Abstract
Background
A severe flare of colitis in patients with IBD treated with immunosuppressive therapy may be complicated by an underlying CMV infection. The aim of this pilot study was to investigate the diagnostic efficacy of quantitative polymerase chain reaction (PCR) to detect CMV DNA in stool samples of IBD patients.
Methods
Twenty-one patients with a severe flare of IBD, incompletely responding or refractory to either steroids or immunosuppressive agents, were included in the study. Nineteen patients completed the study according to the protocol undergoing an endoscopy with biopsies and collection of stool samples. Biopsy and stool samples were qualitatively and quantitatively analyzed for CMV DNA using real-time PCR.
Results
Thirty-two percent (6/19) of the patients had detectable CMV DNA in colonic biopsies and in five (83%) of those patients CMV DNA was detected in the stool. Thirteen patients had negative findings for CMV DNA in biopsy and stool samples. The sensitivity, specificity, and accuracy of the PCR-based stool test for detection of CMV DNA compared to PCR-based detection of CMV in mucosal biopsies were 83, 93, and 90%, respectively.
Conclusions
The pilot study suggests a high accuracy of this non-invasive testing method to detect CMV DNA in stool samples as compared to mucosal biopsies. This approach may offer a non-endoscopic testing modality for underlying CMV infection in patients with a severe flare of IBD, which could also be applied more broadly to determine the prevalence of CMV infections in patients with IBD.
Keywords: Inflammatory bowel disease, Ulcerative colitis, Crohn's disease, Infectious colitis, Cytomegalovirus, Immunosuppression
Introduction
Cytomegalovirus (CMV), a member of the herpesvirus family, is a well-known cause of gastrointestinal pathology in immunocompromised patients, whereas gastrointestinal symptoms in an immunocompetent host are rarely observed [1]. Classically, CMV enterocolitis with large ulcerations visible on upper or lower GI-endoscopy can be observed during CMV viremia in HIV patients and organ transplant patients [2]. A number of reports suggest that CMV infection may be an underlying factor in patients with severe colitis refractory to intravenous steroids and/or immunosuppressive therapy (e.g., cyclosporine) [3–14]. CMV infection is common in the general population with a reported seroprevalence of 40–100% [15]. Therefore, latency of the virus and reactivation during exacerbation of the colitis in patients with IBD is probably the most common cause of detected CMV infection. The prevalence of CMV IgG in patients with ulcerative colitis (UC) was recently described as 70% and a reactivation of CMV detected by CMV antigenemia and plasma quantitative PCR occurred in more than 50% of ulcerative colitis (UC) patients while being treated with steroids or immunosuppressive drugs [12].
With the exception of retinitis, a CMV infection cannot be diagnosed alone using clinical skills. Also, no standardized recommendations exist on how best to diagnose a concurrent CMV infection in the setting of colitis or how to interpret positive or negative findings. Several techniques are available for diagnosing CMV colitis with each one having advantages and disadvantages. CMV infection of the colon can be diagnosed using histology (hematoxylineosin staining) or immunohistochemistry [16], which is considered a gold standard, but due to sampling error may also yield false-negative results. Serology is very limited in detecting active CMV infection, since anti-CMV IgM antibody levels can persist up to 2 years after infection and immunocompromised patients may not mount an IgM response. CMV culture can be performed on a whole variety of body fluids and tissue, but has the disadvantage of long incubation times and a relative low sensitivity [17]. The CMV antigen test detects the internal matrix protein pp65 in circulating leukocytes but the technique is only semiquantitative and less sensitive as compared to CMV PCR [18], which is currently the most sensitive test for detecting systemic CMV infection in plasma or serum samples [17]. PCR has been also applied for the amplification of CMV DNA from mucosal biopsies [19, 20].
During active CMV infection, CMV viral shedding occurs in the urine, respiratory secretions, and stool of infected patients [21, 22]. The analysis of these shed viral particles in bronchoalveolar secretions may increase the sensitivity for the detection of an underlying CMV infection as shown in patients with suspected CMV pneumonitis [23, 24]. Qualitative and/or quantitative stool CMV DNA testing by PCR is a noninvasive technique [22, 25] and may offer the possibility to test for a concurrent CMV infection in patients with refractory IBD without the need for obtaining endoscopic biopsies. Therefore, our aim was to evaluate the sensitivity, specificity, accuracy, and feasibility of the detection of CMV DNA in stool samples compared to the detection of CMV DNA in intestinal biopsies from patients with a severe flare of IBD and suspicion of associated CMV-colitis or CMV-enteritis.
Methods
The prospective study was conducted at the University of North Carolina from July 2007 until November 2008. The local Institutional Review Board (IRB) approved the study. All patients included in the study had been hospitalized due to an exacerbation of a previously proven inflammatory bowel disease (Crohn's disease or ulcerative colitis), and underwent a sigmoidoscopy or colonoscopy with biopsies for histology and qualitative CMV-PCR. The biopsies were taken in the rectosigmoid colon or in case of patients with ileostoma (one patient) in the distal ileum (two biopsies for pathological evaluation, one biopsy for PCR). Clinical follow-up was performed by chart review for 6 months after inclusion in the study.
All biopsies were evaluated by a pathologist, who was not aware of the PCR results of the biopsies or plasma using a regular hematoxylin and eosin (H&E) stain. Large cells containing a basophilic intranuclear inclusion were regarded as diagnostic of CMV infection. At the discretion of the pathologist immunohistochemistry stains were also performed on formalin-fixed and paraffin-embedded tissue using standard immunohistochemical techniques (antibody: DAKO clone CCH2 and DDG9) following a citrate-based epitope antigen retrieval step.
One stool sample of each participating patient was collected in a time period of 12 h after the endoscopic procedure (see Fig. 1). At the discretion of the treating physician, plasma samples for the detection of circulating CMV DNA were also drawn in 16 patients. The stool samples were freshly collected (approx. 1.5 g) and immediately placed and homogenized in a conical tube containing 5 ml of STAR buffer (Stool Transport and Recovery buffer, Roche Diagnostics); chloroform (500 μl) was added and the sample was centrifuged (1 min at 3,300 rpm). Total nucleic acids (TNA) were isolated from the supernatant using MagNA Pure LC (Roche Diagnostics). TNA from the colonic biopsies were extracted from 200 μl of viral transport medium in which the colonic biopsies had been ground and/or from 200 μl plasma using the Roche MagNAPure LC according to the manufacturer's instructions. The elution volume was 50 μl.
Fig. 1.
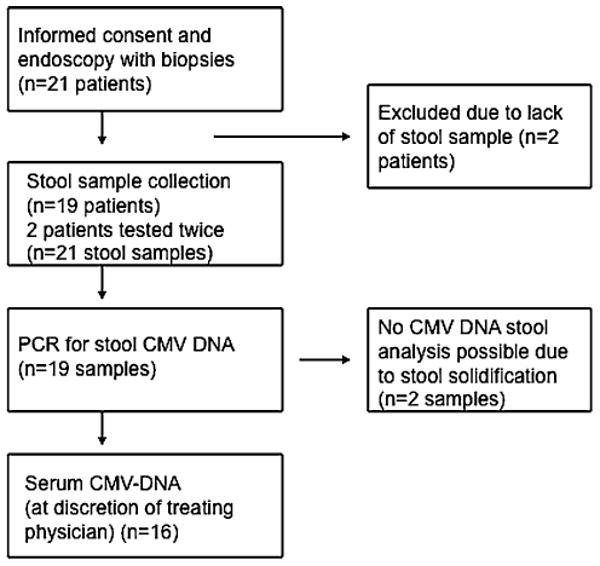
Flow diagram of the study
CMV PCR Assay
The detection of CMV DNA in plasma, biopsies, and stool samples was performed using real-time PCR and previously described primers and TaqMan probe that target the CMV polymerase I gene [26]. The internal control targets were the human albumin gene (qualitative) and TaqMan Exogeneous Internal Positive Control (quantitative; Applied Biosystems, Foster City, CA). The internal control was detected separately for the qualitative assay but as a multiplex in the quantitative assay. The 50-μl reaction mixture contained TaqMan Universal PCR Master Mix (Applied Biosystems), 900-nM (qualitative) or 250-nM (quantitative) primers and 200-nM (qualitative) or 250-nM (quantitative) probe. PCR cycling parameters were 2 min of incubation at 50°C and 10 min at 95°C, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. For the quantitative PCR, a standard curve of CMV Towne strain DNA was assayed. The limit of detection for the qualitative assay is 200 copies/ml and the limit of quantification is 500 copies/ml.
Statistical Analysis
Using endoscopically retrieved mucosal biopsies for PCR-amplification of CMV as the gold standard for diagnosis of active CMV infection, the sensitivity and specificity of stool-based PCR CMV DNA test for the diagnosis of CMV were calculated. The accuracy, defined as total correctly classified over total in population, was also calculated. Finally, the positive predictive value of the test, the probability that the patient has the disease given that a positive test has been obtained, and the negative predictive value (inverse of above) were also calculated.
Results
Twenty-one patients were included in the prospective study and signed the informed consent (Fig. 1). Two of the 21 patients underwent endoscopy, but were not able to provide a stool sample within the study time window of 12 h after sigmoidoscopy and were excluded from further analysis. Two patients were tested twice in the time frame of the study. Therefore, a total 21 stool samples and 21 biopsies from 19 patients were analyzed for CMV DNA. The patient characteristics and concurrent therapies are described in Table 1. A total of 84% of the patients were either treated with oral or intravenous steroids, 26% with azathioprine/6-MP, 10% with methotrexate, and 21% with an anti-TNF agent.
Table 1.
Patient demographics
| Patients (n; M/F) | 10/9 |
| Age (years) median (range) | 34 (20–65) |
| Diagnosis UC/CD | 11/8a |
| Therapy (n) | |
| Oral or intravenous steroids | 16/19 |
| Azathioprine/6-MP | 5/19 |
| Methotrexate | 2/19 |
| Anti-TNF agents | 4/19 |
One patient with ileostomy due to CD had a history of colectomy due to severe colitis. All other seven CD patients had severe Crohn's colitis
CMV DNA Detection in Colonic Biopsies, Stool, and Plasma Specimens
CMV DNA could be amplified by PCR in colonic biopsies (CMVB×PCR+) in 6/19 patients (32%; six biopsies), whereas in 13 patients (15 biopsies) CMV DNA was not detectable in the biopsies (CMVB×PCR−). In 83% (5/6) of the CMVB×PCR+ biopsies either typical histological or immunohistochemical characteristics suggestive of a CMV infection were described by the pathologist.
Two of the 21 stool samples solidified before analysis and the homogenization of the sample in the buffer to extract the viral DNA was technically not possible (in one case the patient was CMVB×PCR+, in the other case the patient was CMVB×PCR−, see Table 2). For the analysis of the sensitivity and specificity of the PCR-based stool test we considered these samples as false-negative and false-positive (Table 3). A total of 5/6 (83%) CMVB×PCR+ cases also had detectable CMV in the corresponding stool sample and 14/15 (93%) CMVB×PCR− cases had no detectable CMV DNA in the corresponding stool sample. The sensitivity and specificity of the stool test compared to PCR of mucosal biopsies were 83 and 93%, respectively. The positive and negative predictive values of the stool-based PCR test for CMV disease were 83 and 93%, respectively (see Table 3). The accuracy of the test was 19/21 or 90%.
Table 2.
CMV DNA copy number in biopsy, stool and plasma specimen in six different study patients
| Case number | Dx | CMV PCR biopsy copies/ml | CMV PCR stool copies/ml | CMV plasma copies/ml |
|---|---|---|---|---|
| 2 | UC | <500 | 2,316 | <500 |
| 4 | UC | <500 | 40,317 | <500 |
| 5 | UC | 1,826 | 40,339 | Negative |
| 9 | UC | 117,775 | NDb | 6,252 |
| 18 | CDa | 91,125 | 2,266 | ND |
| 23 | UC | 834 | 19,672 | <500 |
In the other 13 study patients CMV PCR of 15 biopsies and stool samples were all negative for CMV DNA
ND, not done
Patient with ileostomy due to CD with a history of colectomy due to severe colitis
Determination of CMV DNA technical not possible due to solidification of stool sample prior to homogenization for DNA extraction
Table 3.
2 × 2 table demonstrating the positive or negative findings for CMV DNA based on qualitative PCR in 21 mucosal biopsies and stool samples of 19 patients
| CMV DNA in mucosal biopsies | ||||
|---|---|---|---|---|
| + | - | |||
| CMV DNA in stool samples | + | 5 | 1 | 6 |
| - | 1 | 14 | 15 | |
| 6 | 15 | |||
The detection of systemic CMV DNA by evaluation of plasma by PCR was performed outside the study protocol at the discretion of the treating physician. In 16/21 (76%) of the cases, plasma samples were also obtained. In 67% (4/6) of the CMVB×PCR+ cases plasma CMV DNA could be amplified by PCR, whereas in one case CMV DNA was not detected and in the other case the test was not performed (see Table 2). In the 15 CMVB×PCR− cases, ten analyses of plasma for CMV DNA were performed. In each case, CMV DNA could not be amplified in the plasma.
Clinical Outcome
All six patients with positive CMV biopsy results were treated with oral valganciclovir. Two patients with CMVB×PCR+ UC underwent colectomy due to refractory disease 8 and 9 days after the inclusion in the study, whereas the other three CMVB×PCR+ UC and one CMVB×PCR+ CD patient responded to valganciclovir therapy. However, all three UC patients finally underwent colectomy due to therapy refractory disease after 2.5–5 months. Histology specimen of the colectomy preparations did not reveal active CMV colitis at the time point of resection in any of the cases. Also, 3/13 patients (n = 2 with UC, n = 1 with Crohn's colitis) in the CMV-negative group underwent colectomy 6, 10, and 30 days, respectively, after inclusion in the study.
Discussion
The first case report about a young patient with active UC and a concurrent severe CMV colitis necessitating colectomy at the University of Chicago was reported in 1961 [27]. Since then, numerous other reports and case series have been published. It is currently thought that between 20 and 35% of patients with severe colitis due to IBD, who are either refractory to steroids or to immunosuppressive medications such as cyclosporine, tacrolimus, or an anti-TNF agent, may also suffer from an underlying CMV-enteritis [4, 8, 28, 29]. Multiple techniques are employed to diagnose a concurrent intestinal CMV infection in IBD patients and so far no firm recommendations for the best approach have been established [30–32]. However, the detection of CMV DNA by PCR analysis in mucosal biopsies has been described as the most sensitive method in patients with suspected CMV-enteritis [20, 30, 33].
In this prospective study, we included patients with active IBD not fully responsive or refractory to either steroids and/or other immunosuppressive medications. Thirty-two percentage of these patients had detectable CMV DNA by PCR in colonic biopsies. We demonstrate that fresh stool qualitative and qualitative PCR for CMV is a method with similar sensitivity and specificity to endoscopic collection of mucosal biopsies for PCR amplification of CMV DNA. This is in line with a recent report indicating similar sensitivities and specificities for CMV DNA in stool samples of HIV-infected patients [22]. This report also suggested a high negative predictive value for a concurrent intestinal CMV infection if no CMV shedding could be detected by PCR in the stool sample. In our study, the negative predictive value for a concurrent intestinal CMV infection was 93%. This slightly suboptimal value was due to the fact that we also included patients in the study, who provided stool samples, which solidified shortly after collection, thus excluding these samples from further analysis. We decided not to exclude these samples from the sensitivity and specificity analysis, since we wanted to examine the test given “real-life” conditions. If we had included only stool samples, which could be analyzed for CMV particles by PCR, the positive and negative predictive value of the test would have been 100%.
Histological signs of CMV infection were found on H&E stains (inclusion bodies) or by immunohistochemistry in 83% of the patients (5/6), who were tested positive for CMV DNA by PCR in the stool. One patient with an endoscopic picture highly suggestive of CMV colitis who tested positive for CMV DNA in colonic biopsy and plasma, showed no signs of CMV infection either by H&E stain or by immunohistochemistry. This supports the higher sensitivity of the PCR-based diagnostic approach for the diagnosis of CMV colitis. Additionally we detected CMV DNA in the plasma of 83% of the patients with detectable CMV DNA in the mucosal biopsies. We also observed higher CMV DNA levels in the fecal samples compared to the biopsy and plasma samples. Similar findings have been reported previously [25]. It is not clear whether CMV DNA detected in the stool samples in this study was due to leakage from the blood compartment into the intestinal tract, or if it was derived from intestinal CMV infection. Additionally we would like to point out that currently no specific cut-off levels for the amplified number of CMV virus particles in biopsies or stool are defined indicating, e.g., different histological or clinical severities of a colonic CMV infection. Also, this pilot trial was not designed to compare the multiple different methods for CMV detection such as histology, immunohistochemistry, CMV-pp65 antigen assay, CMV-plasma and biopsy PCR, CMV serology or CMV culture, which has been done in other trials [28, 34]. Another limitation of the present pilot study includes the small number of patients with biopsy-proven CMV colitis. Larger studies also investigating the above-mentioned diagnostic modalities would be of interest.
A total of 40% of the patients with severe colitis and underlying CMV infection underwent colectomy shortly (8 and 9 days) after the additional diagnosis of the CMV infection despite valgancyclovir therapy. In contrast, the colectomy rate in the CMV-negative group was 23% in a time period of 4 weeks after inclusion. The remaining three UC patients with positive CMV DNA PCR first improved with valgancyclovir therapy but underwent a colectomy later on due to refractory disease. The higher likelihood of colectomy due to an underlying CMV-colitis despite antiviral therapy in patients with refractory UC has been demonstrated in the past, however, individual predictive risk factors for non-responsiveness to antiviral therapy and colectomy as well as a strict causality of CMV infection as an only reason for disease deterioration could not yet be determined [29]. In our case series, the resected specimen of all of the patients showed no signs of active CMV infection as assessed by histology ruling out continuing fulminant CMV infection despite valgancyclovir therapy. There was also no association between the amount of CMV-copies in the colonic biopsies, in the stool or plasma samples and the time point of colectomy. Only the CD patient with CMV enteritis, proven not only by PCR, but also by immunohistochemistry, slowly improved after the valgancyclovir therapy, but also had prolonged symptoms of disease activity and was found to have sustained small-bowel ulcerations on a repeated endoscopy. The symptoms and ulcerations improved after the patient was started on an anti-TNF agent. The anti-TNF therapy was only started after a repeat endoscopy ruled out recurrent CMV enteritis. Recent reports also indicate, that it might be safe to start patients with diagnosed CMV colitis on anti-TNF agents [35, 36].
There are several advantages of detecting CMV DNA in fresh stool specimen by PCR. This is a fast, cheap, and uncomplicated procedure, which only involves the collection of a fresh stool specimen. Additionally, it does not involve invasive procedures such as a sigmoidoscopy or colonoscopy to obtain the biopsies necessary for the PCR analysis, which makes this diagnostic approach especially valuable in an outpatient setting. Moreover, since many patients with severe colitis often only undergo a sigmoidoscopy for obtaining the biopsies, the approach of detecting shed CMV viral particles in the stool could theoretically also reveal a CMV infection only limited to the right colon, small bowel, or upper GI tract.
A potential drawback of the method is the need for a fresh and liquid stool specimen. If the stool specimen cannot be homogenized due to solidification or if the stool is solid, DNA cannot be reliably extracted from the stool specimen. This is illustrated by the fact that homogenization of the stool specimen in two of our patients was not possible due to solidification of the sample. The collection and fast extraction of DNA from the stool specimen using a stabilizing buffer is also necessary to prevent DNA degradation.
In summary, this pilot study suggests a high sensitivity, specificity, and accuracy for the detection of CMV DNA in stool samples compared to mucosal biopsies using PCR. This non-invasive diagnostic method offers a fast testing protocol in the clinical in- and outpatient setting and might also present a new modality to broadly analyze the prevalence of intestinal CMV infections in IBD patients.
Acknowledgments
This study was funded by an unrestricted research grant from Procter and Gamble. Dr. Herfarth is supported by National Institutes of Health grant R21 DK080408-01 and the Crohn's and Colitis Foundation of America (CCFA).
Contributor Information
Hans H. Herfarth, Email: hherf@med.unc.edu, Division of Gastroenterology and Hepatology, Department of Medicine, University of North Carolina, Bioinformatics Bldg., CB#7080, Chapel Hill, NC 27599, USA.
Millie D. Long, Division of Gastroenterology and Hepatology, Department of Medicine, University of North Carolina, Bioinformatics Bldg., CB#7080, Chapel Hill, NC 27599, USA
Tara C. Rubinas, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC, USA
Mikki Sandridge, Division of Gastroenterology and Hepatology, Department of Medicine, University of North Carolina, Bioinformatics Bldg., CB#7080, Chapel Hill, NC 27599, USA.
Melissa B. Miller, Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC, USA
References
- 1.Galiatsatos P, Shrier I, Lamoureux E, Szilagyi A. Meta-analysis of outcome of cytomegalovirus colitis in immunocompetent hosts. Dig Dis Sci. 2005;50:609–616. doi: 10.1007/s10620-005-2544-6. [DOI] [PubMed] [Google Scholar]
- 2.Buckner FS, Pomeroy C. Cytomegalovirus disease of the gastrointestinal tract in patients without aids. Clin Infect Dis. 1993;17:644–656. doi: 10.1093/clinids/17.4.644. [DOI] [PubMed] [Google Scholar]
- 3.Minami M, Ohta M, Ohkura T, et al. Cytomegalovirus infection in severe ulcerative colitis patients undergoing continuous intravenous cyclosporine treatment in Japan. World J Gastroenterol. 2007;13:754–760. doi: 10.3748/wjg.v13.i5.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kambham N, Vij R, Cartwright CA, Longacre T. Cytomegalovirus infection in steroid-refractory ulcerative colitis: a case-control study. Am J Surg Pathol. 2004;28:365–373. doi: 10.1097/00000478-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Schulenburg A, Turetschek K, Wrba F, et al. Early and late gastrointestinal complications after myeloablative and non-myeloablative allogeneic stem cell transplantation. Ann Hematol. 2004;83:101–106. doi: 10.1007/s00277-003-0756-4. [DOI] [PubMed] [Google Scholar]
- 6.Shrestha BM, Parton D, Gray A, et al. Cytomegalovirus involving gastrointestinal tract in renal transplant recipients. Clin Transplant. 1996;10:170–175. [PubMed] [Google Scholar]
- 7.Papadakis KA, Tung JK, Binder SW, et al. Outcome of cytomegalovirus infections in patients with inflammatory bowel disease. Am J Gastroenterol. 2001;96:2137–2142. doi: 10.1111/j.1572-0241.2001.03949.x. [DOI] [PubMed] [Google Scholar]
- 8.Cottone M, Pietrosi G, Martorana G, et al. Prevalence of cytomegalovirus infection in severe refractory ulcerative and Crohn's colitis. Am J Gastroenterol. 2001;96:773–775. doi: 10.1111/j.1572-0241.2001.03620.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman HS, Kahn AC, Iacobuzio-Donahue C, Talamini MA, Lillemoe KD, Hamilton SR. Cytomegaloviral enterocolitis: clinical associations and outcome. Dis Colon Rectum. 1999;42:24–30. doi: 10.1007/BF02235178. [DOI] [PubMed] [Google Scholar]
- 10.Wakefield AJ, Fox JD, Sawyerr AM, et al. Detection of herpesvirus DNA in the large intestine of patients with ulcerative colitis and Crohn's disease using the nested polymerase chain reaction. J Med Virol. 1992;38:183–190. doi: 10.1002/jmv.1890380306. [DOI] [PubMed] [Google Scholar]
- 11.Cooper HS, Raffensperger EC, Jonas L, Fitts WT., Jr Cytomegalovirus inclusions in patients with ulcerative colitis and toxic dilation requiring colonic resection. Gastroenterology. 1977;72:1253–1256. [PubMed] [Google Scholar]
- 12.Matsuoka K, Iwao Y, Mori T, et al. Cytomegalovirus is frequently reactivated and disappears without antiviral agents in ulcerative colitis patients. Am J Gastroenterol. 2007;102:331–337. doi: 10.1111/j.1572-0241.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- 13.Kojima T, Watanabe T, Hata K, Shinozaki M, Yokoyama T, Nagawa H. Cytomegalovirus infection in ulcerative colitis. Scand J Gastroenterol. 2006;41:706–711. doi: 10.1080/00365520500408584. [DOI] [PubMed] [Google Scholar]
- 14.Dimitroulia E, Spanakis N, Konstantinidou AE, Legakis NJ, Tsakris A. Frequent detection of cytomegalovirus in the intestine of patients with inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:879–884. doi: 10.1097/01.mib.0000231576.11678.57. [DOI] [PubMed] [Google Scholar]
- 15.Krech U. Complement-fixing antibodies against cytomegalovirus in different parts of the world. Bull World Health Organ. 1973;49:103–106. [PMC free article] [PubMed] [Google Scholar]
- 16.de la Hoz RE, Stephens G, Sherlock C. Diagnosis and treatment approaches of CMV infections in adult patients. J Clin Virol. 2002;25 2:S1–12. doi: 10.1016/s1386-6532(02)00091-4. [DOI] [PubMed] [Google Scholar]
- 17.Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998;11:533–554. doi: 10.1128/cmr.11.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowshani AT, Bemelman FJ, van Leeuwen EM, van Lier RA, ten Berge IJ. Clinical and immunologic aspects of cytomegalovirus infection in solid organ transplant recipients. Transplantation. 2005;79:381–386. doi: 10.1097/01.tp.0000148239.00384.f0. [DOI] [PubMed] [Google Scholar]
- 19.Cotte L, Drouet E, Bailly F, Vitozzi S, Denoyel G, Trepo C. Cytomegalovirus DNA level on biopsy specimens during treatment of cytomegalovirus gastrointestinal disease. Gastroenterology. 1996;111:439–444. doi: 10.1053/gast.1996.v111.pm8690210. [DOI] [PubMed] [Google Scholar]
- 20.Kou T, Nakase H, Tamaki H, Kudo T, Nishio A, Chiba T. Cytomegalovirus infection in patients with ulcerative colitis diagnosed by quantitative real-time PCR analysis. Dig Dis Sci. 2006;51:1052–1055. doi: 10.1007/s10620-006-8006-y. [DOI] [PubMed] [Google Scholar]
- 21.Numazaki K, Chiba S. Current aspects of diagnosis and treatment of cytomegalovirus infections in infants. Clin Diagn Virol. 1997;8:169–181. doi: 10.1016/s0928-0197(97)10005-8. [DOI] [PubMed] [Google Scholar]
- 22.Michel D, Marre E, Hampl W, et al. Intestinal cytomegalovirus disease in immunocompromised patients may be ruled out by search for cytomegalovirus DNA in stool samples. J Clin Microbiol. 1995;33:3064–3067. doi: 10.1128/jcm.33.11.3064-3067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chemaly RF, Yen-Lieberman B, Castilla EA, et al. Correlation between viral loads of cytomegalovirus in blood and bronchoalveolar lavage specimens from lung transplant recipients determined by histology and immunohistochemistry. J Clin Microbiol. 2004;42:2168–2172. doi: 10.1128/JCM.42.5.2168-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chemaly RF, Yen-Lieberman B, Chapman J, et al. Clinical utility of cytomegalovirus viral load in bronchoalveolar lavage in lung transplant recipients. Am J Transplant. 2005;5:544–548. doi: 10.1111/j.1600-6143.2005.00747.x. [DOI] [PubMed] [Google Scholar]
- 25.Boom R, Sol C, Weel J, et al. Detection and quantitation of human cytomegalovirus DNA in faeces. J Virol Methods. 2000;84:1–14. doi: 10.1016/s0166-0934(99)00127-5. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez JL, Storch GA. Multiplex, quantitative, real-time pcr assay for cytomegalovirus and human DNA. J Clin Microbiol. 2002;40:2381–2386. doi: 10.1128/JCM.40.7.2381-2386.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell RD, Warner NE, Levine RS, Kirsner JB. Cytomegalic inclusion disease and ulcerative colitis; report of a case in a young adult. Am J Med. 1961;30:334–340. doi: 10.1016/0002-9343(61)90105-x. [DOI] [PubMed] [Google Scholar]
- 28.Domenech E, Vega R, Ojanguren I, et al. Cytomegalovirus infection in ulcerative colitis: A prospective, comparative study on prevalence and diagnostic strategy. Inflamm Bowel Dis. 2008;14:1373–1379. doi: 10.1002/ibd.20498. [DOI] [PubMed] [Google Scholar]
- 29.Kishore J, Ghoshal U, Ghoshal UC, et al. Infection with cytomegalovirus in patients with inflammatory bowel disease: Prevalence, clinical significance and outcome. J Med Microbiol. 2004;53:1155–1160. doi: 10.1099/jmm.0.45629-0. [DOI] [PubMed] [Google Scholar]
- 30.Kandiel A, Lashner B. Cytomegalovirus colitis complicating inflammatory bowel disease. Am J Gastroenterol. 2006;101:2857–2865. doi: 10.1111/j.1572-0241.2006.00869.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakase H, Matsumura K, Yoshino T, Chiba T. Systematic review: Cytomegalovirus infection in inflammatory bowel disease. J Gastroenterol. 2008;43:735–740. doi: 10.1007/s00535-008-2246-x. [DOI] [PubMed] [Google Scholar]
- 32.Hommes DW, Sterringa G, van Deventer SJ, Tytgat GN, Weel J. The pathogenicity of cytomegalovirus in inflammatory bowel disease: A systematic review and evidence-based recommendations for future research. Inflamm Bowel Dis. 2004;10:245–250. doi: 10.1097/00054725-200405000-00011. [DOI] [PubMed] [Google Scholar]
- 33.Cotte L, Drouet E, Bissuel F, Denoyel GA, Trepo C. Diagnostic value of amplification of human cytomegalovirus DNA from gastrointestinal biopsies from human immunodeficiency virus-infected patients. J Clin Microbiol. 1993;31:2066–2069. doi: 10.1128/jcm.31.8.2066-2069.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshino T, Nakase H, Ueno S, et al. Usefulness of quantitative real-time pcr assay for early detection of cytomegalovirus infection in patients with ulcerative colitis refractory to immunosuppressive therapies. Inflamm Bowel Dis. 2007;13:1516–1521. doi: 10.1002/ibd.20253. [DOI] [PubMed] [Google Scholar]
- 35.D'Ovidio V, Vernia P, Gentile G, et al. Cytomegalovirus infection in inflammatory bowel disease patients undergoing anti-tnfalpha therapy. J Clin Virol. 2008;43:180–183. doi: 10.1016/j.jcv.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Criscuoli V, Mocciaro F, Orlando A, Rizzuto MR, Renda MC, Cottone M. Cytomegalovirus disappearance after treatment for refractory ulcerative colitis in 2 patients treated with infliximab and 1 patient with leukapheresis. Inflamm Bowel Dis. 2009;15:810–811. doi: 10.1002/ibd.20742. [DOI] [PubMed] [Google Scholar]


