Abstract
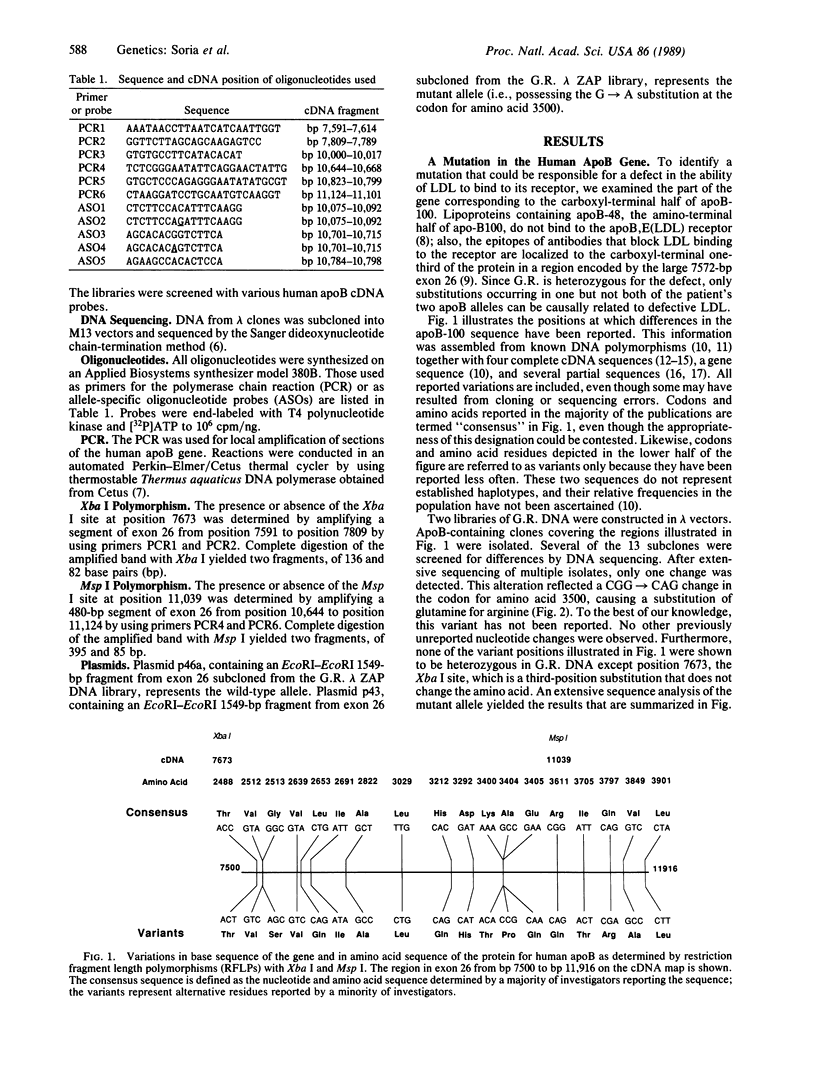
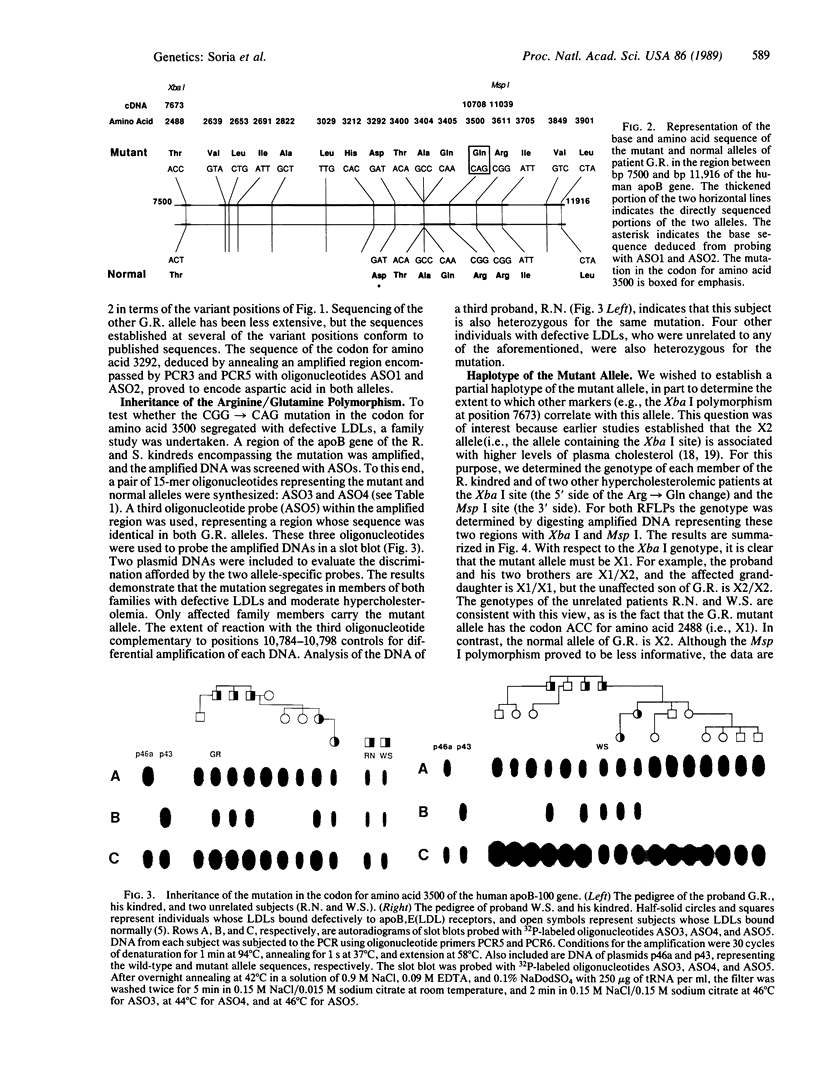
Familial defective apolipoprotein (apo) B-100 is a genetic disease that leads to hypercholesterolemia and to an increased serum concentration of low density lipoproteins that bind defectively to the apoB,E(LDL) receptor. The disorder appears to result from a mutation in the gene for apoB-100. Extensive sequence analysis of the two alleles of one subject heterozygous for the disorder has revealed a previously unreported mutation in the codon for amino acid 3500 that results in the substitution of glutamine for arginine. This same mutant allele occurs in six other, unrelated subjects and in eight affected relatives in two of these families. A partial haplotype of this mutant apoB-100 allele was constructed by sequence analysis and restriction enzyme digestion at positions where variations in the apoB-100 are known to occur. This haplotype is the same in three probands and four affected members of one family and lacks a polymorphic Xba I site whose presence has been correlated with high cholesterol levels. Thus, it appears that the mutation in the codon for amino acid 3500 (CGG----CAG), a CG mutational "hot spot," defines a minor apoB-100 allele associated with defective low density lipoproteins and hypercholesterolemia.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D., Schafer M., White R. Restriction sites containing CpG show a higher frequency of polymorphism in human DNA. Cell. 1984 Jan;36(1):131–138. doi: 10.1016/0092-8674(84)90081-3. [DOI] [PubMed] [Google Scholar]
- Berg K. DNA polymorphism at the apolipoprotein B locus is associated with lipoprotein level. Clin Genet. 1986 Dec;30(6):515–520. doi: 10.1111/j.1399-0004.1986.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Berg K., Hames C., Dahlén G., Frick M. H., Krishan I. Genetic variation in serum low density lipoproteins and lipid levels in man. Proc Natl Acad Sci U S A. 1976 Mar;73(3):937–940. doi: 10.1073/pnas.73.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg K., Powell L. M., Wallis S. C., Pease R., Knott T. J., Scott J. Genetic linkage between the antigenic group (Ag) variation and the apolipoprotein B gene: assignment of the Ag locus. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7367–7370. doi: 10.1073/pnas.83.19.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson P., Darnfors C., Olofsson S. O., Bjursell G. Analysis of the human apolipoprotein B gene; complete structure of the B-74 region. Gene. 1986;49(1):29–51. doi: 10.1016/0378-1119(86)90383-5. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Yang C. Y., Chen P. F., Setzer D., Tanimura M., Li W. H., Gotto A. M., Jr, Chan L. The complete cDNA and amino acid sequence of human apolipoprotein B-100. J Biol Chem. 1986 Oct 5;261(28):12918–12921. [PubMed] [Google Scholar]
- Cladaras C., Hadzopoulou-Cladaras M., Nolte R. T., Atkinson D., Zannis V. I. The complete sequence and structural analysis of human apolipoprotein B-100: relationship between apoB-100 and apoB-48 forms. EMBO J. 1986 Dec 20;5(13):3495–3507. doi: 10.1002/j.1460-2075.1986.tb04675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Elovson J., Jacobs J. C., Schumaker V. N., Puppione D. L. Molecular weights of apoprotein B obtained from human low-density lipoprotein (apoprotein B-PI) and from rat very low density lipoprotein (apoprotein B-PIII). Biochemistry. 1985 Mar 12;24(6):1569–1578. doi: 10.1021/bi00327a042. [DOI] [PubMed] [Google Scholar]
- Hegele R. A., Huang L. S., Herbert P. N., Blum C. B., Buring J. E., Hennekens C. H., Breslow J. L. Apolipoprotein B-gene DNA polymorphisms associated with myocardial infarction. N Engl J Med. 1986 Dec 11;315(24):1509–1515. doi: 10.1056/NEJM198612113152403. [DOI] [PubMed] [Google Scholar]
- Huang L. S., de Graaf J., Breslow J. L. ApoB gene MspI RFLP in exon 26 changes amino acid 3611 from Arg to Gln. J Lipid Res. 1988 Jan;29(1):63–67. [PubMed] [Google Scholar]
- Hui D. Y., Innerarity T. L., Milne R. W., Marcel Y. L., Mahley R. W. Binding of chylomicron remnants and beta-very low density lipoproteins to hepatic and extrahepatic lipoprotein receptors. A process independent of apolipoprotein B48. J Biol Chem. 1984 Dec 25;259(24):15060–15068. [PubMed] [Google Scholar]
- Innerarity T. L., Weisgraber K. H., Arnold K. S., Mahley R. W., Krauss R. M., Vega G. L., Grundy S. M. Familial defective apolipoprotein B-100: low density lipoproteins with abnormal receptor binding. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6919–6923. doi: 10.1073/pnas.84.19.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott T. J., Pease R. J., Powell L. M., Wallis S. C., Rall S. C., Jr, Innerarity T. L., Blackhart B., Taylor W. H., Marcel Y., Milne R. Complete protein sequence and identification of structural domains of human apolipoprotein B. Nature. 1986 Oct 23;323(6090):734–738. doi: 10.1038/323734a0. [DOI] [PubMed] [Google Scholar]
- Law A., Wallis S. C., Powell L. M., Pease R. J., Brunt H., Priestley L. M., Knott T. J., Scott J., Altman D. G., Miller G. J. Common DNA polymorphism within coding sequence of apolipoprotein B gene associated with altered lipid levels. Lancet. 1986 Jun 7;1(8493):1301–1303. doi: 10.1016/s0140-6736(86)91222-5. [DOI] [PubMed] [Google Scholar]
- Law S. W., Grant S. M., Higuchi K., Hospattankar A., Lackner K., Lee N., Brewer H. B., Jr Human liver apolipoprotein B-100 cDNA: complete nucleic acid and derived amino acid sequence. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8142–8146. doi: 10.1073/pnas.83.21.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig E. H., Blackhart B. D., Pierotti V. R., Caiati L., Fortier C., Knott T., Scott J., Mahley R. W., Levy-Wilson B., McCarthy B. J. DNA sequence of the human apolipoprotein B gene. DNA. 1987 Aug;6(4):363–372. doi: 10.1089/dna.1987.6.363. [DOI] [PubMed] [Google Scholar]
- Lusis A. J. Genetic factors affecting blood lipoproteins: the candidate gene approach. J Lipid Res. 1988 Apr;29(4):397–429. [PubMed] [Google Scholar]
- Marcel Y. L., Hogue M., Theolis R., Jr, Milne R. W. Mapping of antigenic determinants of human apolipoprotein B using monoclonal antibodies against low density lipoproteins. J Biol Chem. 1982 Nov 25;257(22):13165–13168. [PubMed] [Google Scholar]
- Marcel Y. L., Innerarity T. L., Spilman C., Mahley R. W., Protter A. A., Milne R. W. Mapping of human apolipoprotein B antigenic determinants. Arteriosclerosis. 1987 Mar-Apr;7(2):166–175. doi: 10.1161/01.atv.7.2.166. [DOI] [PubMed] [Google Scholar]
- Protter A. A., Hardman D. A., Sato K. Y., Schilling J. W., Yamanaka M., Hort Y. J., Hjerrild K. A., Chen G. C., Kane J. P. Analysis of cDNA clones encoding the entire B-26 region of human apolipoprotein B. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5678–5682. doi: 10.1073/pnas.83.15.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmud P. J., Barni N., Kessling A. M., Carlsson P., Darnfors C., Bjursell G., Galton D., Wynn V., Kirk H., Hayden M. R. Apolipoprotein B gene variants are involved in the determination of serum cholesterol levels: a study in normo- and hyperlipidaemic individuals. Atherosclerosis. 1987 Sep;67(1):81–89. doi: 10.1016/0021-9150(87)90267-x. [DOI] [PubMed] [Google Scholar]
- Tikkanen M. J. Ag system re-explored: studies with monoclonal anti-apolipoprotein B antibodies. Adv Exp Med Biol. 1987;210:55–62. doi: 10.1007/978-1-4684-1268-0_9. [DOI] [PubMed] [Google Scholar]
- Vega G. L., Grundy S. M. In vivo evidence for reduced binding of low density lipoproteins to receptors as a cause of primary moderate hypercholesterolemia. J Clin Invest. 1986 Nov;78(5):1410–1414. doi: 10.1172/JCI112729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisgraber K. H., Innerarity T. L., Newhouse Y. M., Young S. G., Arnold K. S., Krauss R. M., Vega G. L., Grundy S. M., Mahley R. W. Familial defective apolipoprotein B-100: enhanced binding of monoclonal antibody MB47 to abnormal low density lipoproteins. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9758–9762. doi: 10.1073/pnas.85.24.9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Antonarakis S. E., Bell W., Griffin A. M., Kazazian H. H., Jr Nonsense and missense mutations in hemophilia A: estimate of the relative mutation rate at CG dinucleotides. Am J Hum Genet. 1988 May;42(5):718–725. [PMC free article] [PubMed] [Google Scholar]





