Abstract
Genes and gene products do not function in isolation but within highly interconnected “interactome” networks, modeled as graphs of nodes and edges representing macromolecules and interactions between them, respectively. We propose to investigate genotype-phenotype associations by methodical use of alleles that lack single interactions, while retaining all others, in contrast to genetic approaches designed to eliminate gene products completely. We describe an integrated strategy based on the reverse yeast two-hybrid system to isolate and characterize such edge-specific, or “edgetic” alleles. We establish a proof-of-concept with CED-9, a C. elegans BCL2 ortholog involved in apoptosis. Using ced-9 edgetic alleles, we uncover a new potential functional link between apoptosis and a centrosomal protein, demonstrating both the interest and efficiency of our strategy. This approach is amenable to higher throughput and is particularly applicable to interactome network analysis in organisms for which transgenesis is straightforward.
Classical “forward” genetics and functional genomics or “reverse” genetics have together assigned potential function(s) to tens of thousands of genes across dozens of organisms. With the availability of genome sequences and the development of automated phenotypic analyses, reverse genetics strategies based on null or nearly null alleles such as gene knockouts and RNA interference-based knockdowns are rapidly becoming a major sour ce of gene function information. However, functional interpretation of (nearly) null alleles is often complicated by the fact that gene products do not operate in isolation but act upon each other within complex and dynamic interaction, or “interactome” networks.
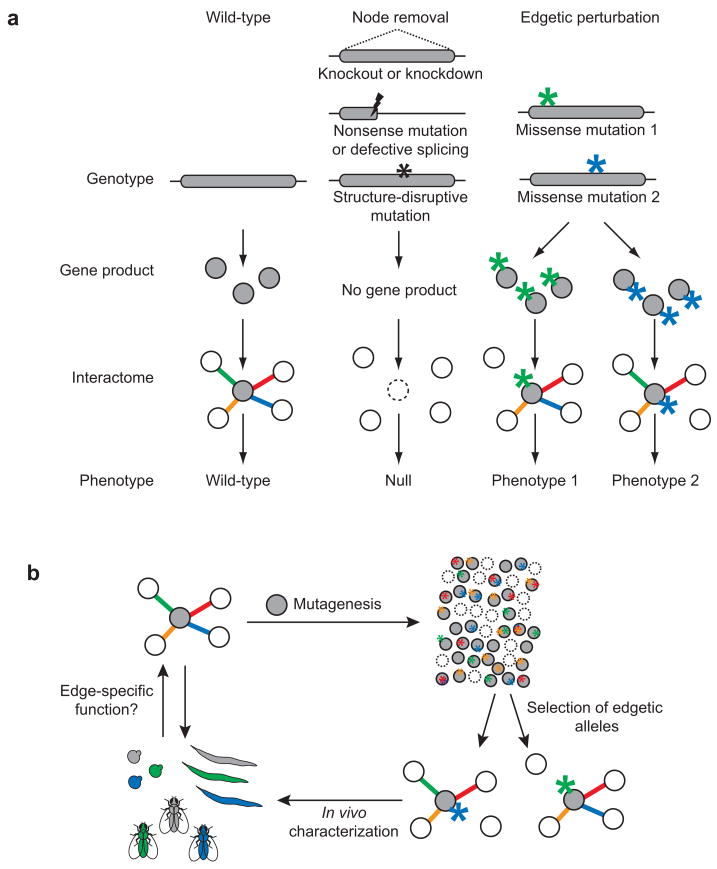
In interactome graphs, where macromolecules and interactions between them are represented by “nodes” and “edges”, respectively, knockouts or knockdowns should be modeled as eliminating a node and all its edges (Fig. 1a)1,2. More precise determination of molecular function(s) should be possible with the development of new systematic strategies to generate alleles that perturb a single interaction or edge at-a-time, while maintaining all others unperturbed. Systematic use of such “edgetic” alleles should be useful to evaluate in vivo roles of individual interactions (Fig. 1b).
Figure 1.
Schematic representations of genotype-phenotype associations. (a) Possible phenotypes resulting from distinct network perturbations caused by different experimental strategies or mutation types. Lightning bolts, nonsense mutations; stars, missense mutations. (b) Edgetic strategy applied to a protein of interest (grey node). Colors represent specific edges, their specific perturbation, and the specific corresponding phenotypes.
The reverse yeast two-hybrid (R-Y2H) and one-hybrid (R-Y1H) systems are powerful tools to identify mutations disrupting protein-protein and DNA-protein interactions using genetic selections3–6. However the efficiency of early versions of the R-Y2H system was somewhat limited by the fact that most R-Y2H interaction-defective alleles correspond to truncating mutations unless a strategy is used to enrich for missense mutations5,7–9. While dual-reporter systems were developed to eliminate missense alleles, these systems only allow assaying two partners of a protein simultaneously and are limited if the two partners studied bind to the same region10,11.
In this report, we describe an integrated strategy to systematically isolate edgetic alleles for subsequent in vivo characterization. As proof-of-concept, we applied this strategy to Caenorhabditis elegans CED-912, an ortholog of the human anti-apoptotic oncoprotein BCL2. We efficiently identified a spectrum of edgetic alleles with various interaction defects caused by specific perturbations of CED-9 binding sites. A subset of ced-9 edgetic alleles reintroduced in vivo caused specific phenotypes clearly distinct from the ced-9 null phenotype, and suggestive of a physical and functional link between apoptosis and the centrosome. Our integrated pipeline interrogates interaction networks by perturbing edges instead of nodes, and therefore complements the technological arsenal provided by gene knockouts and gene knockdowns to systematically investigate gene function.
RESULTS
The C. elegans BCL2 family member CED-9 prevents apoptosis by sequestering the Apaf-1 ortholog CED-413. Apoptosis is triggered when EGL-1 (a BCL2 homology domain 3 (BH3) protein) expression is turned on14. By physically interacting with CED-9, EGL-1 triggers conformational changes in CED-9, releasing CED-4 and allowing CED-4-mediated activation of the CED-3 caspase15,16.
The ced-9 gene was initially identified through the isolation of a dominant allele, ced-9(n1950), which suppresses apoptosis12,17. In this allele a single amino acid change, G169E, prevents EGL-1-induced dissociation of otherwise wild-type CED-9/CED-4 complex formation, and thus CED-9(G169E) can be considered edgetic by our definition. All four additional alleles currently available [ced-9(n2812); ced-9(n2077); ced-9(n2161); ced-9(n1653ts)] result in complete or near complete CED-9 depletion, i.e. CED-9 node removal (Supplementary Fig. 1).
Isolation of ced-9 edgetic alleles insensitive to EGL-1
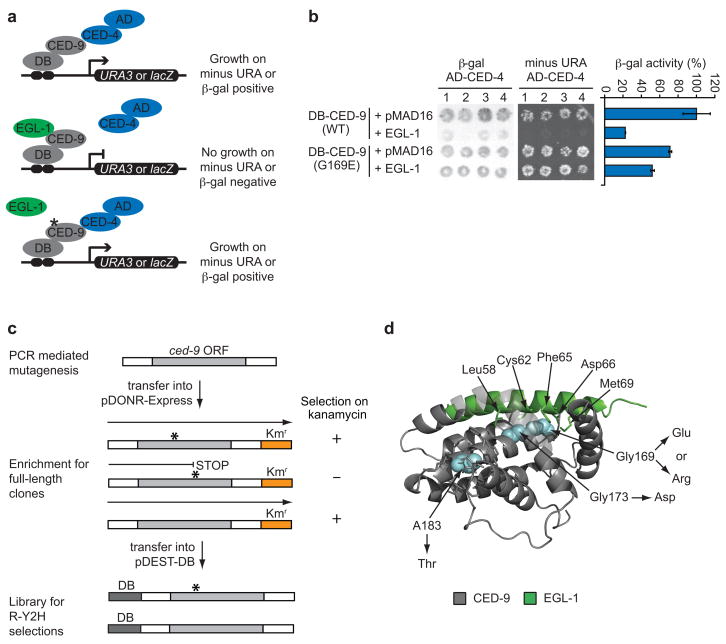
We tested whether our strategy could be used to isolate CED-9(G169E)-like edgetic alleles. We first demonstrated that Y2H is suitable to: (i) detect the CED-9/CED-4 interaction, (ii) reconstitute the EGL-1-induced dissociation of this interaction, and (iii) recapitulate the CED-9(G169E) edgetic profile (Fig. 2a,b; see Supplementary Data 1 for details). We then generated a CED-9ΔTM [CED-9 lacking its C-terminal transmembrane domain] mutant library enriched for full-length ORFs (Fig. 2c and Supplementary Table 1) and used Y2H to select from this library those mutants maintaining the interaction with CED-4 in the presence of EGL-1, as indicated by growth on selective media lacking uracil.
Figure 2.
Isolation of ced-9 alleles insensitive to EGL-1. (a) Schematic of modified Y2H assay used to identify edgetic alleles that maintain CED-9 interaction with CED-4 in presence of EGL-1. DB: Gal4-DNA binding domain. AD: Gal4-activation domain. (b) Y2H phenotypes of the interaction between CED-4 and CED-9 (wild-type or G169E) in the absence or presence of EGL-1. Each combination is shown in quadruplicate. Panels show a filter β-galactosidase assay (left), growth assay on media without uracil (middle) and a quantitative β-galactosidase assay (right). Error bars represent standard error of the mean (n = 4). (c) CED-9ΔTM mutant library generation. ORFs mutagenized by PCR are cloned by Gateway reaction into pDONR-Express, a bacterial expression vector containing a kanamycin (Kan) resistance-encoding gene (Kmr) placed in frame with the ORF cloning site8. The selection of E. coli transformants on Kan-containing plates is designed to eliminate nonsense mutations and out-of-frame changes, enriching the library with full-length ORFs that can then be transferred into the pDEST-DB Y2H vector by Gateway reaction for R-Y2H selections. White boxes surrounding ced-9 ORF represent Gateway recombination sites. (d) Crystal structure of a CED-9 (grey)/EGL-1 (light green) complex (PDB ID code 1TY4)18. Blue, residues mutated in ced-9 edgetic alleles insensitive to EGL-1. Substitutions are indicated. EGL-1 residues less than 4 A away from CED-9 mutated residues and CED-9 residues less than 4 A away from A183 are shown as sticks. For clarity hydrogen atoms have been omitted.
We identified four alleles that maintain CED-9 interaction with CED-4 despite the presence of EGL-1. Strikingly, two of these contain substitutions of G169, the amino acid mutated in ced-9(1950), the dominant allele originally isolated in a forward genetic screen (Fig. 2d). One change corresponds exactly to the previously described ced-9(1950) G169E substitution, demonstrating the power of our Y2H genetic selection. The other change, G169R, is new but similar to G169E, i.e. substitution of a glycine by a bulky charged residue. The third allele is a G173D mutation, a substitution of a glycine close to G169 in the CED-9 sequence. In the CED-9/EGL-1 co-crystal18, G169 and G173 are adjacent and are both in contact with the EGL-1 BH3 peptide (Fig. 2d). The A183T substitution in the fourth allele affects a residue outside of the EGL-1 BH3 binding groove but within the same α-helix as G169 and G173. An A183Y substitution decreases the melting temperature of the CED-9/EGL-1 complex by 5°C19, consistent with our observation that a mutation of the same residue to a threonine affects the CED-9/EGL-1 interaction. Altogether we demonstrated that a genetic selection in the appropriate Y2H yeast strain background can be used to efficiently isolate ced-9 edgetic alleles.
Integrated strategy to isolate edgetic alleles
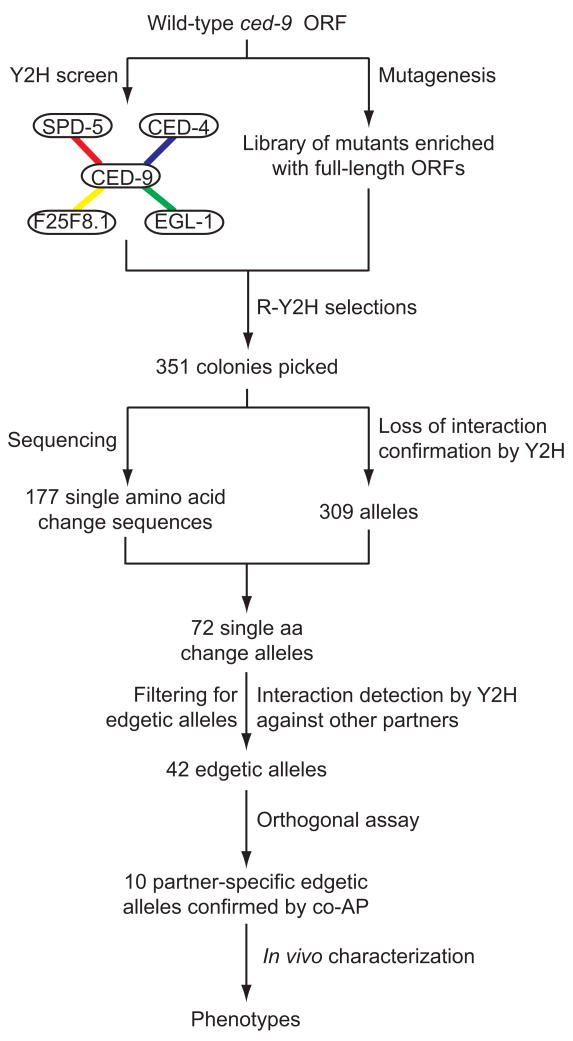
We generalized the approach for less characterized and novel interactions (Fig. 3). We screened CED-9ΔTM against C. elegans cDNA20 and ORFeome21 libraries by Y2H. We recovered both EGL-1 and CED-4 as CED-9ΔTM interactors, validating our Y2H screen14,22. We also identified two novel Y2H interactors: residues 829–1198 of SPD-5 and full-length F25F8.1. SPD-5 is a centriole protein essential for centrosome maturation and mitotic spindle assembly23. F25F8.1 is an uncharacterized protein with no known orthologs outside of the Caenorhabditis genus. We validated all four Y2H interactions by co-affinity purification (co-AP) in human HEK293T cells using a Glutathione-S-Transferase (GST) pull-down protocol21 with CED-9ΔTM and full-length EGL-1, CED-4, SPD-5, and F25F8.1 (Supplementary Fig. 2). Having validated SPD-5 and F25F8.1 as likely bona fide biophysical interactors, we used our edgetic strategy to interrogate the biological relevance of these interactions.
Figure 3.
Schema of the edgetic strategy. The interaction network of CED-9 is mapped by Y2H and confirmed by Co-AP in human HEK293T cells (Supplementary Fig. 2). In parallel, a CED-9ΔTM mutant library enriched for full-length ORFs is generated (Fig. 2c). Interaction defective alleles are isolated by R-Y2H from the CED-9ΔTM mutant library (Supplementary Data 2). PCR amplicons of ced-9 alleles obtained directly from yeast colonies are sequenced to identify potential mutations and reintroduced by gap repair into fresh yeast cells to confirm loss-of-interaction phenotypes by Y2H (Supplementary Figs. 3–5, Supplementary Tables 2–4, and Supplementary Data 2). Interaction-defective alleles are subsequently tested by Y2H against other CED-9 partners to distinguish between edgetic and non-edgetic alleles (Supplementary Figs. 3–5, Supplementary Tables 2–4, and Supplementary Data 2). The interaction profiles of a subset of interaction-specific edgetic alleles are validated by co-AP in human HEK293T cells (Supplementary Fig. 6 and Supplementary Data 2). Validated alleles are expressed in vivo and the phenotypic consequences of their expression examined (Fig. 6).
From the CED-9ΔTM mutant library (Figs. 2c and 3), we used R-Y2H to select mutants unable to interact with either CED-4 or SPD-5, taking advantage of SPAL10::URA36, a counter-selectable reporter gene whose expression causes toxicity in the presence of 5-fluoroorotic acid (5-FOA)24; loss of interaction results in the ability to grow on plates containing 5-FOA. Since the CED-9/F25F8.1 interaction does not confer 5-FOA sensitivity, we screened for ced-9 mutants unable to interact with F25F8.1 by looking for decreased GAL1::lacZ-inducedβ-gal activity (see Supplementary Data 2 for details). Altogether, a total of 351 potential ced-9 alleles were obtained, with 192, 144, and 15 of them unable to interact with CED-4, SPD-5, and F25F8.1, respectively.
After PCR amplification of these potential alleles, sequencing and interaction detection by Y2H against all CED-9 partners to confirm the interaction defects and determine their specificity (Fig. 3, Supplementary Figs. 3–5, Supplementary Tables 2–5, and Supplementary Data 2), we found 42 alleles with an edgetic profile (that is, disrupting one or a subset of interactions), each affecting one of 33 different amino acids along the CED-9 sequence (~13% of the sequence, Supplementary Table 5). In contrast, 30 alleles impair all CED-9 binding capacities and therefore are considered non-edgetic.
To further validate the interaction profiles of selected edgetic alleles, we used co-affinity purification (co-AP) pull-downs in human HEK293T cells as an orthogonal protein interaction assay21,25. We tested 16 partner-specific edgetic alleles, five defective for CED-4, nine for SPD-5, and two for F25F8.1, for their ability to bind all four CED-9 interactors mentioned above (Supplementary Fig. 6). A substantial proportion of edgetic alleles (10 out of 16) obtained using the R-Y2H could be validated by co-AP (Supplementary Data 2).
Structural analysis of edgetic and non-edgetic residues
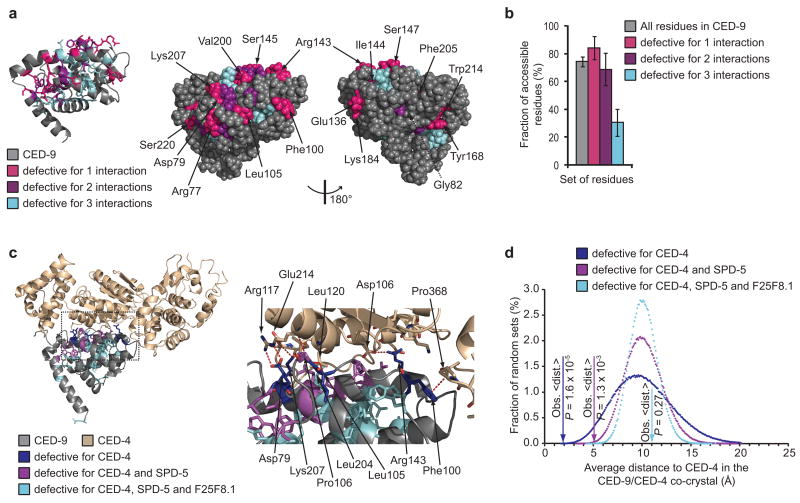
Although our edgetic strategy does not require a priori knowledge of tertiary structure, we reasoned that we could use such information to investigate the properties of residues mutated in our edgetic and non-edgetic alleles (“edgetic and non-edgetic residues”, respectively). To assess whether affected residues are preferentially located in protein binding sites, we quantified their surface exposure in the CED-9 tertiary structure (Fig. 4a). We defined as solvent-accessible those residues that have 10% or more of solvent-accessible surface area in at least one of the three available CED-9 crystal structures16,18,19. This criterion takes into account variations between these three structures. Edgetic residues, especially those mutated in alleles defective for one interaction, are on average more accessible than non-edgetic residues (Fig. 4a,b; see Supplementary Data 3 for details). Noticeably, the average surface exposure observed for the non-edgetic residues is significantly lower than expected by chance (P < 10−6; empirical P-value; Supplementary Fig. 7).
Figure 4.
Edgetic and non-edgetic residues in CED-9 and CED-9/CED-4 structures. (a) Ribbon diagram of CED-9 (PDB ID code 1OHU)19 (left); residues mutated in R-Y2H alleles are shown as sticks (hydrogen atoms omitted). Space filling representation of the same structure in the identical (middle) and opposite (right) orientations. Residues mutated in alleles defective for only one interaction are labeled. G82 (dashed line) is buried. (b) Fraction of residues (all residues versus the residues mutated in the indicated sets of ced-9 alleles) accessible in at least one of the three CED-9 structures16,18,19 using a 10% solvent-accessible surface area cutoff. Error bars represent standard error for a binomial distribution. (c) Ribbon diagram of CED-9 complexed with one CED-4 monomer (PDB ID code 2A5Y)16 (left); residues mutated in CED-4-interaction defective alleles are shown as sticks. In the zoom-in of the same view (right), CED-4 residues that interact with CED-9 residues mutated in CED-4-specific edgetic alleles are also shown as sticks. Red dashed lines, interactions. Oxygen atoms are red and nitrogen atoms blue. Hydrogen atoms are omitted. (d) Distribution of the average distance to CED-4 in the CED-9/CED-4 co-crystal obtained for 1,000,000 random sets of 6, 14 or 24 residues as compared to the average distance (Obs. <dist.>) of the residues mutated in the indicated sets of CED-4-interaction defective alleles.
These observations suggest that edgetic alleles of ced-9 target relatively more accessible residues that are likely part of interaction regions. In contrast, non-edgetic alleles are defective for all three CED-9 interactions because of disruptive substitutions in the CED-9 core. Corroboration comes from the non-conservative nature of these substitutions (Supplementary Table 5). For instance, 10 non-edgetic alleles (~1/3) have an α-helix residue mutated into proline. Strikingly, two non-edgetic alleles that we isolated [CED-9(Y149C) and CED-9(Y149H)] contain a substitution of the tyrosine which is mutated in the ced-9(n1653ts) allele (Supplementary Fig. 1) and was shown to be crucial for CED-9 structure18. Besides demonstrating the efficiency of our strategy, this finding further supports the proposal that non-edgetic alleles are defective for all interactions because of a disrupted CED-9 tertiary structure.
If non-edgetic mutations disrupt CED-9 tertiary structure while edgetic mutations affect specific interaction regions, non-edgetic alleles should tend to encode relatively unstable proteins. To test this, we expressed wild-type CED-9, a set of 14 edgetic alleles defective for one interaction and 14 non-edgetic alleles, all as GST fusion proteins in human cells (Supplementary Fig. 8). Proteins encoded by edgetic alleles were expressed at levels comparable to that of wild-type CED-9 fusion protein. In contrast, the non-edgetic mutant proteins could not be detected or were expressed at much lower levels than wild-type CED-9. As expected, two CED-9 truncated proteins also showed reduced expression levels (STOP1 and STOP2, Supplementary Fig. 8). Non-edgetic mutations at position Y149 (Y149C and Y149H) resulted in decreased stability and poor expression of CED-9 as previously reported for the Y149N mutation18. These data strongly suggest that edgetic and non-edgetic alleles result from distinct molecular defects: destabilization and degradation for the non-edgetic alleles, and more subtle changes that do not affect the overall stability of CED-9 for edgetic alleles.
Edgetic and non-edgetic residues in binding sites
To further test our model for the structural basis of edgetic versus non-edgetic alleles, we took advantage of the CED-9/CED4 co-crystal16, locating in this quaternary structure the residues mutated in all alleles defective for CED-4 interactionFig. 4c). All six distinct residues mutated in the alleles defective for CED-4 interaction only are located at the CED-9/CED-4 interface, significantly more than expected by chance (P = 1.2 × 10−4; hypergeometric test). Half of the 14 residues mutated in the edgetic alleles defective for CED-4 and one additional partner are at the CED-4 binding site, also more than expected by chance (P = 0.022; hypergeometric test). In contrast, only one of the 24 non-edgetic residues is in contact with CED-4, a significantly unlikely occurrence (P = 9.5 × 10−3; hypergeometric test).
We also compared the average distance to CED-4 of the residues in each set to the average distance of sets of residues picked at random (Fig. 4d). While 24 CED-9 residues picked at random have one chance in four to be further away from CED-4 than the non-edgetic residues (P = 0.27; empirical P-value), the edgetic residues are significantly closer to CED-4 than expected by chance (P = 1.3 × 10−3 and 1.6 × 10−5 for the alleles defective for two and one interaction, respectively; empirical P-values). These results argue for fundamentally different properties of edgetic versus non-edgetic perturbations, where mutations of edgetic residues likely result in the alteration of the CED-9/CED-4 interface, whereas mutations of non-edgetic residues likely disrupt the CED-9/CED-4 interaction by altering CED-9 structure.
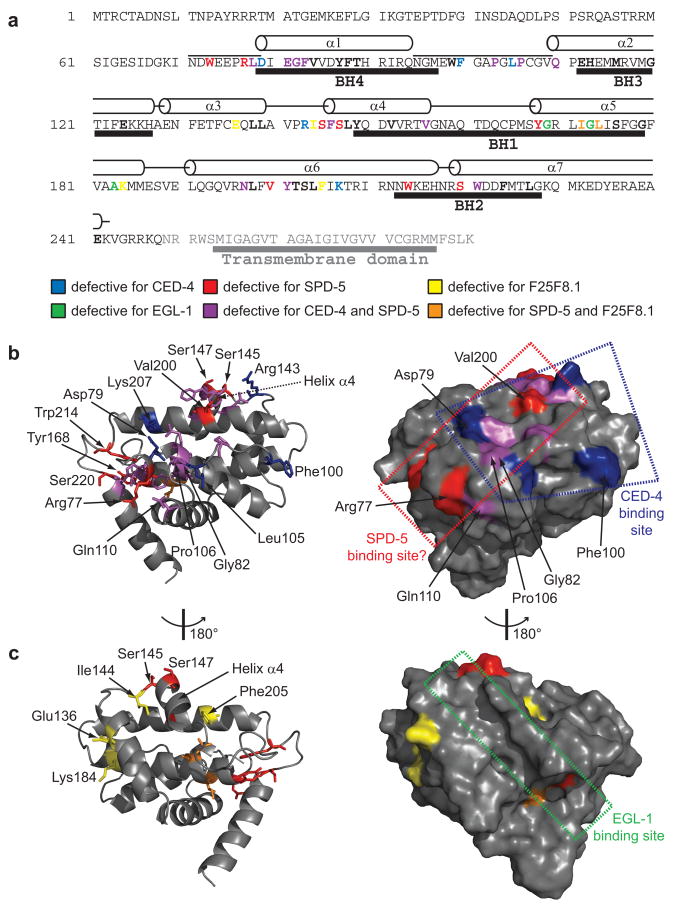
Since there is no obvious clustering of edgetic residues for any specific partner on the CED-9 primary sequence (Fig. 5a), suggesting that the binding sites for SPD-5, F25F8.1 and CED-4 are conformational, we used sets of edgetic residues to map the putative binding sites for SPD-5 and F25F8.1 (Fig. 5b,c, Supplementary Figs. 9 and 10; see Supplementary Data 4 for detail). Our edgetic strategy enabled the isolation of partner-specific edgetic alleles for each CED-9 partner even though the CED-9 interaction surfaces seem intricate, with partly overlapping sites.
Figure 5.
Positioning edgetic residues in CED-9 structures. (a) Positions of edgetic residues in the CED-9 sequence. The portion of CED-9 present in the crystal (PDB ID code 1OHU)19 and the α-helices observed in the corresponding structure are indicated above the sequence; BCL2 Homology (BH) domains19 are indicated under the sequence. Edgetic and non-edgetic residues are in bold font, edgetic residues are colored as indicated. (b) Ribbon diagram of the CED-9 structure (PDB ID code 2A5Y)16 (left); residues mutated in edgetic alleles defective for CED-4 and/or SPD-5 interaction are shown as sticks. Helix α4 (the region undergoing EGL-1-induced conformational changes) is indicated18. Van der Waals surface of the same structure in identical orientation (right). The CED-4 binding site and the hypothetical SPD-5 binding site are shown. (c) Ribbon diagram of the CED-9 structure (PDB ID code 2A5Y)16 (left) at opposite orientation with respect to (b). Residues mutated in edgetic alleles defective for SPD-5 and/or F25F8.1 interactions are shown as sticks. Van der Waals surface of the same structure in identical orientation (right). The EGL-1 binding site is shown.
Node removal and edgetic perturbation in vivo
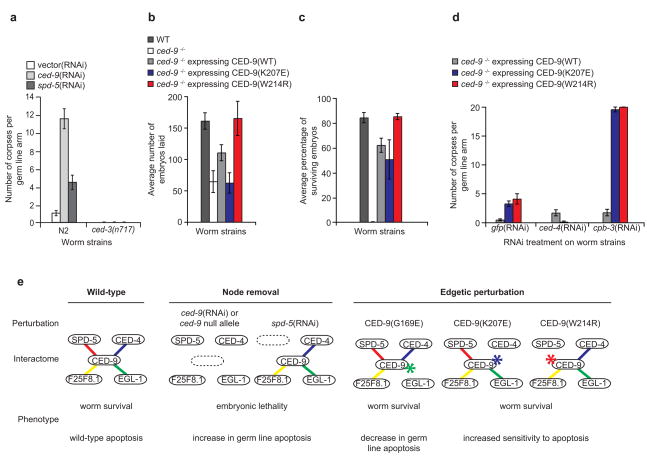
RNAi of ced-9 results in apoptosis-triggered embryonic lethality26 due to increased germ cell death (Fig. 6a). In addition to previously described defects in mitotic spindle assembly resulting in embryonic lethality23, RNAi of spd-5 appeared to also increase apoptosis in the germ line, approximately half as much as did ced-9(RNAi) (Fig. 6a). This observation is consistent with our identification of SPD-5 as a biophysical interactor of CED-9 and suggests that SPD-5, in addition to its role in mitosis, is also involved in apoptosis regulation. As with ced-9(RNAi), spd-5(RNAi)-induced germ cell apoptosis was suppressed in a ced-3 null background, indicating that spd-5 and ced-9 act within the same genetic pathway as ced-3.
Figure 6.
Node removal and edgetic perturbation in vivo. (a) Corpse count per germ line arm in wild-type (N2) or apoptosis-defective ced-3(n717) worms treated with the indicated RNAi. Vector(RNAi) is the negative control. (b) Average number of embryos laid in the indicated strains (n~10 worms for each strain). Error bars represent standard error of the mean (c) Fraction of worm broods reaching adulthood in the indicated strains. Error bars represent standard error for a binomial distribution. (d) Corpse count per germ line arm in the indicated strains treated with RNAi targeting gfp (negative control), ced-4, or cpb-3. Error bars represent standard error of the mean. (e) Schematic of phenotypic consequences of selected network perturbations. Node removal is induced either by ced-9 or spd-5(RNAi), while edgetic perturbation is caused by the CED-9(G169E)12,17, CED-9(W214R), or CED-9(K207E) mutations. WT: wild-type [unc-69(e587)] worm strain. ced-9−/−: worm strain carrying a ced-9 null allele [ced-9(n1950n2161)]. CED-9 wild-type, CED-9(K207E) and CED-9(W214R) are expressed from constructs integrated into the ced-9 null allele worm strain genetic background.
To evaluate if the CED-9/SPD-5 interaction directly contributes to the embryonic lethality and germ cell death observed upon spd-5(RNAi), we characterized worms carrying CED-9(W214R), a SPD-5-specific edgetic allele. In parallel we analyzed worms carrying CED-9(K207E), a CED-4-specific edgetic allele, to evaluate the phenotypic consequences of perturbing the CED-9/CED-4 interaction.
We generated transgenic lines carrying CED-9(K207E) and CED-9(W214R) by microparticle bombardment, and crossed them into worms carrying the ced-9(n2161) null allele. Compared to worms rescued with a wild-type ced-9 transgene, CED-9(K207E) worms laid fewer embryos (P = 0.04; Student t-test), similar to ced-9 null mutants (Fig. 6b). However wild-type ced-9 transgene and both edgetic alleles were able to rescue the embryonic lethality conferred by the ced-9 null allele (Fig. 6c). Even though we cannot exclude that CED-9(K207E) could retain some residual capacity to bind CED-4 in vivo while being impaired for this interaction ex vivo, the rescue observed with this transgene suggests that the anti-apoptotic action of CED-9 during embryonic development is not exclusively correlated to CED-4 sequestration. These data also show that the embryonic lethality that occurs in animals subjected to spd-5(RNAi) is not necessarily due to loss of the CED-9/SPD-5 interaction as worms expressing CED-9(W214R) are viable.
The viability of transgenic worms expressing CED-9(K207E) or CED-9(W214R) allowed investigation of the role of CED-9/CED-4 and CED-9/SPD-5 interactions in germ line apoptosis. We subjected animals to apoptotic challenges induced by ced-4(RNAi) and cpb-3(RNAi), which suppress and mildly increase germ cell apoptosis, respectively27,28. Without an apoptotic challenge [gfp(RNAi)], CED-9(K207E) and CED-9(W214R) worms exhibited a small increase in germ line apoptosis compared to worms rescued with a wild-type ced-9 allele (~3, ~4 and <1 dead cells respectively, P = 8.2 × 10−5 and 0.05; Student t-test) (Fig. 6d), but germ cell apoptosis in these animals was much less pronounced relative to ced-9(RNAi)-induced apoptosis (Fig. 6a). As anticipated, ced-4(RNAi) of CED-9(K207E) and CED-9(W214R) mutants suppressed apoptosis in the germ line. RNAi treatment of cpb-3 strongly increased germ cell apoptosis in CED-9(K207E) and CED-9(W214R) worms, compared to the small increase observed in animals rescued with a wild-type ced-9 allele (~18, ~20 and ~2 dead cells respectively, P = 5.2 × 10−12 and 5.8 × 10−8; Student t-test). This finding argues that, similarly to CED-9/CED-4, the CED-9/SPD-5 interaction also protects germ cells from apoptosis.
Node and edge removal can result in diverse phenotypic profiles, uncovering different aspects of the apoptosis module (Fig. 6e). RNAi experiments presented here implicate ced-9 and spd-5 in ced-3-mediated germ line apoptosis, and show genetic links between these actors. As both ced-9 and spd-5 are essential genes, knockdowns lead to embryonic lethality, precluding further characterization. Partner-specific edgetic alleles described in this study, which restore viability of mutant worms, underscore that in contrast to the CED-9/EGL-1 interaction both CED-9/CED-4 and CED-9/SPD-5 protein-protein interactions contribute to negative control of germ line apoptosis, especially in response to particular apoptotic triggers.
DISCUSSION
We present here an integrated strategy to select ex vivo a series of edgetic alleles specifically defective for one or a few protein interactions, as a way to better understand their role in complex interaction networks. Applying this strategy to CED-9, we provide evidence that we can identify edgetic alleles that: (i) lack only a subset of interactions, as supported both by Y2H experiments with two distinct reporter genes and by co-AP data, (ii) have interaction defects that are likely due to specific changes in or close to protein interaction sites, as revealed by structural analyses of CED-9 alone, or in complex with CED-4 or with the EGL-1 BH3 domain, and (iii) have in vivo phenotypes different from those caused by null or near-null perturbations (Fig. 6e). An edgetic mutation that only affects the interaction between CED-9 and SPD-5 results in increased sensitivity to apoptotic stimuli. In contrast, the null phenotype for the corresponding genes is embryonic lethality. Hence, our platform represents one alternative to define functions for essential genes beyond their null, complete loss-of-function phenotype.
Though the normal biological function of the CED-9/SPD-5 interaction is not fully defined, there are two likely possibilities. The CED-9/SPD-5 interaction may be required to suppress apoptosis during spindle assembly, during centrosome assembly, or at other times during cell division when SPD-5 is present. Alternatively SPD-5 may “moonlight”29 in the apoptotic pathway, as supported by the fact that loss of the CED-9/SPD-5 interaction sensitizes cells to apoptosis caused by loss of CPB-3, an RNA binding protein with no known function in spindle assembly or cell division.
Biological systems consist of interaction networks in which many types of macromolecules associate with and act upon each other, and biological properties of living organisms reflect the local and global properties of these networks. For example, several well-characterized inherited human disease alleles associated to particular disease phenotypes have been shown to correspond to edgetic perturbations30. Among the many biophysical interactions identified so far, a critical step is to identify the biologically relevant ones31 and understand how they contribute to cellular systems. To address such questions, tools that probe interactions (edges) rather than macromolecules themselves are needed. Considering that tens of thousands of interactions have been mapped for an increasing number of organisms, such edgetic perturbation strategies must be compatible with high-throughput settings. Our platform is a reverse genetics strategy to interrogate protein-protein interactions in the context of interactome networks. We propose the systematic use of “edgetic perturbation” reagents, whether as alleles or small compounds, to analyze the properties of interaction networks.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Stan Korsmeyer. We thank the members of the Vidal Lab and the Dana-Farber Cancer Institute Center for Cancer Systems Biology (CCSB) and particularly Anne-Ruxandra Carvunis for helpful discussions. This work was supported by National Institutes of Health grants R01 HG001715 from the NHGRI and NIGMS and R33 CA105405, R33 CA132073 and R21/R33 CA081658 from the NCI (M.V.), U01 CA105423 from the NCI (PI, Stuart Orkin, project leader, M.V.), and by Institute Sponsored Research funds from the Dana-Farber Cancer Institute Strategic Initiative awarded to CCSB. M.D. and G.L. were supported by a “Research Fellow” fellowship from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium). B.C. was supported by the Belgian Program on Interuniversity Attraction Poles initiated by the Federal Office for Scientific, Technical and Cultural Affairs (IAP P6/19 PROFUSA). S.M. was supported by an NIH NRSA training grant fellowship (T32CA09361). P.-O.V. was supported by EMBO long-term fellowship No. 61-2002. Support was provided by the Leukemia Research Foundation to M.B. M.V. and R.B. are “Honorary Research Associate” and “Research Director” from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium), respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI, NHGRI, NIGMS or the NIH.
Footnotes
AUTHOR CONTRIBUTIONS
M.D., B.C., S.M., P.-O.V., M.A.Y., Q.Z., R.B., J.V., M.B., and M.V. conceived the experiments and analyses. S.M. and P-O.V generated the ced-9 mutant library and performed Y2H screens and R-Y2H selections. M.D. and P-O.V. cloned the alleles. P-O.V. and M.D. performed the co-APs. M.D. developed and implemented the modified Y2H assay. B.C. performed the structural analyses. B.C. and M.A.Y. performed the statistical analyses. N.S. generated the transgenic animals under the supervision of S.M. and M.B. S.M. performed survival and apoptosis challenge experiments. M.D. and V.R. performed the mutant alleles stability experiment. M.D., B.C., S.M., P-O.V., M.E.C. and M.V. wrote the manuscript. All authors discussed the results. D.E.H. and M.V. conceived and co-directed the project.
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 2.Vidal M. Interactome modeling. FEBS Lett. 2005;579:1834–1838. doi: 10.1016/j.febslet.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 3.Shih HM, et al. A positive genetic selection for disrupting protein-protein interactions: identification of CREB mutations that prevent association with the coactivator CBP. Proc Natl Acad Sci USA. 1996;93:13896–13901. doi: 10.1073/pnas.93.24.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leanna CA, Hannink M. The reverse two-hybrid system: a genetic scheme for selection against specific protein/protein interactions. Nucleic Acids Res. 1996;24:3341–3347. doi: 10.1093/nar/24.17.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vidal M, Braun P, Chen E, Boeke JD, Harlow E. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc Natl Acad Sci USA. 1996;93:10321–10326. doi: 10.1073/pnas.93.19.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endoh H, et al. Integrated version of reverse two-hybrid system for the postproteomic era. Methods Enzymol. 2002;350:525–545. doi: 10.1016/s0076-6879(02)50983-6. [DOI] [PubMed] [Google Scholar]
- 8.Gray PN, Busser KJ, Chappell TG. A novel approach for generating full-length, high coverage allele libraries for the analysis of protein interactions. Mol Cell Proteomics. 2007;6:514–526. doi: 10.1074/mcp.T600023-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Kritikou EA, et al. C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival. Genes Dev. 2006;20:2279–2292. doi: 10.1101/gad.384506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inouye C, Dhillon N, Durfee T, Zambryski PC, Thorner J. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics. 1997;147:479–492. doi: 10.1093/genetics/147.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeder MK, Serebriiskii IG, Golemis EA, Chernoff J. Analysis of small GTPase signaling pathways using p21-activated kinase mutants that selectively couple to Cdc42. J Biol Chem. 2001;276:40606–40613. doi: 10.1074/jbc.M103925200. [DOI] [PubMed] [Google Scholar]
- 12.Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Chang HY, Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 14.Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 15.del Peso L, Gonzalez VM, Nunez G. Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation. J Biol Chem. 1998;273:33495–33500. doi: 10.1074/jbc.273.50.33495. [DOI] [PubMed] [Google Scholar]
- 16.Yan N, et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- 17.Hengartner MO, Horvitz HR. Activation of C. elegans cell death protein CED-9 by an amino-acid substitution in a domain conserved in Bcl-2. Nature. 1994;369:318–320. doi: 10.1038/369318a0. [DOI] [PubMed] [Google Scholar]
- 18.Yan N, et al. Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Woo JS, et al. Unique structural features of a BCL-2 family protein CED-9 and biophysical characterization of CED-9/EGL-1 interactions. Cell Death Differ. 2003;10:1310–1319. doi: 10.1038/sj.cdd.4401303. [DOI] [PubMed] [Google Scholar]
- 20.Walhout AJ, et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 21.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spector MS, Desnoyers S, Hoeppner DJ, Hengartner MO. Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature. 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 23.Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 24.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 25.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 26.Lettre G, et al. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ. 2004;11:1198–1203. doi: 10.1038/sj.cdd.4401488. [DOI] [PubMed] [Google Scholar]
- 27.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 28.Boag PR, Nakamura A, Blackwell TK. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- 29.Jeffery CJ. Moonlighting proteins--an update. Mol Biosyst. 2009;5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Q, et al. Edgetic perturbation models of human genetic disorders. Mol Syst Biol. 2009 doi: 10.1038/msb.2009.80. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walhout AJ, Vidal M. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- 33.Walhout AJ, et al. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 34.Reboul J, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 35.Xu L, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 36.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 37.Lins L, Thomas A, Brasseur R. Analysis of accessible surface of residues in proteins. Protein Sci. 2003;12:1406–1417. doi: 10.1110/ps.0304803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmann ER, et al. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol. 2002;12:1908–1918. doi: 10.1016/s0960-9822(02)01262-9. [DOI] [PubMed] [Google Scholar]
- 41.Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 43.Lehner B, Tischler J, Fraser AG. RNAi screens in Caenorhabditis elegans in a 96-well liquid format and their application to the systematic identification of genetic interactions. Nat Protoc. 2006;1:1617–1620. doi: 10.1038/nprot.2006.245. [DOI] [PubMed] [Google Scholar]
- 44.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.