Abstract
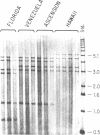
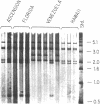
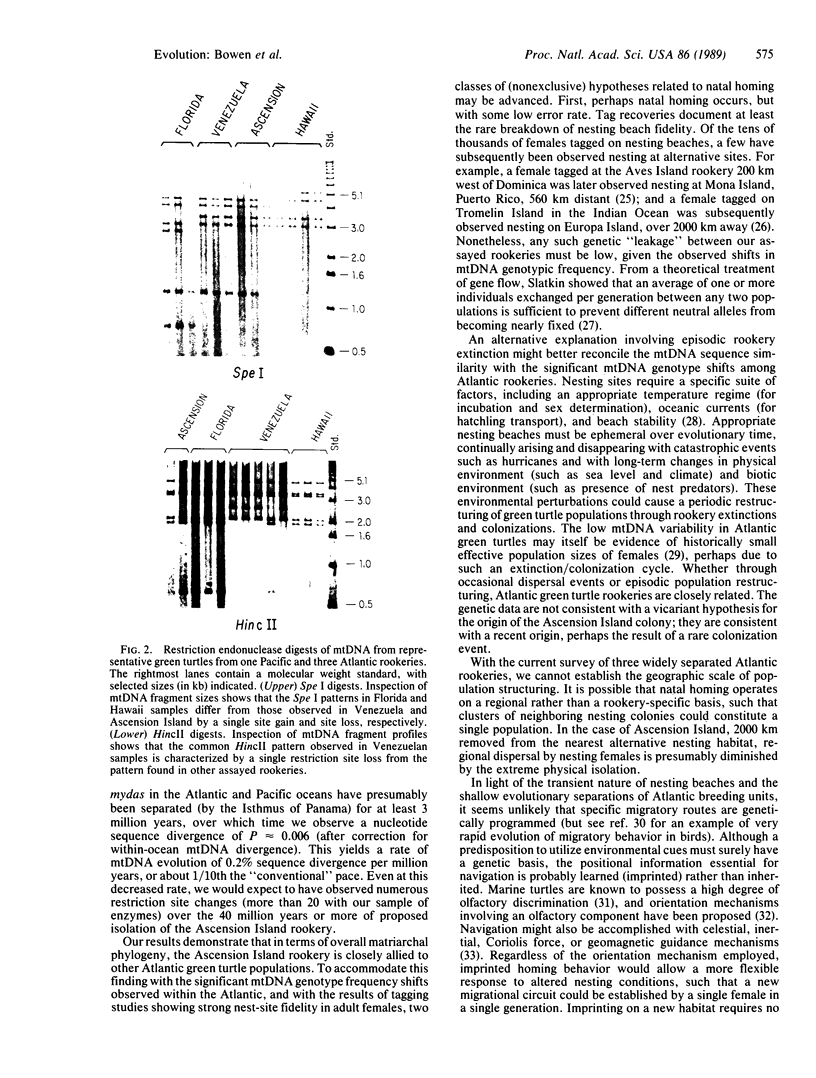
Green turtles (Chelonia mydas) that nest on Ascension Island, in the south-central Atlantic, utilize feeding grounds along the coast of Brazil, more than 2000 km away. To account for the origins of this remarkable migratory behavior, Carr and Coleman [Carr, A. & Coleman, P. J. (1974) Nature (London) 249, 128-130] proposed a vicariant biogeographic scenario involving plate tectonics and natal homing. Under the Carr-Coleman hypothesis, the ancestors of Ascension Island green turtles nested on islands adjacent to South America in the late Cretaceous, soon after the opening of the equatorial Atlantic Ocean. Over the last 70 million years, these volcanic islands have been displaced from South America by sea-floor spreading, at a rate of about 2 cm/year. A population-specific instinct to migrate to Ascension Island is thus proposed to have evolved gradually over tens of millions of years of genetic isolation. Here we critically test the Carr-Coleman hypothesis by assaying genetic divergence among several widely separated green turtle rookeries. We have found fixed or nearly fixed mitochondrial DNA (mtDNA) restriction site differences between some Atlantic rookeries, suggesting a severe restriction on contemporary gene flow. Data are consistent with a natal homing hypothesis. However, an extremely close similarity in overall mtDNA sequences of surveyed Atlantic green turtles from three rookeries is incompatible with the Carr-Coleman scenario. The colonization of Ascension Island, or at least extensive gene flow into the population, has been evolutionarily recent.
Keywords: mitochondrial DNA, intraspecific phylogeny, gene flow, genetic distance
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Ball R. M., Arnold J. Current versus historical population sizes in vertebrate species with high gene flow: a comparison based on mitochondrial DNA lineages and inbreeding theory for neutral mutations. Mol Biol Evol. 1988 Jul;5(4):331–344. doi: 10.1093/oxfordjournals.molbev.a040504. [DOI] [PubMed] [Google Scholar]
- Avise J. C. Mitochondrial DNA and the evolutionary genetics of higher animals. Philos Trans R Soc Lond B Biol Sci. 1986 Jan 29;312(1154):325–342. doi: 10.1098/rstb.1986.0011. [DOI] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M. Polymorphism in mitochondrial DNA of humans as revealed by restriction endonuclease analysis. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3605–3609. doi: 10.1073/pnas.77.6.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten U., Wharton D., Wilson A. C. Maternal inheritance of mitochondrial DNA during backcrossing of two species of mice. J Hered. 1985 Sep-Oct;76(5):321–324. doi: 10.1093/oxfordjournals.jhered.a110103. [DOI] [PubMed] [Google Scholar]
- Hughes D. M., Newton-John H., Chay O. M., Landau L. I. Lung function after pertussis. Aust Paediatr J. 1987 Oct;23(5):277–282. doi: 10.1111/j.1440-1754.1987.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Koch A. L., Carr A., Ehrenfeld D. W. The problem of open-sea navigation: the migration of the green turtle to ascension island. J Theor Biol. 1969 Jan;22(1):163–179. doi: 10.1016/0022-5193(69)90085-x. [DOI] [PubMed] [Google Scholar]
- Lansman R. A., Shade R. O., Shapira J. F., Avise J. C. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. III. Techniques and potential applications. J Mol Evol. 1981;17(4):214–226. doi: 10.1007/BF01732759. [DOI] [PubMed] [Google Scholar]
- Manton M. L., Karr A., Ehrenfeld D. W. An operant method for the study of chemoreception in the green turtle, Chelonia mydas. Brain Behav Evol. 1972;5(2):188–201. doi: 10.1159/000123747. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields G. F., Wilson A. C. Calibration of mitochondrial DNA evolution in geese. J Mol Evol. 1987;24(3):212–217. doi: 10.1007/BF02111234. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987 May 15;236(4803):787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]