Abstract
Among other signals, cell growth is particularly controlled by the target of rapamycin (TOR) pathway that includes the tuberous sclerosis complex genes (TSC1/2), and through transcriptional effects regulated by c-myc. Overexpression of Drosophila Myc and TSC1/2 cause opposing growth and proliferation defects. Despite this relationship, direct regulatory connections between Myc and the TSC have only recently been evaluated. Other than studies of p53 regulation, little consideration has been given to transcriptional regulation of the TSC genes. Here we review evidence that transcriptional controls are potentially important regulators of TSC2 expression, and that Myc is a direct repressor of its expression. Since tuberin loss de-represses Myc protein, the connection between these two growth regulators is positioned to act as a feed-forward loop that would amplify the oncogenic effects of decreased tuberin or increased Myc. Further experiments will be needed to clarify the mechanisms underlying this important connection, and evaluate its overall contribution to cancers caused by TSC loss or Myc gain.
Keywords: c-Myc, translation initiation regulation, rapamycin, tuberin
Growth Regulation in Human Cancer—Opposing Roles of c-Myc and the Tuberous Sclerosis Complex
The unique metabolism of cancer cells offers a currently under-exploited target for oncogene-specific therapies.1 Indeed, enhanced anabolism in its broadest sense is one of many key events that are required for a cell to become malignant.2,3 Cell growth is generally considered to be upstream of and required for cell division. This principle was first described in yeast where DNA synthesis is initiated and a cell divides only after it grows beyond a minimum size threshold.4–6 The c-myc oncogene was one of the first transcription factors found to control transcription of processes that add mass to a cell. An early hint of myc’s control of growth was provided by evidence that it regulates the eIF4F translation initiation complex,7,8 which controls the rate limiting step in translation. Genetic experiments in Drosophila quickly confirmed dMyc’s central role in growth control.9 dMyc gain or loss had primary effects on cell growth with secondary effects on cell division. 10 This result was recapitulated in rat fibroblast cells lacking c-myc, which exhibited a growth and consequent proliferation defect.11
If there is a part of the cell that needs to increase as a cell moves toward cell division, myc is known to regulate its synthesis (Fig. 1).12 Importantly, all four components of the translation apparatus are regulated by myc: (1) It directly stimulates rRNA synthesis by RNA Polymerase I (Pol I);13–15 (2) Chromatin immunoprecipitation, conditional Myc expression, Myc overexpression in primary cells and/or expression loss in Myc null cells all implicate ribosomal proteins as Myc targets;16–18 (3) Myc activates RNA Polymerase III (Pol III) through TFIIIB;19 (4) Translation initiation factors are the key rate-limiting components of protein synthesis, and several are myc targets.18 While there is little evidence that rRNA, tRNA or ribosomal protein overexpression plays a primary role in malignancy, accumulating evidence highlights the potential importance of translation initiation factors as drivers in cancer,20–23 especially eIF4E and eIF3S6 (Int-6).24–27 On the other hand, myc’s control of glycolysis at multiple entry points suggests connections to the Warburg effect,28 showing that even general metabolic pathways can function in a specifically abnormal manner in cancer cells. Decreased cell size is considered a signature for a pathway that regulates growth, and the small cell size of dMyc null cells stands out as a strong confirmation of its importance in growth.
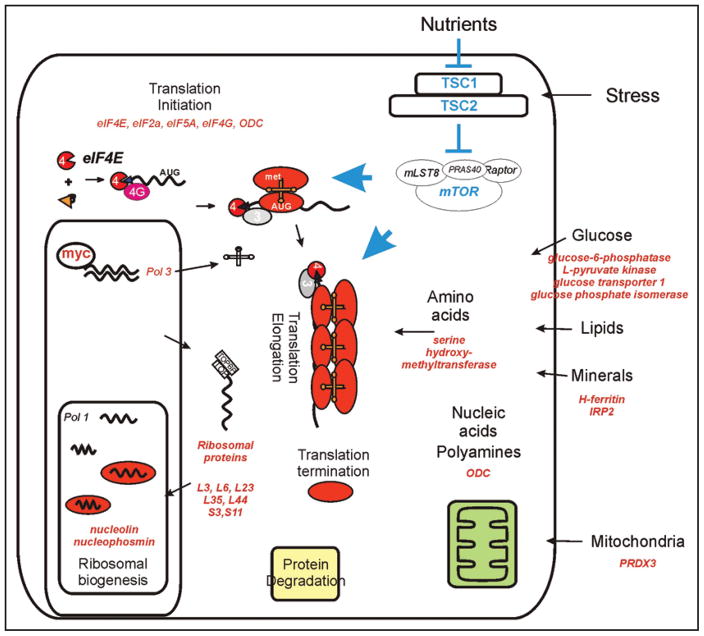
Figure 1.
A schematic diagram listing the anabolic paths containing genes regulated by c-myc. Gene products listed in red have been shown to be myc targets. Myc transcriptional regulation connects with mammalian target of rapamycin (mTOR) nutrient sensing pathway through their intersecting effects on translation initiation and elongation. This connection was first shown in Drosophila experiments,36 and mechanisms that might directly regulate their interactions are explored in this manuscript.
In comparison, the Target of Rapamycin (TOR) is a central integrator of signaling pathways whose output controls cell growth.29 Like dMyc, loss of Drosophila TOR (dTOR) also causes a small cell phenotype.30 Indeed cell size effects were a prominent readout of the various experiments that revealed the components of the TOR pathway. The small cell size of dTOR null cells was immediately noted to phenocopy loss of insulin signaling. The small size of ribosomal protein S6 kinase (S6 kinase) null flies also suggested that S6 kinase was the end target of TOR signaling, which was confirmed by S6 kinase mutant’s synthetic lethality with dTOR null flies.31 These components of the TOR signaling pathway had obvious connections to the anabolic processes important to general cell growth. However, the realization that the tuberous sclerosis (TSC1 and TSC2) genes were essential components of the TOR pathway firmly connected control of cell growth to cancer. The identification, positional cloning and characterization of tuberin first revealed its cell signaling function as a GTPase activating protein (GAP).32,33 The large size of Drosophila mutants in TSC2 then prompted the genetic studies that connected dTsc1 and dTsc2 upstream to Akt and downstream to ribosomal protein S6 kinase (S6K).34,35
Loss of the Drosophila homolog of TSC2 (dTSC2) causes enhanced growth, and this mutation was consequently named gigas.36 In an initial publication describing dTSC2 this cell size phenotype was directly compared with dMyc. dTsc1 and 2 were found to counteract Myc in both cell size and proliferation control. Mammalian experiments confirmed this antagonism between myc and the TSC genes. While studying cell size regulation by the TSC proteins, Hengstschlager and colleagues demonstrated antagonism between Myc and tuberin in mammalian cells.37 Vectors that increased expression of tuberin slowed cell proliferation and decreased cell size. Rat 1 cells expressing a chimeric construct containing the estrogen receptor hormone binding domain fused to c-myc (Rat1-MycER) together with TSC2 was activated using hydroxyl-tamoxifen, which stimulates proliferation. Importantly, myc activation reversed tuberin’s inhibition of proliferation. Although we have long understood that translation initiation control was an end output of both c-myc and TOR signaling, direct connections between the two pathways have only recently been considered.38
Do Transcriptional Controls of the Tuberous Sclerosis Complex Genes Exist?
The TOR pathway has largely been studied as a classic signal transduction path. Consequently, the vast majority of TSC studies have been performed in cell culture systems where the expression of the TSCs and other TOR pathway members is generally assumed to be constitutive. In fact, TSC2 expression varies between cell lines of different tissue origins,32 between adult and fetal tissues in mice,39 and between various tissues in mice,40 rats41,42 and humans.43,44 Analysis of the murine transcriptome emphasizes this point (Fig. 2A).45 Expression patterns of a beta-galactosidase reporter gene integrated into the TSC2 genomic locus in knockout constructs in mice confirmed that these heterogeneous tissue-specific expression patterns are controlled at the transcriptional level since the β-gal reporter protein expression patterns recapitulate TSC2 mRNA expression patterns determined by in situ analyses. While a large number of explanations are available for the relative tissue specificity of disease manifestations in patients with tuberous sclerosis, it is nevertheless interesting to note that the highest levels of TSC2 expression occur in portions of the brain most affected by the disease.39
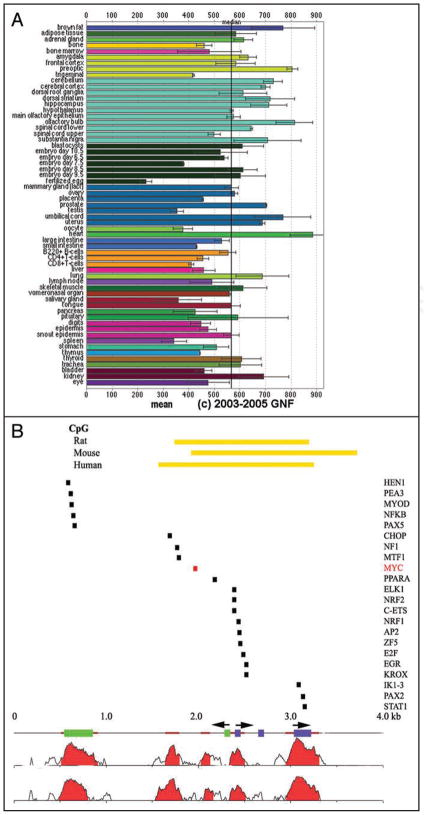
Figure 2.
(A) Tissue-specific expression of TSC2 mRNA was assessed using the transcriptome database at http://symatlas.gnf.org/SymAtlas/.112 Shown are the normalized expression levels of TSC2 in the indicated tissues, presented as mean+/− standard error. (B) An evolutionary analysis of conserved transcription factor binding sites performed using Genome rVista between the human, mouse and rat tuberous sclerosis 2 (TSC2) promoters identifies Myc binding sites as one of several candidate regulators of TSC2 expression.47,59 NTHL1 exons are identified as green rectangles and TSC2 exons as purple rectangles. The two major start sites of TSC2 are identified by arrowheads over the TSC2 exons, and the NTHL1 start site by one positioned over the green rectangles.44,46,57 Homology plots highlight conservation between mouse and human (just below the exon bars) and rat and human (at bottom). Highly conserved, non-coding regions are identified as brown bars and the CpG islands for all three promoters are identified by yellow-orange bars at top. rVista analysis identifies the listed transcription factors as those conserved between all three promoters.
Although its transcriptional control has received relatively little attention compared with studies of TSC2 in signaling pathways, the TSC2 promoter has been partially characterized using standard approaches.46 The TSC2 promoter is remarkable because it spans only 733 nucleotides and it is bi-directionally shared with the nth endonuclease III-like 1 gene (NTHL1).46 In its TSC2 orientation, the promoter controls initiation at as many as five different transcription start sites.44 The strong sequence conservation of this promoter region between human, mouse and rat strongly supports the idea that transcriptional controls of TSC2 expression are likely to be important for its proper expression.44 A conservation plot and transcription factor binding site analysis using genome rVista identifies a number of conserved transcriptional control elements in the TSC2 promoter (Fig. 2B).47 One conserved ets target site is particularly interesting because it controls expression in both the TSC2 and NTHL1 orientations.48 Taken together, these data suggest that the TSC2 promoter deserves increased attention.
Logically, one would expect that transcriptional loss of expression of the TSC tumor suppressor genes might be found in some tumor systems. Loss of tumor suppressor genes expression can often be accomplished using multiple different mechanisms in different tumors. For example, decreased TSC2 mRNA has been demonstrated in a set of brain tumors.49 Although TSC2 expression loss or promoter inactivation through methylation have not yet been generally identified,50,51 recent studies have suggested that TSC2 is a p53 regulated gene.52,53 Activation of a temperature sensitive p53 controls S6 kinase phosphorylation and sensitivity to rapamycin.52 By examining TOR pathway components, both PTEN and TSC2 mRNA levels were found to be increased by activation of p53. This induction was thought to be functionally significant because gamma-radiation induces p53, and gamma radiation induced expression of TSC2 and PTEN in a large number of cell lines in a p53-dependent manner.53 Despite these functional studies of TSC2 expression, neither chromatin immunoprecipitation nor promoter mapping has yet implicated a direct interaction between p53 and the TSC2 promoter, and it lacks a conserved p53 binding site in the rVista promoter analysis (Fig. 2B).
Evidence for Direct Controls of Tuberin by myc, and Myc by Tuberin
Both c-myc and mTOR signaling converge on gene products that control translation initiation, the rate limiting step in growth control (Fig. 1). We and others have shown that myc regulates eIF4E.7,54 The eIF4E binding protein (4EBP1) that antagonizes eIF4E is as important a downstream target of the mTOR pathway as S6 kinase.55 Since Drosophila experiments had shown antagonism between myc and tuberin, we evaluated translation initiation capacity in Myc−/− cells (HO15) to better understand the molecular basis for a possible direct interaction between myc and the mTOR pathway.11 We first evaluated the contribution of translation initiation control to their growth defect by challenging them with rapamycin. Arrested rat myc−/− and wild type cells were serum stimulated, and DNA synthesis was measured using tritiated thymidine incorporation. Rat myc−/− cells were defective in serum-stimulated DNA synthesis compared to wild types, and the mTOR inhibitor rapamycin markedly increased this effect (Fig. 3A). This differential rapamycin affect was independent of cell cycle position (Fig. 3B). Moreover, rapamycin sensitivity was at least somewhat specific for loss of myc function because murine embryonic cyclin D1 null fibroblasts showed no difference in rapamycin effect (Fig. 3C).
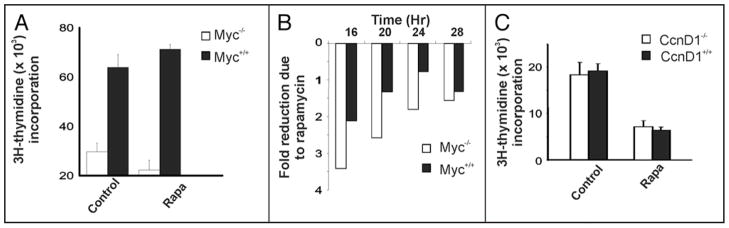
Figure 3.
DNA synthesis shows greater serum-induction and less rapamycin sensitivity in Myc null cells. Cells were arrested by confluence followed by serum starvation for 48 hours. Cells were then stimulated to re-enter the cell cycle with 10% fetal bovine serum in the presence of DMSO control, or rapamycin as described.113 Cells were then pulsed with tritiated thymidine for 2 hours, and harvested for scintillation counting as described.114 (A) Myc null (myc−/−—HO15) and wild type (myc+/+—TGR) cells stimulated with serum for 20 hours. (B) Increased sensitivity of Myc null cells to Rapamycin is independent of cell cycle position. Cells were arrested by confluence followed by serum starvation for 48 hours. Cells were stimulated to re-enter the cell cycle with 10% fetal bovine serum in the presence DMSO or rapamycin. The cells were then grown for the indicated times, pulsed with tritiated thymidine for 2 hours, and harvested for scintillation counting. Shown is the fold-reduction in thymidine uptake comparing the rapamycin treated samples to the DMSO treated control samples at each indicated time point for each cell type. (C) Same as in (A), above, except that cyclin D1 null (D) mouse embryonic cells, and their wild type counterparts, were used.
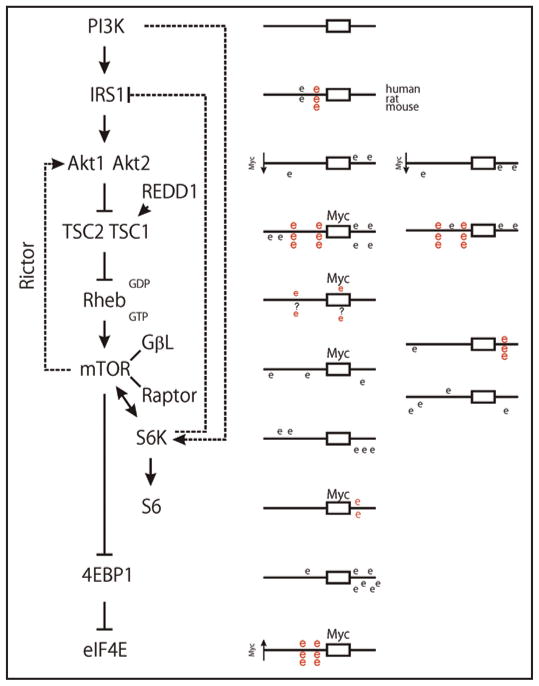
Since cells lacking c-myc were sensitive to rapamycin’s effects, we reasoned that some component(s) of the mTOR pathway might be regulated by c-myc. This idea was reinforced by the presence of a highly conserved Myc target site in the first intron of the NTHL1 gene, which is therefore positioned to control the TSC2 promoter. We further evaluated the genomic regions of all of the canonical mTOR pathway genes for Myc target sites (CACGTG) (Fig. 4) since myc often affects multiple components of a single signaling pathway. Reference cDNA sequences for human, rat and mouse mTOR pathway gene products were identified using the NCBI sequence database.56 Genomic promoter sequences and transcription initiation sites were then identified using the Database of Human Transcription Start sites,57 the Advanced Biomedical Computing Center promoter analysis software,58 and the 5′ end of the reference sequences. 5,000 nucleotides of sequence upstream and downstream of the transcription initiation sites were downloaded and manipulated using Clone Manager Suite (Scientific and Educational Software, Durham, NC). Myc target sites that were conserved between the three species were identified in the aligned regions using rVISTA.59 Importantly, in addition to TSC2 conserved myc sites were identified in the IRS1, TSC1, GβL and ribosomal protein S6 promoter regions. We then demonstrated that Myc protein bound the TSC2, GβL and rpS6 promoter sites in chromatin immunoprecipitation studies.38
Figure 4.
A TOR pathway diagram is shown, along with schematic diagrams of the CACGTG Myc target sites found within 5 kb of their transcriptional initiation sites that were either conserved in human, mouse and rat (red E) promoter regions or not conserved (black E). (Details in Ravitz et al.38) These genomic analyses were used as a starting point to test candidate proteins for changes in expression between myc null and wild type cells.
In further studies, we then directly tested the functional significance of Myc’s binding to the promoters of these mTOR signaling components.38 Translation initiation rates were significantly decreased in myc−/− cells as evaluated using polysomal profiles. 40S/60S ribosomal subunit ratios were especially affected. This decrease in the smaller 40S ribosomal subunit that contains ribosomal protein S6 seemed consistent with the potential control of the overall mTOR pathway by the three genes whose promoters were bound by myc. We therefore tested these genes for further evidence of myc regulation. Among mTOR signaling components, quantitative mRNA analysis demonstrated that Myc directly affected expression levels of the TSC2, GβL and rpS6 mRNAs. While myc activated GβL and rpS6 expression, it repressed TSC2 expression.
Importantly, conditional mycER activation increased Myc binding to the TSC2 promoter while decreasing its expression. Myc repression is essential to its capacity to transform cells, although repression through a Myc target site is not a classic mechanism.60 Repression at Myc binding sites can be accomplished by competitive binding with Mnt,61 an E box binding competitor that interacts with max. However, increased myc binding should titrate Mnt off the TSC2 promoter making Mnt an unlikely mediator. Alternative mechanisms involving Myc box 3 are under consideration.62,63 However, Myc recruitment of repressors to the initiator regions seems the most likely mechanism for this effect and the presence of a conserved YY1 site in the TSC2 promoter makes this an attractive possible mechanism.
The TSC2 repression potentially contributes to myc transformation since TSC2 mRNA levels were markedly reduced in myc-transformed RAT1A cells. The physiologic significance of Myc’s impact on mTOR signaling was further demonstrated by the effects of a TSC2 siRNA, which reversed TSC2’s effects and increased S6 kinase activity in myc null cells. Furthermore, overexpression of Myc and TSC2 was antagonistic in a soft agar colony formation assay, demonstrating a potentially significant role in myc-induced tumors. Together, these findings demonstrate that regulation of TSC2 and other components of mTOR signaling may participate in Myc’s ability to transform cells.
Oncologic Significance of Tuberin-Myc Connection
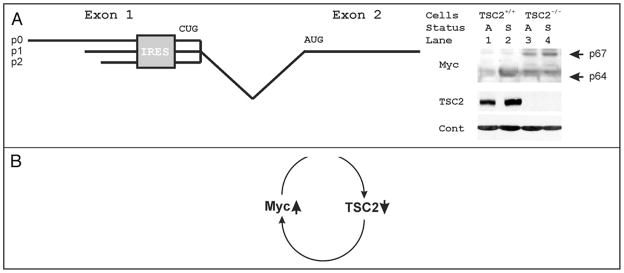
Just as Myc may control eIF4E, a feed-forward loop has been shown where increased eIF4E increases Myc protein levels.54 A variety of genes are known to be translationally controlled by eIF4E and the translation initiation apparatus.64–67 Myc is particularly controlled by the translation apparatus, largely through alterations in interactions with its internal ribosomal entry sequence.67–70 The human c-myc locus encodes two proteins, c-Myc1 and c-Myc2, that are translated from a CUG and an AUG codon respectively (Fig. 5A).71 In addition, its mRNAs initiate from three different promoters that yield different 5′ mRNAs, p0, p1 and p2. Importantly, an internal ribosomal entry site (IRES) lies just upstream of the CUG codon.68 Importantly, activation of TOR controls Myc translation and isoform expression.72 Furthermore, rapamycin inhibits both cap-dependent translation,73 and IRES dependent translation of c-myc.67 Similarly, altered c-myc regulation correlated with rapamycin resistance in cells derived from childhood cancers.74 To assess a potential feed forward effect of tuberin loss on Myc, we evaluated c-Myc protein expression in TSC2 null cells (Fig. 5B). We found that loss of TSC2 increased expression levels of the longer p67 Myc isoform. In addition, TSC2 loss also increased the smaller p64 Myc isoform in quiescent cells (compare lanes 1 and 4, Fig. 5). Thus, TSC2 levels have the potential to participate in a feed-forward loop whereby a gain of Myc would be reinforced by its translational enhancement as tuberin decreases, and a loss of TSC2 would be reinforced by a resulting increase in Myc protein levels (Fig. 5C. This feed-forward loop is positioned to enhance the known cooperative interactions between Akt and Myc transformation,75 where their dual effects on TSC2 would combine to further de-repress Rheb’s activation of mTOR (Fig. 4). Tumors caused by a c-myc transgene indeed develop more quickly in mice that are also TSC2+/−, documenting the potential importance of such a feed forward mechanism.76
Figure 5.
(A) Shown is a diagram of the first two exons of c-myc. Three initiation sites at P0, P1 and P2 result in incorporation of three different portions of exon 1 of myc in different mRNAs. An internal ribosomal entry site (IRES) is then positioned upstream of a non-canonical CUG initiation codon. The canonical initiation AUG codon is positioned in the 5′ proximal end of c-myc exon 2. (B) Protein lysates from growing wild type and TSC2 null mouse cells were probed in standard immunoblots for Myc, TSC2 and actin. Immunoblots demonstrate that TSC2 null cells lack tuberin expression, and the actin controls demonstrate equal loading. The larger and smaller Myc isoforms are indicated in contact and serum-deprivation arrested cells (A), and cells stimulated by the addition of serum for 6 hours (S). (C) Diagram of a proposed feed-forward loop whereby increased Myc causes decreased tuberin, and decreased tuberin causes increased Myc.
The tuberin gene was identified using five TSC-associated chromosomal deletions at 16p13.3.32 Loss of a second TSC2 allele was then swiftly demonstrated in renal tumors.77 Strong evidence for LOH is found in lymphangioleiomyomatosis,78–81 as well as in TS complex-associated and sporadic kidney tumors.82–86 Surprisingly, however, tumorigenesis is not always accompanied by loss of heterozygosity in tuberous sclerosis.87–89 For example, loss of TS function is harder to document in brain tumors.83,90–95 Loss of significant tumor suppressor loci or inactivation through other means can also be an important part of the pathogenesis of non-inherited cancers as was first demonstrated for the retino-blastoma gene product (pRb) in sporadic breast cancers.96 In this light, TS LOH involvement has been suggested for some sporadic tumors including bladder,97–99 breast,100 lung101,102 and pancreas.103,104 Finally, at least two recent reports have linked loss of tuberin expression with poor clinical outcome in breast and lung cancer.50,105 On the other side, the translational targets of loss of TSC1/2 are not well characterized and Myc is an important potential candidate as such a critical target.
Supporting the genetic evidence linking the TS complex with Myc, several assays for target genes of c-myc revealed connections to TSC2.106–108 A first microarray experiment compared lymphomas containing and lacking myc translocations, and neural tumors containing and lacking N-myc amplifications.106 This study suggested that TSC2 is actually induced by Myc, although this conclusion was largely the consequence of the low expression of TSC2 in the single Farage line used as the non-Myc control, and no normal cells were included for comparison. On the other hand, this publication demonstrated a classic Myc E box binding site (CACGTG) in the TSC2 promoter that was confirmed as a high affinity Myc binding site in subsequent chromatin immunoprecipitation arrays.107 More importantly, Ren and colleagues performed a cluster analysis to identify mRNAs that were associated with increased Myc levels comparing 46 tumor and normal tissues.108 They identified clusters of myc upregulated and downregulated genes whose expression was most changed in the four Burkitt lymphoma cell lines and three additional cell lines (HL60, K562 and MOLT4) known to express high levels of c-myc. TSC2 signals were markedly downregulated by myc along with a small cluster of about 15 additional genes that included the p27 tumor suppressor.
Overall, the myc-TSC2 connection now links the two best characterized growth pathways. The biggest dilemma facing myc studies is its relatively huge number of candidate targets.109 By focusing on a single signaling pathway, we identified TSC2 as the key target within the TOR signaling cascade that could be assisted by GβL, S6K and rpS6 to amplify myc’s relatively modest individual transcriptional effects, which is similar to Myc’s effects on several steps within the glycolysis pathway.28 Further work will be needed to dissect the actual mechanisms that make the TSC2 promoter a repression target where GβL and rpS6 are activation targets in that path. In that light, Pam is a protein that associates with Myc that functions as a ubiquitin ligase for tuberin,110,111 which will need to be considered. Finally, changes in Myc protein levels and isoforms associated with loss of tuberin will also need further mechanistic evaluation. Given the importance of Myc and TOR in growth regulation, the existence of a direct regulatory connection between them is likely an important part of global growth regulation.
References
- 1.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–23. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchison JM. The biology of the cell cycle. Cambridge [Eng.]: University Press; 1971. [Google Scholar]
- 5.Hartwell LH. Genetic control of the cell division cycle in yeast II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–94. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 6.Baserga R. The biology of cell reproduction. Cambridge, Mass: Harvard University Press; 1985. [Google Scholar]
- 7.Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2alpha in response to growth induction by c-myc. Proc Natl Acad Sci USA. 1993;90:6175–8. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt EV. The role of c-myc in cellular growth control. Oncogene. 1999;18:2988–96. doi: 10.1038/sj.onc.1202751. [DOI] [PubMed] [Google Scholar]
- 9.Gallant P, Shiio Y, Cheng PF, Parkhurst SM, Eisenman RN. Myc and Max homologs in Drosophila. Science. 1996;274:1523–7. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- 10.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–90. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–48. [PubMed] [Google Scholar]
- 12.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–10. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 14.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–8. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 15.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 16.Dang CV. 2005 http://www.myccancergene.org/
- 17.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–21. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–4. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 20.Rosenwald IB. The role of translation in neoplastic transformation from a pathologist’s point of view. Oncogene. 2004;23:3230–47. doi: 10.1038/sj.onc.1207552. [DOI] [PubMed] [Google Scholar]
- 21.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E—from translation to transformation. Oncogene. 2004;23:3172–9. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 22.Holland EC, Sonenberg N, Pandolfi PP, Thomas G. Signaling control of mRNA translation in cancer pathogenesis. Oncogene. 2004;23:3138–44. doi: 10.1038/sj.onc.1207590. [DOI] [PubMed] [Google Scholar]
- 23.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–99. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 24.De Benedetti A, Rhoads RE. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8212–6. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–7. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 26.Asano K, Merrick WC, Hershey JW. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997;272:23477–80. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- 27.Watkins SJ, Norbury CJ. Cell cycle-related variation in subcellular localization of eIF3e/INT6 in human fibroblasts. Cell Prolif. 2004;37:149–60. doi: 10.1111/j.1365-2184.2004.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–36. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proud CG. Role of mTOR signalling in the control of translation initiation and elongation by nutrients. Curr Top Microbiol Immunol. 2004;279:215–44. doi: 10.1007/978-3-642-18930-2_13. [DOI] [PubMed] [Google Scholar]
- 30.Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–94. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 2002;16:2627–32. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Identification and characterization of the tuberous sclerosis gene on chromosome 16. The European Chromosome 16 Tuberous Sclerosis Consortium. Cell. 1993;75:1305–15. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 33.Wienecke R, Konig A, DeClue JE. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. J Biol Chem. 1995;270:16409–14. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- 34.Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation and organ size. Cell. 2001;105:357–68. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 35.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–92. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–55. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- 37.Rosner M, Hofer K, Kubista M, Hengstschlager M. Cell size regulation by the human TSC tumor suppressor proteins depends on PI3K and FKBP38. Oncogene. 2003;22:4786–98. doi: 10.1038/sj.onc.1206776. [DOI] [PubMed] [Google Scholar]
- 38.Ravitz MJ, Chen L, Lynch M, Schmidt EV. c-myc Repression of TSC2 contributes to control of translation initiation and Myc-induced transformation. Cancer Res. 2007;67:11209–17. doi: 10.1158/0008-5472.CAN-06-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsson PG, Schofield JN, Edwards YH, Frischauf AM. Expression and differential splicing of the mouse TSC2 homolog. Mamm Genome. 1996;7:212–5. doi: 10.1007/s003359900057. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi T, Minowa O, Kuno J, Mitani H, Hino O, Noda T. Renal carcinogenesis, hepatic hemangiomatosis and embryonic lethality caused by a germ-line Tsc2 mutation in mice. Cancer Res. 1999;59:1206–11. [PubMed] [Google Scholar]
- 41.Fukuda T, Kobayashi T, Yasui H, Tsutsumi M, Konishi Y, Hino O. Distribution of Tsc2 protein in various normal rat tissues and renal tumours of Tsc2 mutant (Eker) rat detected by immunohistochemistry. Virchows Arch. 1999;434:341–50. doi: 10.1007/s004280050350. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda T, Kobayashi T, Momose S, Yasui H, Hino O. Distribution of Tsc1 protein detected by immunohistochemistry in various normal rat tissues and the renal carcinomas of Eker rat: detection of limited colocalization with Tsc1 and Tsc2 gene products in vivo. Lab Invest. 2000;80:1347–59. doi: 10.1038/labinvest.3780143. [DOI] [PubMed] [Google Scholar]
- 43.Wienecke R, Maize JC, Jr, Reed JA, de Gunzburg J, Yeung RS, DeClue JE. Expression of the TSC2 product tuberin and its target Rap1 in normal human tissues. Am J Pathol. 1997;150:43–50. [PMC free article] [PubMed] [Google Scholar]
- 44.Imai K, Sarker AH, Akiyama K, Ikeda S, Yao M, Tsutsui K, et al. Genomic structure and sequence of a human homologue (NTHL1/NTH1) of Escherichia coli endonuclease III with those of the adjacent parts of TSC2 and SLC9A3R2 genes. Gene. 1998;222:287–95. doi: 10.1016/s0378-1119(98)00485-5. [DOI] [PubMed] [Google Scholar]
- 45.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi T, Urakami S, Cheadle JP, Aspinwall R, Harris P, Sampson JR, Hino O. Identification of a leader exon and a core promoter for the rat tuberous sclerosis 2 (Tsc2) gene and structural comparison with the human homolog. Mamm Genome. 1997;8:554–8. doi: 10.1007/s003359900502. [DOI] [PubMed] [Google Scholar]
- 47.Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–9. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Honda S, Kobayashi T, Kajino K, Urakami S, Igawa M, Hino O. Ets protein Elf-1 bidi-rectionally suppresses transcriptional activities of the tumor suppressor Tsc2 gene and the repair-related Nth1 gene. Mol Carcinog. 2003;37:122–9. doi: 10.1002/mc.10123. [DOI] [PubMed] [Google Scholar]
- 49.Wienecke R, Guha A, Maize JC, Jr, Heideman RL, DeClue JE, Gutmann DH. Reduced TSC2 RNA and protein in sporadic astrocytomas and ependymomas. Ann Neurol. 1997;42:230–5. doi: 10.1002/ana.410420215. [DOI] [PubMed] [Google Scholar]
- 50.Kataoka K, Fujimoto K, Ito D, Koizumi M, Toyoda E, Mori T, et al. Expression and prognostic value of tuberous sclerosis complex 2 gene product tuberin in human pancreatic cancer. Surgery. 2005;138:450–5. doi: 10.1016/j.surg.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 51.Chakraborty S, Mohiyuddin SM, Gopinath KS, Kumar A. Involvement of TSC genes and differential expression of other members of the mTOR signaling pathway in oral squamous cell carcinoma. BMC cancer. 2008;8:163. doi: 10.1186/1471-2407-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–9. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2 and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–53. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 54.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–34. doi: 10.1158/0008-5472.CAN-07-5876. [DOI] [PubMed] [Google Scholar]
- 55.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–87. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Information NCfB. 2006 http://www.ncbi.nlm.nih.gov/
- 57.Yamashita R, Suzuki Y, Wakaguri H, Tsuritani K, Nakai K, Sugano S. DBTSS: DataBase of Human Transcription Start Sites, progress report 2006. Nucleic Acids Res. 2006;34:86–9. doi: 10.1093/nar/gkj129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Center ABC. 2006 http://grid.abcc.ncifcrf.gov/promoters.php.
- 59.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:217–21. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- 61.Hurlin PJ, Queva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes & Development. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 62.Sankar N, Baluchamy S, Kadeppagari RK, Singhal G, Weitzman S, Thimmapaya B. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene. 2008;27:5717–28. doi: 10.1038/onc.2008.181. [DOI] [PubMed] [Google Scholar]
- 63.Kurland JF, Tansey WP. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–9. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- 64.Rosenwald IB, Lazaris-Karatzas A, Sonenberg N, Schmidt EV. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Molecular & Cellular Biology. 1993;13:7358–63. doi: 10.1128/mcb.13.12.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelsen CJ, Rickheim DG, Tucker MM, Hansen LK, Albrecht JH. Evidence that cyclin D1 mediates both growth and proliferation downstream of TOR in hepatocytes. J Biol Chem. 2003;278:3656–63. doi: 10.1074/jbc.M209374200. [DOI] [PubMed] [Google Scholar]
- 66.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3′UTR. J Cell Biol. 2005;169:245–56. doi: 10.1083/jcb.200501019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi Y, Sharma A, Wu H, Lichtenstein A, Gera J. Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J Biol Chem. 2005;280:10964–73. doi: 10.1074/jbc.M407874200. [DOI] [PubMed] [Google Scholar]
- 68.Nanbru C, Lafon I, Audigier S, Gensac MC, Vagner S, Huez G, Prats AC. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–6. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 69.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–8. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 70.Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–9. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–95. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 72.West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–80. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 73.Gera JF, Mellinghoff IK, Shi Y, Rettig MB, Tran C, Hsu JH, et al. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279:2737–46. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- 74.Hosoi H, Dilling MB, Liu LN, Danks MK, Shikata T, Sekulic A, et al. Studies on the mechanism of resistance to rapamycin in human cancer cells. Mol Pharmacol. 1998;54:815–24. doi: 10.1124/mol.54.5.815. [DOI] [PubMed] [Google Scholar]
- 75.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–7. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 76.Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci USA. 2008;105:10853–8. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Green AJ, Smith M, Yates JR. Loss of heterozygosity on chromosome 16p13.3 in hamartomas from tuberous sclerosis patients. Nat Genet. 1994;6:193–6. doi: 10.1038/ng0294-193. [DOI] [PubMed] [Google Scholar]
- 78.Crooks DM, Pacheco-Rodriguez G, DeCastro RM, McCoy JP, Jr, Wang JA, Kumaki F, et al. Molecular and genetic analysis of disseminated neoplastic cells in lymphangioleiomyomatosis. Proc Natl Acad Sci USA. 2004;101:17462–7. doi: 10.1073/pnas.0407971101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sato T, Seyama K, Fujii H, Maruyama H, Setoguchi Y, Iwakami S, et al. Mutation analysis of the TSC1 and TSC2 genes in Japanese patients with pulmonary lymphangioleiomyomatosis. J Hum Genet. 2002;47:20–8. doi: 10.1007/s10038-002-8651-8. [DOI] [PubMed] [Google Scholar]
- 80.Carsillo T, Astrinidis A, Henske EP. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc Natl Acad Sci USA. 2000;97:6085–90. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smolarek TA, Wessner LL, McCormack FX, Mylet JC, Menon AG, Henske EP. Evidence that lymphangiomyomatosis is caused by TSC2 mutations: chromosome 16p13 loss of heterozygosity in angiomyolipomas and lymph nodes from women with lymphangiomyomatosis. Am J Hum Genet. 1998;62:810–5. doi: 10.1086/301804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Henske EP, Neumann HP, Scheithauer BW, Herbst EW, Short MP, Kwiatkowski DJ. Loss of heterozygosity in the tuberous sclerosis (TSC2) region of chromosome band 16p13 occurs in sporadic as well as TSC-associated renal angiomyolipomas. Genes Chromosomes Cancer. 1995;13:295–8. doi: 10.1002/gcc.2870130411. [DOI] [PubMed] [Google Scholar]
- 83.Henske EP, Scheithauer BW, Short MP, Wollmann R, Nahmias J, Hornigold N, et al. Allelic loss is frequent in tuberous sclerosis kidney lesions but rare in brain lesions. Am J Hum Genet. 1996;59:400–6. [PMC free article] [PubMed] [Google Scholar]
- 84.Longa L, Scolari F, Brusco A, Carbonara C, Polidoro S, Valzorio B, et al. A large TSC2 and PKD1 gene deletion is associated with renal and extrarenal signs of autosomal dominant polycystic kidney disease. Nephrol Dial Transplant. 1997;12:1900–7. doi: 10.1093/ndt/12.9.1900. [DOI] [PubMed] [Google Scholar]
- 85.Parry L, Maynard JH, Patel A, Clifford SC, Morrissey C, Maher ER, et al. Analysis of the TSC1 and TSC2 genes in sporadic renal cell carcinomas. Br J Cancer. 2001;85:1226–30. doi: 10.1054/bjoc.2001.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karbowniczek M, Yu J, Henske EP. Renal angiomyolipomas from patients with sporadic lymphangiomyomatosis contain both neoplastic and non-neoplastic vascular structures. Am J Pathol. 2003;162:491–500. doi: 10.1016/S0002-9440(10)63843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–93. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 88.Jozwiak J. Hamartin and tuberin: Working together for tumour suppression. Int J Cancer. 2006;118:1–5. doi: 10.1002/ijc.21542. [DOI] [PubMed] [Google Scholar]
- 89.Jozwiak J, Jozwiak S. Giant cells: contradiction to two-hit model of tuber formation? Cell Mol Neurobiol. 2005;25:795–805. doi: 10.1007/s10571-005-4932-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henske EP, Wessner LL, Golden J, Scheithauer BW, Vortmeyer AO, Zhuang Z, et al. Loss of tuberin in both subependymal giant cell astrocytomas and angiomyolipomas supports a two-hit model for the pathogenesis of tuberous sclerosis tumors. Am J Pathol. 1997;151:1639–47. [PMC free article] [PubMed] [Google Scholar]
- 91.Yeung RS, Katsetos CD, Klein-Szanto A. Subependymal astrocytic hamartomas in the Eker rat model of tuberous sclerosis. Am J Pathol. 1997;151:1477–86. [PMC free article] [PubMed] [Google Scholar]
- 92.Al-Saleem T, Wessner LL, Scheithauer BW, Patterson K, Roach ES, Dreyer SJ, et al. Malignant tumors of the kidney, brain and soft tissues in children and young adults with the tuberous sclerosis complex. Cancer. 1998;83:2208–16. [PubMed] [Google Scholar]
- 93.Przkora R, Meyer-Puttlitz B, Schmitt O, Berthold F, Nothen M, Krauss J, et al. Analysis of the TSC2 gene in human medulloblastoma. Acta Neuropathol (Berl) 2001;102:380–4. doi: 10.1007/s004010100393. [DOI] [PubMed] [Google Scholar]
- 94.Uhlmann EJ, Apicelli AJ, Baldwin RL, Burke SP, Bajenaru ML, Onda H, et al. Heterozygosity for the tuberous sclerosis complex (TSC) gene products results in increased astrocyte numbers and decreased p27-Kip1 expression in TSC2+/− cells. Oncogene. 2002;21:4050–9. doi: 10.1038/sj.onc.1205435. [DOI] [PubMed] [Google Scholar]
- 95.Mizuguchi M, Mori M, Nozaki Y, Momoi MY, Itoh M, Takashima S, Hino O. Absence of allelic loss in cytomegalic neurons of cortical tuber in the Eker rat model of tuberous sclerosis. Acta Neuropathol (Berl) 2004;107:47–52. doi: 10.1007/s00401-003-0778-y. [DOI] [PubMed] [Google Scholar]
- 96.Lee EY, To H, Shew JY, Bookstein R, Scully P, Lee WH. Inactivation of the retinoblastoma susceptibility gene in human breast cancers. Science. 1988;241:218–21. doi: 10.1126/science.3388033. [DOI] [PubMed] [Google Scholar]
- 97.Knowles MA, Habuchi T, Kennedy W, Cuthbert-Heavens D. Mutation spectrum of the 9q34 tuberous sclerosis gene TSC1 in transitional cell carcinoma of the bladder. Cancer Res. 2003;63:7652–6. [PubMed] [Google Scholar]
- 98.Hornigold N, Devlin J, Davies AM, Aveyard JS, Habuchi T, Knowles MA. Mutation of the 9q34 gene TSC1 in sporadic bladder cancer. Oncogene. 1999;18:2657–61. doi: 10.1038/sj.onc.1202854. [DOI] [PubMed] [Google Scholar]
- 99.Habuchi T, Devlin J, Elder PA, Knowles MA. Detailed deletion mapping of chromosome 9q in bladder cancer: evidence for two tumour suppressor loci. Oncogene. 1995;11:1671–4. [PubMed] [Google Scholar]
- 100.Lininger RA, Park WS, Man YG, Pham T, MacGrogan G, Zhuang Z, Tavassoli FA. LOH at 16p13 is a novel chromosomal alteration detected in benign and malignant microdissected papillary neoplasms of the breast. Hum Pathol. 1998;29:1113–8. doi: 10.1016/s0046-8177(98)90422-1. [DOI] [PubMed] [Google Scholar]
- 101.Takamochi K, Ogura T, Suzuki K, Kawasaki H, Kurashima Y, Yokose T, et al. Loss of heterozygosity on chromosomes 9q and 16p in atypical adenomatous hyperplasia concomitant with adenocarcinoma of the lung. Am J Pathol. 2001;159:1941–8. doi: 10.1016/S0002-9440(10)63041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suzuki K, Ogura T, Yokose T, Nagai K, Mukai K, Kodama T, et al. Loss of heterozygosity in the tuberous sclerosis gene associated regions in adenocarcinoma of the lung accompanied by multiple atypical adenomatous hyperplasia. Int J Cancer. 1998;79:384–9. doi: 10.1002/(sici)1097-0215(19980821)79:4<384::aid-ijc13>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 103.Francalanci P, Diomedi-Camassei F, Purificato C, Santorelli FM, Giannotti A, Dominici C, et al. Malignant pancreatic endocrine tumor in a child with tuberous sclerosis. Am J Surg Pathol. 2003;27:1386–9. doi: 10.1097/00000478-200310000-00012. [DOI] [PubMed] [Google Scholar]
- 104.Verhoef S, van Diemen-Steenvoorde R, Akkersdijk WL, Bax NM, Ariyurek Y, Hermans CJ, et al. Malignant pancreatic tumour within the spectrum of tuberous sclerosis complex in childhood. Eur J Pediatr. 1999;158:284–7. doi: 10.1007/s004310051073. [DOI] [PubMed] [Google Scholar]
- 105.Jiang WG, Sampson J, Martin TA, Lee-Jones L, Watkins G, Douglas-Jones A, et al. Tuberin and hamartin are aberrantly expressed and linked to clinical outcome in human breast cancer: the role of promoter methylation of TSC genes. Eur J Cancer. 2005;41:1628–36. doi: 10.1016/j.ejca.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 106.Schuldiner O, Benvenisty N. A DNA microarray screen for genes involved in c-MYC and N-MYC oncogenesis in human tumors. Oncogene. 2001;20:4984–94. doi: 10.1038/sj.onc.1204459. [DOI] [PubMed] [Google Scholar]
- 107.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–9. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 110.Murthy V, Han S, Beauchamp RL, Smith N, Haddad LA, Ito N, Ramesh V. Pam and its ortholog highwire interact with and may negatively regulate the TSC1. TSC2 complex. J Biol Chem. 2004;279:1351–8. doi: 10.1074/jbc.M310208200. [DOI] [PubMed] [Google Scholar]
- 111.Han S, Witt RM, Santos TM, Polizzano C, Sabatini BL, Ramesh V. Pam (Protein associated with Myc) functions as an E3 ubiquitin ligase and regulates TSC/mTOR signaling. Cell Signal. 2008;20:1084–91. doi: 10.1016/j.cellsig.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–36. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 114.Ravitz MJ, Yan S, Dolce C, Kinniburgh AJ, Wenner CE. Differential regulation of p27 and cyclin D1 by TGFbeta and EGF in C3H 10T1/2 mouse fibroblasts. J Cell Physiol. 1996;168:510–20. doi: 10.1002/(SICI)1097-4652(199609)168:3<510::AID-JCP3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]