Abstract
Individual pairs of polymer tethered silver nanoparticles, so called silver plasmon rulers, enable distance and orientation measurements on the nanoscale. The reduced linear dichroism and the spectrum of the light scattered from individual plasmon rulers encode information about their orientation and average interparticle separation, respectively. We took advantage of the gain in information silver plasmon rulers offer as probes in optical tracking and analyzed the translational and rotational motions as well as the extension of individual silver plasmon rulers diffusing on the plasma membrane of lysed HeLa cells. Consistent with a compartmentalization of the cell surface on the length scales of the plasmon rulers, most rulers were either immobilized or performed a confined lateral diffusion. Structural details of a plasmon ruler’s confinement region became accessible utilizing the orientation and interparticle separation dependent optical response of the plasmon rulers. This approach, which we refer to as polarization resolved plasmon coupling microscopy, enabled a detailed structural characterization of individual membrane compartments and provided a quantitative metrics to characterize the structural lateral heterogeneity of cell membranes on sub-micron length scales. In combination with adequate tracking methods, the “dance” performed by membrane confined dimers of flexibly linked noble metal nanoparticles revealed deep insight into the underlying membrane morphology.
I. Introduction
The plasma membrane of mammalian cells is a complex hybrid material that contains approximately equal contributions in weight from both proteins and lipids. It represents a two dimensional diffusion system, in which the spatial organization and dynamics of both lipids and proteins plays an important role in regulating signal transduction and membrane traffic.1 Some of the biological functionality of the membrane is enabled by its spatial organization and different elements have been identified to play a role in structuring the membrane.2 Single particle tracking experiments have shown that the plasma membrane is compartmentalized; phospholipids3 and membrane proteins4 perform a “hop” diffusion between different compartments in the membrane. The observed compartmentalization of the membrane on sub-micron length scales was attributed to the underlying cortical actin network that defines fences or corrals in the membrane and that can serve as scaffold for anchored transmembrane proteins in an anchored membrane-protein picket model3. Specialized scaffolding molecules associated with the membrane have also been identified to organize various components on the plasma membrane involved in important cellular activities such as exocytosis and endocytosis.5
Another potential cause for lateral heterogeneity in biological membranes arises from the self-organization of the lipids in the membrane.6–9 There is evidence that the coexistence of liquid ordered (lo) and liquid-disordered (ld) domains provides additional spatial organization of biological membranes.10 A partitioning of the membrane in lo and ld domains and a selective recruitment of membrane proteins into these domains could play an active role in signaling processes by facilitating direct interactions between the receptors in areas of high local concentration.11 The size, lifetime, and mechanism of the formation of the lo domains, remain, however, matters of intense debate. The latter is partly due to a current lack of adequate tools for probing the dynamic membrane structure on nanometer length scales and thus motivates the development of dynamic high resolution cell membrane profiling approaches.12
Important insights into the organization of cellular surfaces has been deduced from particle tracking experiments.13–17 In fact, it was only due to the fast temporal resolution accessible in single particle tracking that it became possible to detect the hop motion of individual lipid molecules and membrane proteins in the plasma membrane of mammalian cells. These observations eventually led to the formulation of the anchored membrane-protein picket model.3
One limitation of conventional high-speed particle tracking is that individual nanoparticle labeled surface species can no longer be resolved once they approach each other to within the diffraction limit of ~400 nm in the visible. We have recently shown that this limitation can be overcome by utilizing the distance dependence of the plasmon coupling between individual noble metal nanoparticles.18 If two noble metal nanoparticles approach each other to within ~2 particle diameters the individual particle plasmons start to couple, and the resonance wavelength red-shifts with decreasing interparticle separation.19, 20 Spectral shifts that result from direct near-field interactions between individual diffusing particles at constant refractive index can be detected as a ratiometric shift, provided that the particles are simultaneously tracked on two color channels. We have used this approach, which we called plasmon coupling microscopy (PCM), to resolve close contacts between individual nanoparticle labeled fibronection-integrin complexes.18 Since the optical signal of individual nanoparticles in PCM is based on light scattering, the particles don’t blink or bleach. In contrast to fluorescence based optical superresolution methods,21–24 PCM is not limited by the photophysical instabilities of fluorescent dyes and can - in principle - monitor nanometer length scale distances between individual nanoparticle labelled surface groups with high temporal resolution for an unlimited time.25–27
PCM is not limited to probe interactions between individual nanoparticle probes. Very recently Aaron et al. used a plasmon coupling based imaging approach to monitor global receptor regulation states in living cells.28 The ability to study interactions between individual nanoparticle labeled surface groups with PCM is, however, extremely useful for many mechanistic studies, especially if one considers that plasmon coupling does not only affect the spectrum of the scattered light but also its polarization.29 Unlike for individual spherical particles, which scatter light of all polarizations with equal probability, the light scattered from strongly coupled noble metal nanoparticles is polarized with the polarization of the scattered light pointing into the direction of the long dimer axis.
In this manuscript we take advantage of the distance and orientation dependent optical properties of pre-assembled dimers of flexibly linked silver nanoparticles, so called silver plasmon rulers,30, 31 as probes in particle tracking. Our aim is to apply these active nanostructures to map the two-dimensional organization of mammalian plasma membranes on sub-micron length scales. Plasma membranes are compartmentalized and the individual compartments of a cell membrane represent potential traps for the silver plasmon ruler probes. We use spherical silver nanoparticles with an average diameter of ~30 nm and an equilibrium separation in solution of ~14 nm in this work. The average compartment size in HeLa cells has been reported to be on the order of ~70 nm.32 Since the average interparticle separation and rotational freedom of the plasmon rulers in compartments of this size is expected to depend on structural details of the confinement (see Figure 1), the probes represent promising tools for a two-dimensional profiling of individual membrane compartments.
Figure 1.
Pairs of individual polymer tethered nanoparticles serve as probes in polarization resolved plasmon coupling microscopy (PRPCM) to probe the spatial organization of the plasma membrane. Analysis of the motion of diffusing nanoparticles on a cell membrane through conventional particle tracking provides information about the membrane compartmentalization. For dimers that are trapped in attractive sites on the membrane surface, the wavelength and the polarization of the scattered light provide additional information about the interparticle separation and the rotational freedom of the dimers in these sites. The average interparticle separation and mobility of the plasmon rulers is expected to decrease with increasing confinement as illustrated for examples a) and b).
II. Results and Discussion
Noble metal nanoparticles with diameters > 20 nm are efficient light scatters25, 26 and can be localized with high spatial precision in very short integration times, 27,33,34 provided that the scattering background is low. This requirement is not necessarily fulfilled for individual particles bound to living cells. Different cellular components, in particular the nucleus, create a high scattering background which makes high-speed tracking (in this study with frame rates up to 500 Hz) of small nanoparticle probes challenging. To maximize the signal-to-noise in single plasmon ruler tracking, all experiments in this work were therefore performed on membranes of lysed cells. Removal of the cell nucleus led to a significant reduction of the scattering background.
We prepared plasma membranes of cervical cancer (HeLa) cells immobilized on a glass support through acoustic rupturing35 of cells grown on No1 coverglass slides. In Figure 2 we show fluorescence and darkfield scattering images of a resulting membrane after treatment with fluorescently-tagged phalloidin. Phalloidin binds selectively at the interface of F-actin subunits and is commonly used as stain for actin filaments.36 The cortical actin, which plays a prominent role for the spatial organization of the membrane, is clearly visible in Figure 2a as dense network of branched filaments below the inner leaflet of the plasma membrane.
Figure 2.

a) Fluorescence and b) darkfield images of a cell membrane after acoustic rupturing and staining with fluorescently tagged phalloidin. The phalloidin binds specifically to F-actin subunits and stains the actin network supporting the membrane in the fluorescent image.
Our cell membrane in vitro model has two important advantages over conventional membrane models: i) it includes the underlying actin network that supports and structures the membrane and ii) the membranes have the same composition as intact cellular membranes. However, like for any in vitro system, our model system does not have the full functionality of the living cell. Despite this limitation, the lysed cells remain a useful model for the development of the proposed cell surface profiling approach and provide valuable insight into the compartmentalization of cell membranes on sub-micron length scales.
The signal-to-noise in particle tracking depends on the brightness and contrast of the individual particles. Ideal probes for PCM have large scattering cross-sections in spectral ranges with low background from the cellular membrane, sharp resonances and steep resonance wavelength (λres) vs. interparticle separation (S) relationships. We decided to use silver and not gold plasmon rulers in our studies since silver nanoparticles of ~30 nm diameter have their resonances in the blue region of the electromagnetic spectrum where the scattering background from cellular membranes is lower (see Figure S1) than in the green where the resonance of gold nanoparticles with comparable size lie. Silver plasmon rulers also have a larger dynamic range in the distance dependent spectral response than gold plasmon rulers and are therefore more sensitive for detecting small distance changes.30
Silver nanoparticles are in general less stable in salt buffers than gold nanoparticles and undergo oxidative corrosion.37, 38 Chemical self-assembly procedures for silver nanoparticles require therefore sufficient stabilization of the particles through appropriate surface ligands. We used a monolayer of thiolated alkyl-[polyethylene glycol(PEG)]-acetate (HS-(CH2)5-(OCH2CH2)6-OCH2-COOH) to render the silver nanoparticles stable in Hank’s buffered solution (137.93 mM NaCl, 5.33 mM KCl, 4.13 mM NaHCO3, 0.441 mM KH2PO4, 0.338 mM Na2HPO4, 5.56 mM D-Glucose). The stabilized silver particles were then assembled into dimers using a DNA programmed self-assembly strategy. Details regarding the assembly and characterization of silver plasmon rulers containing nanoparticles with an average diameter of d = 30 ± 4 nm (as obtained by transmission electron microscopy (TEM)) are summarized in the Methods Section of the Supporting Information. The resulting preparations contained typically ~75% plasmon rulers.
The PEG stabilized silver plasmon rulers attached efficiently and non-reversibly to freshly prepared plasma membranes and could be tracked with frame rates of up to 500 Hz with signal-to-noise ratios > 3. All experiments were performed in home-built flowchambers at 25°C.
Polarization Resolved and Ratiometric Tracking of Silver Plasmon Rulers
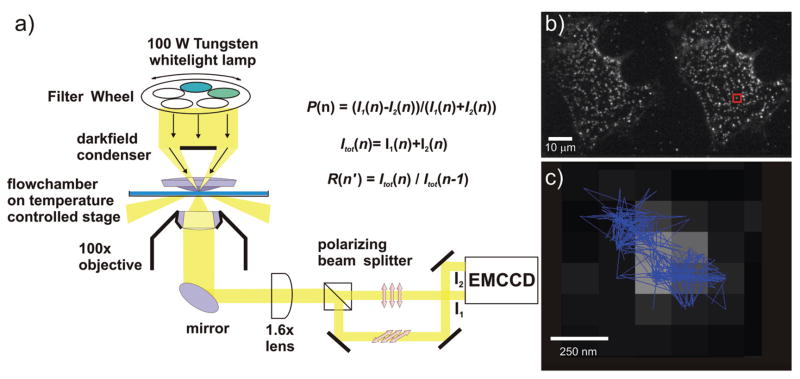
Our aim was to implement a tracking scheme that yields information not only about the plasmon rulers’ spatial coordinates but also about the polarization as well as the resonance wavelength of the light scattered from individual silver plasmon rulers. Our experimental set-up is based on a conventional inverted darkfield microscope (Olympus IX71) and is illustrated in Figure 3a. Unpolarized whitelight from a 100 W tungsten lamp passes a filter wheel (Lambda 10–3, Sutter Instrument), which can toggle between two different monochromatic filters. The light is then injected into the specimen plane at oblique angles using an oil darkfield condenser (Numerical Aperture (NA) = 1.2). This illumination geometry ensures that only light that is scattered in the specimen plane is collected by the 100× oil immersion objective (NA = 0.6). The collected light passes another 1.6× magnification lens and is then split into two images with orthogonal light polarizations using a polarizing beam splitter and finally re-imaged on two translated areas on the same electron multiplying charge coupled device (EMCCD, Andor iXon+). The effective pixel size in this imaging detector is 160 nm.
Figure 3.
Experimental set-up for polarization resolved plasmon coupling microscopy (PRPCM). a) In a microscope with darkfield configuration the samples are illuminated with unpolarized light with alternating excitation wavelengths. The light scattered from individual nanoparticle dimers is collected with a 100x objective and then split into two othogonal polarization channels which are reimaged on two translated regions of an electron multiplying charged coupled device (EMCCD). The intensities I1 and I2 on the two polarization channels in each frame n are used to calculate the reduced linear dichroism P and the total intensities of two subsequent frames are used to calculate the intensity ratio R. b) Image of silver plasmon rulers bound to a HeLa membrane on two orthogonal polarization channels. c) Trajectory of an individual plasmon ruler marked in b). The figure shows the scattering image at t = 0 and the fitted position of the maximum as function of time as blue trace.
Figure 3b shows an image of silver dimers on a cell membrane simultaneously recorded on two orthogonal polarization channels. Individual dimers were simultaneously tracked on both channels by fitting39 the dimer image on two polarization channels I1 and I2. Each channel was background corrected by the scattering intensity of the membrane in that channel, and we used the maxima of the curve-fitted39 images, or point-spread-functions, of the individual dimers to determine their coordinates. Under typical experimental conditions we achieve a localization precision of σ = 60 nm.
The trajectories of the individual dimers were obtained by linking the coordinates of the maxima over time using a custom written MATLAB program that was based on the nearest-neighbor-tracking algorithm developed by Wieser and Schuetz.40 The intensities, I1(n) and I2(n) of the plasmon rulers were then obtained by integrating over the point-spread-functions on the two polarization channels in each frame n.
The blow-up in Figure 3c displays the trajectory of a single plasmon ruler diffusing on the cell membrane recorded on one of the polarization channels. A second trajectory of the same plasmon ruler was simultaneously recorded on the second polarization channel. The relative intensity of the plasmon rulers on the two channels encodes information about the orientation of the long dimer axis as a function of space and time. The integrated intensities I1(n) and I2(n) of a dimer on the two polarization channels are used to calculate the reduced linear dichroism41 P(n) = (I1(n)−I2(n))/(I1(n)+I2(n)) for a dimer in each frame n. In anisotropic noble metal nanostructures such as dimers29 and nanorods42 P depends on the orientation of the long dimer axis and can therefore be used to monitor the rotation motion of those nanostructures on the membrane surface.
The possibility of alternating the excitation wavelength between subsequent frames through toggling between different bandpass filters allows a ratiometric tracking of individual plasmon rulers. Ratiometric spectral information are obtained by computing the intensity ratios R(n′) = Itot(n)/Itot(n−1) from the total intensities Itot(n)=I1(n)+I2(n) of individual plasmon rulers in two subsequent frames n−1 and n for the entire movie. The new index n′ is defined as n′ = n/2; n was chosen to refer to the frame recorded with the longer wavelength. This approach generates R values for all plasmon rulers tracked on the cell surface as a function of time. The sensitivity of the ratiometric tracking approach for detecting distance changes in plasmon rulers depends on the interparticle separation and the choice of the filter combination. We used combinations of bandpass filters that covered the spectral range between 430 nm (corresponding to ~λres of an individual particle) and 530 nm (corresponding to ~λres of a strongly coupled dimer).
The implemented ratiometric tracking scheme using alternating excitation wavelengths is intended to augment a simultaneous polarization resolved tracking of silver plasmon rulers and is referred to as polarization resolved plasmon coupling microscopy (PRPCM). Polarization resolved tracking was performed with temporal resolutions of up to 500 Hz. This temporal resolution was accessible for measurements of P only without acquiring additional spectral information. The maximum temporal resolution accessible in PRPCM for measurements of R was significantly lower and limited by the mechanics of the filterwheel to ~5 Hz. This is, however, not an intrinsic limitation of the technology; replacement of the filterwheel, for instance, with two shuttered monochromatic lasers would allow significantly higher frame rates. In this work we limited ourselves, however, to the temporal resolution available with the instrumentation readily available in our laboratory. We emphasize that the current implementation of PRPCM is not well suited to monitor rapid fluctuations in the interparticle separation. Instead, it provides reliable estimates of the time-averaged interparticle separation in individual diffusing plasmon rulers.
Diffusion Modes of Silver Plasmon Rulers on HeLa Cell Membranes
In “normal” Brownian lateral diffusion the mean square displacement (MSD) of an object grows linearly as a function of time (t): MSD ~ t. A cell membrane is, however, heterogenous and contains traps for the nanoparticles with varying potential depths spatially and temporally distributed over its entire surface. These traps influence the dimer motion and can result in an anomalous diffusion in which the diffusion coefficient becomes a function of time. In this case the mean square displacement (MSD) grows nonlinearly and is described by a power law in t.43, 44
To characterize the translational mobility of silver nanoparticle dimers on different time scales we apply here the moment scaling spectrum introduced by Ferrari et al.45 This analysis is based on the calculated moments of displacements,45, 46 where a moment μ of order ν for a specific frame shift Δn corresponding to a time shift δt = Δn Δt (Δt is the fixed time interval between two frames), is defined as:
where x⃗l (n) is the position vector (xl(n),yl(n)) on trajectory l at t = nΔt. Assuming a power law of the form μν(δt) ∝ δtγν,45 the scaling coefficients γν are determined through linear fits to double-logarithmic plots of μν versus δt and the two dimensional diffusion coefficients (Dν) of order ν > 0 are obtained from the ordinate intercepts It as Dv = (2v)−1 exp(It).46 The linear diffusion coefficient is a special case in this treatment and obtained for ν = 2.
The γν versus ν relationship is the moment scaling spectrum and its slope (SMSS) characterizes the mobility of the silver nanoparticle dimers on the membrane. SMSS values of 0, 0.5 and 1 correspond to immobilized, free and directed (ballistic) diffusion, respectively.46 SMSS values between 0 and 0.5 indicate confined motions, whereas slopes in the range 0.5 – 1 indicate motions that are in the super-diffusive regime.46
The moment scaling approach provides a quantitative measure that enables a systematic comparison of a large number of single dimer trajectories with each other.47 In Figure 4a we plot the fitted SMSS values against the respective linear diffusion coefficients D2 for a total of 70 trajectories. The plot summarizes the results obtained for polarization resolved tracking (temporal resolution 500 Hz, black filled circles) as well as for polarization resolved ratiometric tracking (temporal resolution 5 Hz, red filled circles).
Figure 4.
a) Slope of the moment scaling spectrum (SMSS) versus linear diffusion coefficient (D2). Black filled circles were obtained from tracks of individual plasmon rulers recorded in a polarization resolved fashion with high temporal resolution (500 Hz). The red filled circles belong to trajectories obtained through polarization resolved ratiometric tracking with a temporal resolution of 5 Hz. Plasmon rulers PR1-3 are analyzed in more detail in the text. b) Rotational correlation time (τ) versus D2 for the data set recorded with 500 Hz.
The silver plasmon rulers on the cellular membrane show a range of different lateral diffusion modes. For D2 < 2×10−3 μm2/s the measured SMSS values are low and independent of the diffusion coefficient but for D2 > 2×10−3 μm2/s, SMSS grows with increasing D2. Silver plasmon rulers with diffusion coefficients in the range D2 = 1×10−5 – 2×10−3 μm2/s have very low SMSS values (SMSS < 0.05) and are considered to be immobilized. Plasmon rulers with D2 = 2×10−3 – 7×10−2 μm2/s and SMSS = 0.05 – 0.40 perform confined diffusion and plasmon rulers with D2 = 7×10−2 – 2×10−1 μm2/s and SMSS = 0.40 – 0.52 perform quasi-free diffusion. Overall, our analysis of the lateral mobility of the silver plasmon rulers on the cell membrane shows that most of the probes are either immobilized or perform a confined diffusion.
The performed polarization resolved tracking studies enabled us to evaluate the rotational mobility of the plasmon rulers as a function of D2, as well. To that end we fitted a single exponential to the autocorrelation of P and obtained a characteristic rotational correlation time τ from the fit (see Figure S2). The resulting τ values are plotted against D2 in Figure 4b. While for D2 < 2×10−3 μm2/s most of the measured diffusion times are below 2.5×10−2 s, for D2 > 2×10−3 μm2/s we find a broader distribution of τ values with τ > 2.5×10−2 s. The essential absence of autocorrelation in P for D2 < 2×10−3 μm2/s suggests that these plasmon rulers are immobilized and that the long dimer axis is fixed in space. The higher τ values for plasmon rulers with D2 > 2×10−3 μm2/s, on the other hand, indicate two-dimensional rotational motions that are characterized by non-zero rotational correlation times.
The broad spread of the τ values for D2 > 2×10−3 μm2/s shows that the rotational dynamics differs substantially between individual plasmon rulers. Our analysis of the plasmon rulers’ lateral diffusion revealed that SMSS increases continuously with growing D2 for D2 > 2×10−3 μm2/s indicating a steady increase in the lateral mobility with growing linear diffusion coefficient. There is no comparable strong correlation between τ and D2 for the rotational dynamics. Instead, the τ values are broadly distributed for D2 > 2×10−3 μm2/s. In principle, this distribution could arise from impurities of our plasmon ruler preparations, which always contain some monomers. However, since the light scattered from an individual spherical particle is not polarized, a contamination with monomers cannot account for the differences in the rotational dynamics in the range τ = 0.025 – 0.38 s for D2 > 2×10−3 μm2/s. Instead, it is more probable that the distribution of the τ values arises from the heterogeneity of the cell membrane due to local differences in the chemical or structural nano-environment of the individual plasmon rulers. Both of these factors would directly affect the rulers’ rotational dynamics.
Monitoring Orientation and Interparticle Separation of Two-dimensional Confined Silver Plasmon Rulers: Watching a Nanoparticle Minuet in Real Time
Deeper insight into the lateral heterogeneity of the plasma membrane is potentially available through PRPCM. For those plasmon rulers that were tracked by polarization resolved ratiometric tracking (red filled circles in Figure 4a), the measured P and R values offer additional information about the interparticle separation and the orientation of the dimer axis on the membrane. The R values can be converted into approximate interparticle separations S, provided that an S(R) calibration relationship is available. We derived such relationships for different filter combinations from scattering spectra of silver dimers with known interparticle separation S (see Figure S3). Our experimental approach of correlating single dimer Rayleigh scattering spectra of individual silver plasmon rulers with their interparticle separation as obtained by TEM followed essentially the experimental procedures described in ref.48. In this work we only apply the S(R) calibrations shown in Figure S3, a detailed analysis of the distance dependent plasmon coupling will be published elsewhere.49
The S(R) calibration was performed with silver nanoparticles that were immobilized on formvar (nr ≈ 1.45) and immersed in a refractive index of nr = 1.47, whereas the membranes (nr ≈ 1.40) in the tracking experiments are sandwiched between glass (nr ≈ 1.50) and buffer solution (nr ≈ 1.34). The difference in the refractive index between these two experimental conditions will affect the S(R) calibration and lead to a systematic error in the derived distances.50 Another source of error for the conversion of R into S values are particle-to-particle variations in particle size and shape, which also directly influence the resonance wavelength.51 The above potential error sources currently limit the accuracy of PRPCM. Nevertheless, even the ability to estimate interparticle separations and to correlate these approximates with the silver plasmon ruler orientation provides valuable insight into the organization of the membrane as a whole as we demonstrate in the following.
Figure 5 illustrates the information content accessible in a typical PRPCM experiment. The figure contains the diffusion tracks and the corresponding P, R, and S values as a function of time for the plasmon rulers labeled as PR1-3 in Figure 4a. We chose PR1-3 for the following analyses since they cover a broad range of translational diffusion modes observed in our experiments. PR1 was recorded with a bandpass (BP) filter combination comprising 430BP10 (center wavelength: 430 nm, spectral width: 10 nm) and 470BP10, PR2&3 were recorded using a combination of 450BP10 and 490BP10. The plotted P values refer to the reduced linear dichroism obtained in the long wavelength channel.
Figure 5.
Polarization resolved ratiometric tracking of three different silver plasmon rulers: a) PR1, b) PR2 c) PR3. The panel on the left shows the track of the plasmon ruler on the cell membrane. The right panels contain the intensity ratio of the monitored wavelength channels (R, red), the derived approximate interparticle separation (S, blue) and the reduced linear dichroism (P, black) as function of time during the plasmon ruler’s diffusion on the cell membrane. For S we included a 5-point sliding average as solid blue line.
PR1 (Figure 5a) is confined but exhibits the highest degree of translational mobility among PR1-3. The approximate average interparticle separation for PR1 is obtained as the average of all individual S measurements in the trajectory as Sav = 13.4 ± 2.8 nm, which is in agreement with the expected end-to-end distance of the DNA tether (S0 = 13.7 nm, see Methods Section in the Supporting Information). On shorter time scales we observe larger systematic changes in the 5-point sliding average of S resulting from variations in the interparticle separation during the plasmon ruler’s diffusion in the confinement region.
The recorded P values for PR1 show a large relative spread and fluctuate seemingly randomly in the range between −0.4 and +0.5. PR1-3 were tracked with temporal resolutions of 5 Hz, but even at this relatively low temporal resolution the orientation dependence of the polarization is not “averaged out”. The observation of a large dynamic range in P indicates a rotational motion in which longer static phases with fixed orientation of the long dimer axis alternate with rotational phases in which the plasmon rulers rapidly find a new orientation on the surface. We refer to this rotational motion as “hop” rotation in the following.
PR2 in Figure 5b explores a smaller area of the cell membrane than PR1. The stronger spatial confinement in case of PR2 coincides with a decrease in the average interpartice separation to Sav = 11.7 ± 2.3 nm. The fluctuations in P observed in the first 80 seconds of the trajectory of PR2 in Figure 5b are again characteristic of a rotational motion in which the long dimer axis hops between discrete angles on the cell surface. This behavior changes during the interval t ≈ 80 – 92 s in which the P values remain close to zero. Since plasmon rulers have non-zero scattering cross-sections along both the long and the perpendicular short dimer axis, the observation of P ≈ 0 is attributed to a rotation of the long dimer axis that is fast on the timescale of the measurement. In this case all plasmon ruler orientations contribute with equal probability to the measurement resulting in no net polarization. The onset of the rapid rotational motion at t = 80 s is accompanied by a systematic increase in S by approximately 2 nm and a decrease in the point-to-point fluctuations of S. These observations indicate an increase in the translational and rotational mobility of the plasmon rulers which could result from heterogeneities within the confinement region or changes of its structure as a function of time.
The diffusion track of PR3 in Figure 5c is spatially more confined than those of PR1&2 and the average interparticle separation obtained from the measured R values is further decreased to Sav = 9.7 ± 1.3 nm. PR3 still exhibits some rotational dynamics, most prominently in the interval t = 58 – 115 s, during which plasmon ruler rotation is correlated with a change in the interparticle separation. Additional more transient orientation changes can be identified at other times throughout the trajectory. All of the recorded P values lie, however, in the range of P ≈ 0 – 0.6. The lower magnitude and frequency of the P fluctuations for PR3 when compared with PR1&2 indicates a reduction of the rotational freedom for PR3.
Overall, we conclude that for PR1-3 in Figure 5 an increasing spatial confinement is correlated with a decreasing rotational mobility and decreasing average interparticle separation.
Next, we want to analyze the confinement of PR3 in more detail. We already know from Figure 5c that PR3 explores a total area on the order of 300 × 300 nm2 during our observation time and that the orientation of the long plasmon ruler axis is constrained to P ≈ 0 – 0.6 during the entire observation time. Since S and P were obtained from tracking experiments they can be analyzed as a function of time as shown in Figure 5 and as a function of their location on the cell surface. In Figure 6 we performed such a spatial analysis and created S and P maps for two different definition ranges. Whereas Figure 6a1 includes all S values of the trajectory, Figure 6a2 contains only the first S quartile, corresponding to the shortest interparticle separations. Similarly, Figure 6b1 includes all P values, whereas Figure 6b2 contains only the fourth P quartile.
Figure 6.
a) S and b) P maps for one plasmon ruler (PR3) that is performing a confined diffusion on a HeLa membrane. The maps were created from a PRPCM trajectory of 120 s length. The S map in a1) includes all data points whereas a2) only shows the shortest interparticle separation (first quartile). The P map in b1) includes all data, the P map in b2) contains only the fourth quartile (P = 0.3 – 0.6). The P and S distributions show that PR3 preferentially resides in three sub-regions marked as C1 – C3.
An inspection of the S and P maps in Figures 6a1&b1 shows that the data points are not equally distributed across the entire confinement region but clustered in three subregions, which are labeled as C1-C3 in Figure 6a. C1-C3 are not strictly associated with distinct times in the trajectory but get populated throughout the observation, and we conclude that PR3 shuttles between these sub-regions. Despite this translational movement, the reduced linear dichroism remains between P ≈ 0 – 0.6 due to a constrained rotational mobility in the entire confinement region. We point out that, although the drawn circumferences around C1-C3 in Figure 6 are somewhat arbitrary, our tracking experiments indicate that each of the sub-regions C1-C3 has dimensions on the order of ~70 nm, which is consistent with previous findings of the compartmentalization of HeLa cell surfaces.32
Interestingly, the less populated region of the green shaded confinement region between x = −150 nm and x = −50 nm is enriched in data points with longer interparticle separations suggesting less spatial constraints. It is unclear why this region is much less populated than C1-C3. One possible explanation could be a chemical heterogeneity of the membrane that favors a localization of PR3 in C1-C3. Correlation of the S and P maps in Figure 6 offers further insight into the sub-regions C1-C3. Close inspection of Figures 6a2 and b2 reveals that nearly 2/3 of the data points in C1 in Figure 6a2 superimpose with the data points in the same area in Figure 6b2 (for a magnification of C1, please see Figure S4). In C1 the shortest interparticle separations are correlated with the highest P values indicative of a preferential alignment of the long plasmon ruler axis in the compressed plasmon ruler. One structural model that could account for the more stringent orientation of the compressed plasmon ruler configuration is an anisotropic shape of C1. Assuming that the extension of C1 is shorter along one specific direction so that it can only accommodate the long plasmon ruler axis in this orientation if the ruler adjusts (i.e. decreases) its interparticle separation, the geometry of C1 would induce the observed preferential orientation of PR3 with short S values. Independent of the exact nature of the preferential orientation, the absence of a comparable correlation in C2 and C3 underlines the heterogeneity of the membrane on deeply subdiffraction limit length scales.
Overall, our analyses of Figures 5 and 6 have demonstrated that PRPCM of silver plasmon rulers is highly complementary to conventional single particle tracking in cell membrane profiling. PRPCM provides additional information about the orientation and interparticle separation of the plasmon rulers and the resulting S and P maps contribute to a systematic characterization the cell membrane morphology on sub-micron length scales. Our studies have also indicated a rich dynamics in S and P for cell surface confined silver plasmon rulers which motivates further studies with improved temporal resolution in the future.
III. Conclusions
Using a polarization resolved tracking approach we monitored the translational and rotational mobility of individual silver plasmon rulers on lysed HeLa cells with a temporal resolution of 500 Hz. A moment scaling spectrum analysis of the translational motion of the individual plasmon rulers revealed that most of the plasmon rulers are either immobilized or undergo a confined diffusion on the cell surface. Those plasmon rulers that undergo confined diffusion show large differences in the rotational dynamics which was attributed to the lateral heterogeneity of the membrane. To obtain additional information about the compartmentalization of the membrane, we implemented a polarization resolved plasmon coupling microscopy (PRPCM) that enabled us to track individual silver plasmon rulers diffusing on the surface of lysed HeLa cells and simultaneously monitor the plasmon rulers’ orientation and interparticle separation. PRPCM provides a quantitative metrics (D2, τ, P, R, S) for a detailed characterization of the mobility, orientation and extension of plasmon rulers at different locations on the cell surface as a function of time. Consistent with a compartmentalization of the membrane on length scales of the plasmon rulers (dimers of 30 nm particles with center to center separation of ~44 nm) we find that confinement of the plasmon rulers on the cell surface affects their rotational freedom and average interparticle separation. We demonstrated that by mapping S and P on the cell surface, detailed insight into the cell membrane organization with nanometer scale spatial resolution becomes accessible. The gain in information in polarization resolved ratiometric tracking when compared with conventional particle tracking makes PRPCM a promising tool for analyzing the lateral heterogeneity of complex cellular surfaces.
Supplementary Material
Acknowledgments
This work was financially supported by the National Institute of Health through grants 5 R21 EB008822-02 and 1 R01 CA138509-01.
Footnotes
Supporting Information Available. Figures S1–S4 and Methods. This material is available free of charge via the internet at http://pubs.acs.org
References
- 1.Whitty A. Nature Chemical Biology. 2008;4:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- 2.Edidin M. Trends in Cell Biology. 2001;11:492–496. doi: 10.1016/s0962-8924(01)02139-0. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. The Journal of Cell Biology. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki K, Ritchie K, Kajikawa E, Fujiwara T, Kusumi A. Biophysical Journal. 2005;88:3659–3680. doi: 10.1529/biophysj.104.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gundelfinger ED, Kessel MM, Qualmann B. Nature Reviews Molecular Cell Biology. 2003;4:127–139. doi: 10.1038/nrm1016. [DOI] [PubMed] [Google Scholar]
- 6.Simons K, Vaz WLC. Annual Reviews of Biophysics and Biomolecular Structure. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 7.London E. Biochimica et Biophysica Acta. 2005;1746:203–220. doi: 10.1016/j.bbamcr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 8.McIntosh TJ, Soimons SA. Reviews of Biophysics and Biomolecular Structure. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 9.Pike L. Biochemical Journal. 2004;378:281–292. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock JF. Nature Reviews Molecular Cell Biology. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond AT, Heberle FA, Baumgart T, Holowka D, Baird B, Feigenson GW. Proceedings of the National Academy of Sciences of the USA. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw AS. Nature Immunology. 2006;11:1139–1142. doi: 10.1038/ni1405. [DOI] [PubMed] [Google Scholar]
- 13.Kwik J, Boyle S, Fooksman D, Margolis L, Sheetz MP, Edidin M. Proceedings of the National Academy of Sciences of the USA. 2003;100:13964–13969. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheetz MP, Turney S, Qian H, Elson EL. Nature. 1989;340:284–288. doi: 10.1038/340284a0. [DOI] [PubMed] [Google Scholar]
- 15.Qian H, Sheetz MP, Elson EL. Biophysical Journal. 1991;60:910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Annual Review of Biophysics and Biomolecular Structure. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 17.Edidin M, Kuo SC, Sheetz MP. Science. 1991;254:1379–1382. doi: 10.1126/science.1835798. [DOI] [PubMed] [Google Scholar]
- 18.Rong G, Wang H, Skewis LR, Reinhard BM. Nano Letters. 2008;8:3386–3393. doi: 10.1021/nl802058q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su KH, Wei QH, Zhang X, Mock JJ, Smith DR, Schultz S. Nano Letters. 2003;3:1087–1090. [Google Scholar]
- 20.Reinhard BM, Siu M, Agarwal H, Alivisatos AP, Liphardt J. Nano Letters. 2005;5:2246–2252. doi: 10.1021/nl051592s. [DOI] [PubMed] [Google Scholar]
- 21.Hell SW, Wichmann J. Optics Letters. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 22.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 23.Rust MJ, Bates M, Zhuang XW. Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hell SW. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 25.Yguerabide J, Yguerabide EE. Analytical Biochemistry. 1998;262:137–156. doi: 10.1006/abio.1998.2759. [DOI] [PubMed] [Google Scholar]
- 26.Yguerabide J, Yguerabide EE. Analytical Biochemistry. 1998;262:157–176. doi: 10.1006/abio.1998.2760. [DOI] [PubMed] [Google Scholar]
- 27.Nan XL, Sims PA, Xie XS. ChemPhysChem. 2008;9:707–712. doi: 10.1002/cphc.200700839. [DOI] [PubMed] [Google Scholar]
- 28.Aaron J, Travis K, Harrison N, Sokolov K. Nano Letters. 2009;9:3612. doi: 10.1021/nl9018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Reinhard BM. The Journal of Physical Chemistry C. 2009;113:11215–11222. doi: 10.1021/jp900874n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nature Biotechnology. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 31.Gunnarsson L, Rindzevicius T, Prikulis J, Kasemo B, Kall M, Zou SL, Schatz GC. The Journal of Physical Chemistry B. 2005;109:1079–1087. doi: 10.1021/jp049084e. [DOI] [PubMed] [Google Scholar]
- 32.Murase K, Fujiwara T, Umemura Y, Suzuki K, Iino K, Yamashita H, Saito M, Murakoshi H, Ritchie K, Kusumi A. Biophysical Journal. 2004;86:4075–4093. doi: 10.1529/biophysj.103.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelles J, Schnapp BJ, Sheetz MP. Nature. 1988;33:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 34.Thompson RE, Larson DR, Webb WW. Biophysical Journal. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown RB, Audet J. Journal of the Royal Society Interface. 2008;5:S131–S138. doi: 10.1098/rsif.2008.0009.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wulf E, Deboden E, Bautz FA, Faulstich H, Wieland TH. Proceedings of the National Academy of Sciences of the USA. 1979;76:4498–4502. doi: 10.1073/pnas.76.9.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doty RC, Tshikhudo TR, Brust M, Fernig DG. Chemistry of Materials. 2005;17:4630–4635. [Google Scholar]
- 38.Lee JS, Lytton-Jean AKR, Hurst SJ, Mirkin CA. Nano Letters. 2007;7:2112–2115. doi: 10.1021/nl071108g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Errico J. [accessed Dec 1, 2009];Matlab Central. http://www.mathworks.com/matlabcentral/fileexchange/8998-surface-fitting-using-gridfit.
- 40.Wieser S, Schuetz GJ. Methods. 2008;46:131–140. doi: 10.1016/j.ymeth.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Wei CY, Lu CY, Kim Y, Vanden Bout D. Journal of Fluorescence. 2007;17:797–804. doi: 10.1007/s10895-007-0234-9. [DOI] [PubMed] [Google Scholar]
- 42.Pierrat S, Hartinger E, Faiss S, Janshoff A, Soennichsen C. The Journal of Physical Chemistry C. 2009;113:11179–11183. [Google Scholar]
- 43.Bouchaud JP, Georges A. Physics Report. 1990;195:127–293. [Google Scholar]
- 44.Feder TJ, Brust-Mascher I, Slattery JP, Baird B, Webb WW. Biophysical Journal. 1996;70:2767–2773. doi: 10.1016/S0006-3495(96)79846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferrari R, Manfroi AJ, Young WR. Physica D. 2001;154:111–137. [Google Scholar]
- 46.Sbalzarini IF, Koumoutsakos P. Journal of Structural Biology. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Ewers H, Smith AE, Sbalzarini IF, Lilie H, Koumoutsakos P, Helenius A. Proceedings of the National Academy of Sciences of the USA. 2005;102:15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Yan B, Reinhard BM. Journal of Physical Chemistry C. 2008;112:15989–15996. doi: 10.1021/jp804790p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L, Wang H, Yan B, Reinhard BM. Journal of Physical Chemistry C. 2010;114:4901–4908. doi: 10.1021/jp911858v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jain PK, el-Sayed MA. Nano Letters. 2008;8:4347–4352. doi: 10.1021/nl8021835. [DOI] [PubMed] [Google Scholar]
- 51.Kelly KL, Coronado E, Zhao LL, Schatz GC. Journal of Physical Chemistry B. 2003;107:668–677. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.