCR-Blunt vector (Invitrogen). Positive clones containing gene inserts were selected by colony PCR analysis. Colonies containing plasmids shown to contain inserts were posted to a commercial company and the cDNA inserts sequenced (GATC Biotech, Constance, Germany). The primer sequences for HA and non-structural (NS) gene cloning, colony PCR and for deep amplicon sequencing can be made available on request.
(e). Sequence analysis
Sequences were aligned alongside a reference (inoculum) sequence using the Gap4 application from the Staden Package (Staden 1996). The Gap4 ‘report mutations’ function was then used to generate a text report of all the base changes, plus their effects on the predicted protein sequence. The mutation report obtained was then parsed with a perl script to obtain the number/proportions of synonymous/non-synonymous changes occurring for each base position in the sequence; this information was then imported into Excel (MS Office) for further manipulation. Further analysis was performed using the SSAHA (Sequence Search and Alignment by Hashing Algorithm) (Ning et al. 2001) version ssaha2, the SNP_analysis.pm Perl module (http://sourceforge.net/projects/snpanalysis/) and our own Perl scripts.
3. Results
We sought to examine the intrahost nucleotide sequence variation following infection of birds of three species with avian influenza viruses. The degree of sequence variation was established by the analysis of cDNA clones prepared directly from swabs of turkeys, chickens and ducks infected experimentally with H5N1 or H7N1 avian influenza virus.
Birds were sampled daily, and swabs were then prepared for analysis by a protocol of reverse transcription and PCR using a high fidelity reverse transcriptase to generate amplified cDNA products and these were cloned into plasmids for analysis of the sequence with no intermediate amplification in tissue culture or fowl's eggs. In the results presented here, a total of approximately 1800 individual cDNA clones have been analysed from the three avian hosts. The size of the amplicon varied depending on the gene: for the HA gene, approximately 1200 nucleotides were amplified leading to 1150 (H5 viruses) or 1112 (H7 viruses) nucleotides being analysed; for the NS protein-encoding segment, 690 nucleotides were analysed. These regions covered the coding sequence for the HA signal sequence, HA1 and the amino terminal of HA2 for the HA gene and NS1 and part of NS2 for RNA segment 8. We calculated the overall nucleotide substitution frequency observed and compared with a consensus sequence derived from the samples, the ratio of synonymous to non-synonymous changes and the localization and the frequency of the nucleotide changes in individual clones.
The overall frequency (table 1) was within a similar order of magnitude, varying from 8.89 × 10−4 to 1.69 × 10−4 substitutions per nucleotide. H7N1 viruses varied across this range; the H5N1 viruses showed a substitution frequency similar to each other at 3.3 × 10−4 for England/50-92 and 5.0 × 10−4 for Turkey/05, a representative of the currently circulating H5N1 viruses (clade 2.2).
Table 1.
Sequence variability of H5N1 and H7N1 influenza viruses during the course of infection of three avian host species.
| number of substitutions |
substitution frequency |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| virus | host species | gene | sequence analysed | number of clones | synonymous | non-synonymous | synonymous | non-synonymous | overall substitution frequency |
| A/turkey/Turkey/1/05 (H5N1-HPAI) | chickens | HA | 29–1178 | 650 | 56 | 318 | 7.49 × 10−5 | 4.25 × 10−4 | 5.00 × 10−4 |
| A/turkey/England/50-92/91 (H5N1-HPAI) | chickens | HA | 29–1178 | 360 | 48 | 101 | 1.16 × 10−4 | 2.44 × 10−4 | 3.60 × 10−4 |
| A/ostrich/Italy/984/00 (H7N1-HPAI) | chickens | HA | 22–1133 | 156 | 53 | 32 | 3.06 × 10−4 | 1.84 × 10−4 | 4.90 × 10−4 |
| A/ostrich/Italy/984/00 (H7N1-HPAI) | turkeys | HA | 22–1133 | 290 | 92 | 135 | 2.85 × 10−4 | 4.19 × 10−4 | 7.04 × 10−4 |
| A/ostrich/Italy/984/00 (H7N1-HPAI) | ducks | HA | 22–1133 | 170 | 6 | 26 | 3.17 × 10−5 | 1.38 × 10−4 | 1.69 × 10−4 |
| A/chicken/Italy/1279/99 (H7N1-LPAI) | turkeys | NS | 27–718 | 83 | 1 | 50 | 1.74 × 10−5 | 8.72 × 10−4 | 8.89 × 10−4 |
One virus, Italy/984, was replicated in three species, and a similar nucleotide substitution frequency was found in each species, viz. 1.69 × 10−4 in ducks, 4.90 × 10−4 in chickens and 7.04 × 10−4 in turkeys. However, the levels of virus in the swabs from each species were different: turkeys show increased susceptibility to virus infection and shed higher levels of virus than chickens, and both turkeys and chickens shed more virus than ducks following infection by this H7N1 virus (I. Essen, L. Brookes, I. H. Brown & J. W. McCauley 2004–2008, unpublished data). We cannot be sure whether the replication levels correlate with mutant frequency; further experiments need to be carried out to determine this.
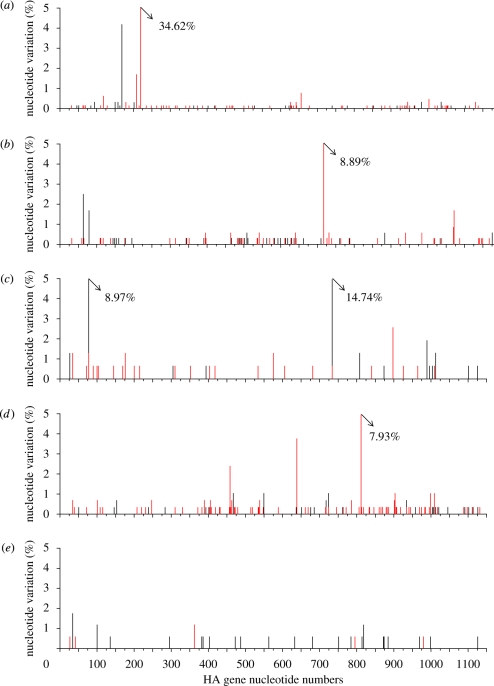
The distributions of the observed mutations across the regions analysed for each set of samples are shown in figures 1 and 2, and several sites can be seen to show a high degree of heterogeneity between clones. Examples are seen at a single but different position in the two H5N1 viruses; one site present in one-third of the clones corresponded to variation at amino acid residue 64 in Turkey/05, and in England/50-92, a high degree of heterogeneity at position 230 was seen with just under 9 per cent of the clones showing variation. In turkeys infected with Italy/984, a similar degree of heterogeneity was seen at position 264. Less amino acid variation was seen in the duck samples. A considerable level of heterogeneity was seen in cDNA clones corresponding to RNA segment 8 prepared from swabs from turkeys infected with Italy/1279 at position 163 of NS1, the substitution of Ile and Leu being highly conservative. Two other sites with 5 per cent variation were seen at positions 48 and 164 of NS1, and heterogeneity was between amino acids S48N and P164S. The significance of these minor variants is not known.
Figure 1.
Distribution of synonymous (black lines) and non-synonymous (red lines) nucleotide substitutions in the HA gene of cDNA clones amplified from swabs taken from infected birds. cDNA clones prepared from swabs taken from chickens infected with (a) A/turkey/Turkey/1/2005 H5N1 and (b) A/turkey/England/50-92/91H5N1, which were analysed between nucleotides 29–1178 of RNA segment 4; and the HA gene from swabs of birds inoculated with A/ostrich/Italy/984/2000 H7N1; (c) chickens, (d) turkeys, and (e) ducks were analysed between nucleotides 22–1133 of RNA segment 4.
Figure 2.
Distribution of synonymous (black lines) and non-synonymous (red lines) nucleotide substitutions in the NS gene nucleotides 27–718 of cDNA clones amplified from swabs taken from birds infected with (a) A/chicken/Italy/1279/99 and (b) A/turkey/Italy/3466/99, H7N1 LPAI influenza viruses.
An additional reason to examine the variation in these two LPAI viruses from the epizootic of avian influenza in Italy between 1999 and 2000 was that as the epizootic developed from one of LPAI to HPAI, viruses encoding a carboxyl-terminal deletion in the NS1 of six, and subsequently ten, amino acids emerged and predominated (Dundon et al. 2006). The two LPAI viruses examined in this study were sampled early in the epizootic (February 1999 for Italy/1279) or in the middle of the epizootic (September 1999 for Italy/3466), and both viruses retained a full-length NS1 polypeptide when isolated. In our experimental infections, we observed no cDNA clones that encoded a premature stop codon to result in a truncated NS1 polypeptide in samples taken from turkeys infected with either virus.
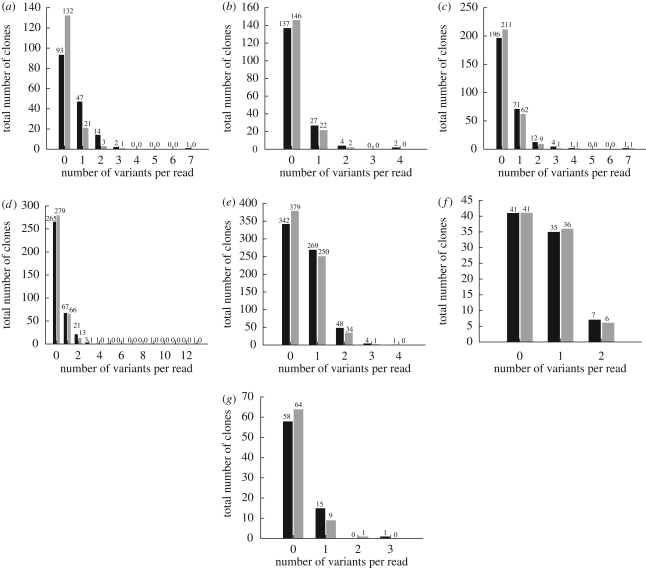
Figure 3 shows the distribution of the number of clones with alterations from the consensus sequences to estimate the proportion of clones that were identical to the consensus sequence. As discussed earlier, a high level of sequence polymorphism was seen in the amplicons from some of the infected birds and this polymorphism affects the frequency distribution plots. This is particularly conspicuous in turkeys infected with Turkey/05, but is also present but less obvious in the examples in which single sites of heterogeneity are below the 10 per cent level. It is striking that in the case of chickens infected with England/50-92, the vast majority of individual cDNA clones represented the consensus sequence, similarly for ducks infected with Italy/984. A very small number of cDNA clones show a markedly increased variation compared with the consensus: one clone from chickens infected with Italy/984 showed seven nucleotide changes, all were synonymous and there were two out of 360 cDNAs produced from swabs from England/50-92 that showed an increased number of variants. Few clones from ducks showed much variation from the consensus, but fewer cDNA clones were derived from swabs taken from this species.
Figure 3.
Graphs generated from the Ssaha2 analysis of cDNA clones of amplicons prepared from swabs taken from infected birds. The number of clones that differed from the consensus and the number of nucleotide changes (black bars) and amino acid changes (grey bars) observed in each clone are indicated. cDNA clones of the HA gene prepared from birds infected with A/ostrich/Italy/984/2000 in (a) chickens, (b) ducks and (c) turkeys. cDNA clones of the HA gene from chickens infected with (d) A/turkey/Turkey/1/2005 and (e) A/turkey/England/50-92/91. cDNA clones of RNA segment 8 prepared from turkeys infected with (f) A/chicken/Italy/1279/99 and chickens infected with (g) A/turkey/Italy/3466/99.
4. Discussion
The analysis described here set out to determine how much intrahost variation might occur with avian influenza viruses in a natural infection of birds. The importance of these observations is that as the virus evolves in one or more bird species, variant viruses may emerge rapidly from a large pool of heterogeneous viruses generated during an infection. Should the divergent pool be subject to selective pressures, such as by immune selection in immune or partially immune individuals or be exposed to antiviral drugs, then antigenic drift of viruses or drug-resistant viruses may rapidly accumulate. Our study here did not measure selective drift in immunologically experienced hosts or in the presence of antiviral drugs, but can serve as a reference to the background variation that might be observed in studies involving natural infection: the work described here may have been influenced by the infectious dose used, the route of infection or prior passage of the virus prior to experimental inoculation. We observed that, in many cases, few nucleotide changes were seen, although viruses exhibited some degree of heterogeneity at specific positions; we have not studied the significance of the changes that we have observed thus far. It is noticeable that some individual clones showed more than one to three nucleotide differences from the consensus sequence of the amplicons.
We have to address whether the limited degree of variation observed reflects the true variation seen in the sample or whether the observations of heterogeneity may be increased as an artefact of manipulation of the RNAs. We cannot comprehensively address this point, but have tried to minimize the degree of artefactual error by using an improved procedure for RT and PCR, with an optimized RT step followed by PCR using a proof-reading DNA polymerase. We have not estimated the error rate of reverse transcription in our process, but did estimate that the errors in the PCR steps are low (H. Xiao & J. W. McCauley 2009, unpublished data). Should sequence variation be caused by errors introduced during manipulation of the genomes, then the real frequency of variation may be considerably lower than our estimates and reflect much less heterogeneity of avian influenza viruses in the infected avian host.
The degree of variation found, taken without consideration of whether the variant was introduced during preparation or was present in the virus particle, resulted in approximately 50 per cent of the cDNA clones containing a variant nucleotide and the variant frequency was of the order of 8.9 × 10−4 to 1.6 × 10−4. If these estimates are similar across the genome, the numbers indicate that within a genome of virus grown in vivo, there may be only one or two nucleotides variant from the consensus in each virus particle; should the population be considered as a quasi-species, then the degree of variation within the proposed virus swarm may be very limited.
The cDNA clones we observed at low frequency with a high number of mutations were, we believe, not caused by an artefact, but may represent minor species of virus in the population. Relevant to this are observations described by Hulse-Post et al. (2005), who found that following infection of ducks with viruses isolated early in the H5N1 panzootic in 2003 and 2004, the viruses showed varying pathogenicity for ducks and that distinctly different viruses emerged with alterations in the HA polypeptide during the course of infection. The samples from which variants emerged were from ducks, humans and chickens. The authors considered the possibility that the original inoculum was mixed, but concluded that prolonged shedding may be an important factor in selection when ducks are infected with a mixed population of viruses. In the cases studied here, prolonged shedding was seen only in ducks infected with Italy/984, the HPAI H7N1 virus, and in chickens infected with the two LPAI H7N1 viruses. The variants observed in the two H5N1 viruses and Italy/984 in chickens and turkeys were detected very quickly after infection, as the birds succumbed to infection rapidly and so we conclude that the variants were present in the inoculum; some of which were confirmed by in-depth sequence analysis of the cDNA produced from the virus inoculum (data not shown).
The methods described in this study for the analysis of RNA extracted directly from swabs were used on experimentally infected birds and provide a baseline estimate of the natural variability of viruses in the field during an outbreak. In an outbreak, especially in an enzootic area, the situation may be very different from our observation of limited virus heterogeneity, and several different variants of influenza virus may simultaneously infect any individual in a flock. This situation of mixed infections may not be easy to investigate retrospectively since swabs may have been pooled during investigations of outbreaks, the focus usually being on the status of the flock rather than an individual bird. In addition, during an outbreak, several different host species may be infected, resulting in selection pressures that may be influenced by the direction of transmission between different host populations. Methods are now available to investigate the micro-evolution of avian influenza within a flock and within an area. As new methods using the next generation sequencing technologies become more extensively used in RNA virus studies, even more analysis of the diversity of viruses within an infected individual can be initiated. Heterogeneity between complete genome sequences can be studied within a single infected individual, and many aspects of virus adaptation to a new host, to anti-viral drug treatment or to immunological pressure may soon be able to be easily quantified.
Acknowledgements
All animal experiments and procedures using live virus were carried out in Defra-approved SAPO-4/ Biosafety Level 3+ (BSL 3+) facilities at VLA-Weybridge.
This work was supported by BBSRC (including grants nos BBS/B/00093 and BB/E010806), the Wellcome Trust, the Medical Research Council and Defra project SE 0776; the authors would also like to offer thanks to the VLA staff involved in the in vivo work required to generate the samples and carry out the preliminary Real Time RT/PCRs.
Footnotes
One contribution of 12 to a Theme Issue ‘Livestock diseases and zoonoses’.
References
- Chen H., et al. 2006Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl Acad. Sci. USA 103, 2845–2850 (doi:10.1073/pnas.0511120103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas E. C., Osterhaus A. D., van Beek R., De Jong J. C., Rimmelzwaan G. F., Senne D. A., Krauss S., Shortridge K. F., Webster R. G.1998Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351, 472–477 (doi:10.1016/S0140-6736(97)11212-0) [DOI] [PubMed] [Google Scholar]
- Duan L., et al. 2008The development and genetic diversity of H5N1 influenza virus in China, 1996–2006. Virology 380, 243–254 (doi:10.1016/j.virol.2008.07.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez M. F., Olinger C. M., Owoade A. A., De Landtsheer S., Ammerlaan W., Niesters H. G., Osterhaus A. D., Fouchier R. A., Muller C. P.2006Avian flu: multiple introductions of H5N1 in Nigeria. Nature 442, 37 (doi:10.1038/442037a) [DOI] [PubMed] [Google Scholar]
- Ducatez M. F., et al. 2007aMolecular and antigenic evolution and geographical spread of H5N1 highly pathogenic avian influenza viruses in western Africa. J. Gen. Virol. 88, 2297–2306 (doi:10.1099/vir.0.82939-0) [DOI] [PubMed] [Google Scholar]
- Ducatez M. F., et al. 2007bGenetic characterization of HPAI (H5N1) viruses from poultry and wild vultures, Burkina Faso. Emerg. Infect. Dis. 13, 611–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundon W. G., Milani A., Cattoli G., Capua I.2006Progressive truncation of the non-structural 1 gene of H7N1 avian influenza viruses following extensive circulation in poultry. Virus Res. 119, 171–176 (doi:10.1016/j.virusres.2006.01.005) [DOI] [PubMed] [Google Scholar]
- European Union 2006Diagnostic manual for avian influenza. See http://eur-lex.europa.eu/LexUriServ/site/en/oj/2006/l_237/l_23720060831en00010027.pdf (accessed 1 April 2009) [Google Scholar]
- Hars J., et al. 2008The epidemiology of the highly pathogenic H5N1 avian influenza in Mute Swan (Cygnus olor) and other Anatidae in the Dombes region (France), 2006. J. Wildl. Dis. 44, 811–823 [DOI] [PubMed] [Google Scholar]
- Hesterberg U., Harris K., Stroud D., Guberti V., Busani L., Pittman M., Piazza V., Cook A., Brown I.2009Avian influenza surveillance in wild birds in the European Union in 2006. Influenza Other Respir. Viruses 3, 1–14 (doi:10.1111/j.1750-2659.2008.00058.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C., Moya A.2002Is the quasispecies concept relevant to RNA viruses? J. Virol. 76, 460–465 (doi:10.1128/JVI.76.1.460-462.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse-Post D. J., et al. 2005Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc. Natl Acad. Sci. USA 102, 10 682–10 687 (doi:10.1073/pnas.0504662102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keawcharoen J., van Riel D., van Amerongen G., Bestebroer T., Beyer W. E., van Lavieren R., Osterhaus A. D., Fouchier R. A., Kuiken T.2008Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14, 600–607 (doi:10.3201/eid1404.071016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K., et al. 2008Pathogenicity and vaccine efficacy of different clades of Asian H5N1 avian influenza A viruses in domestic ducks. J. Virol. 82, 11 374–11 382 (doi:10.1128/JVI.01176-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. S., et al. 2004Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430, 209–213 (doi:10.1038/nature02746) [DOI] [PubMed] [Google Scholar]
- Londt B. Z., Nunez A., Banks J., Nili H., Johnson L. K., Alexander D. J.2008Pathogenesis of highly pathogenic avian influenza A/turkey/Turkey/1/2005 H5N1 in Pekin ducks (Anas platyrhynchos) infected experimentally. Avian Pathol. 37, 619–627 (doi:10.1080/03079450802499126) [DOI] [PubMed] [Google Scholar]
- Maynard Smith J.1989Evolutionary genetics, 1st edn Oxford, UK: Oxford University Press [Google Scholar]
- Monne I., et al. 2008Co-circulation of two sublineages of HPAI H5N1 virus in the Kingdom of Saudi Arabia with unique molecular signatures suggesting separate introductions into the commercial poultry and falconry sectors. J. Gen. Virol. 89, 2691–2697 (doi:10.1099/vir.0.2008/004259-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Holmes E. C., Gonzalez-Candelas F.2004The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2, 279–288 (doi:10.1038/nrmicro863) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z., Cox A. J., Mullikin J. C.2001SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (doi:10.1101/gr.194201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE 2009Update on avian influenza in animals (type H5). See http://www.oie.int/downld/AVIAN%20INFLUENZA/A_AI-Asia.htm (accessed 01 April 2009) [Google Scholar]
- Smith D. B., McAllister J., Casino C., Simmonds P.1997Virus ‘quasispecies’: making a mountain out of a molehill? J. Gen. Virol. 78, 1511–1519 [DOI] [PubMed] [Google Scholar]
- Smith G. J., et al. 2009Characterization of avian influenza viruses A (H5N1) from wild birds, Hong Kong, 2004–2008. Emerg. Infect. Dis. 15, 402–407 (doi:10.3201/eid1503.081190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R.1996The Staden sequence analysis package. Mol. Biotechnol. 5, 233–241 (doi:10.1007/BF02900361) [DOI] [PubMed] [Google Scholar]
- Sturm-Ramirez K. M., et al. 2005Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J. Virol. 79, 11 269–11 279 (doi:10.1128/JVI.79.17.11269-11279.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D., Bahl J., Riley S., Duan L., Zhang J. X., Chen H., Peiris J. S., Smith G. J., Guan Y.2008Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 4, e1000161 (doi:10.1371/journal.ppat.1000161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO 2005WHO global influenza preparedness plan. See http://www.who.int/csr/resources/publications/influenza/GIP_2005_5Eweb.pdf [DOI] [PubMed] [Google Scholar]
- WHO 2009H5N1 avian influenza: timeline of major events. See http://www.who.int/csr/disease/avian_influenza/Timeline_09_03_23.pdf [Google Scholar]
- WHO/OIE/FAO, H5N1 Evolution Working Group 2008Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1) [conference summary]. Emerg. Infect. Dis. 14 See http://www.cdc.gov/EID/content/14/7/e1.htm (doi:10.3201/eid1407.071681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO/OIE/FAO, H5N1 Evolution Working Group 2009Continuing progress towards a unified nomenclature for the highly pathogenic H5N1 avian influenza viruses: divergence of clade 2·2 viruses. Influenza and Other Respiratory Viruses 3, 59–62 (doi:10.1111/j.1750-2659.2009.00078.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasué M., Feare C. J., Bennun L., Feidler W.2006The epidemiology of H5N1 avian influenza in wild birds: why we need better ecological data. Bioscience 56, 1–7 (doi:10.1641/0006-3568(2006)56[923:TEOHAI]2.0.CO;2) [Google Scholar]