Abstract
Lentiviral gene transfer vectors have a number of potential advantages over gammaretroviral vectors including more efficient transduction of nondividing cells, a more favorable integration site profile, and the ability to accommodate large transgenes. Here, we present long-term follow-up data of animals that received lentivirus-transduced CD34-enriched cells. Six long-term surviving dogs were available for analysis. Transgene expression was analyzed from at least 12 months to more than 5 years after transplantation in peripheral blood cells and multiple cell lineages. All animals demonstrated long-term stable transgene expression in peripheral blood myeloid, lymphoid, and red blood cells as well as in platelets. Vector integration sites were analyzed by linear amplification-mediated polymerase chain reaction and showed a polyclonal repopulation pattern in all animals. There was no evidence of any development of monoclonality or leukemia in the animals. The stable long-term multilineage transgene expression, together with detection of the same integration site in myeloid and lymphoid cells, strongly suggests the transduction of long-term repopulating stem cells. Our data demonstrate safe and efficient transduction of multilineage long-term repopulating cells with lentiviral vectors and support the use of such vectors for gene therapy studies in patients.
Introduction
Retrovirus-mediated hematopoietic stem cell (HSC) gene transfer is a powerful tool to treat diseases affecting the hematopoietic system (Stocking and Baum, 1997). For most clinical HSC gene therapy studies, gammaretroviruses have been the vector system used, due in large part to extensive periods of study (Edelstein et al., 2007). Delivery of therapeutic transgenes by gammaretroviral vectors has been successful in treating patients with a variety of monoallelic diseases including X chromosome-linked severe combined immune deficiency (X-SCID) (Hacein-Bey-Abina et al., 2002), chronic granulomatous disease (CGD) (Ott et al., 2006), and severe combined immunodeficiency caused by mutated adenosine deaminase (SCID-ADA) (Aiuti et al., 2002). Unfortunately, in several instances the gene-modified cells have transformed into frank leukemia (Hacein-Bey-Abina et al., 2003a,b, 2008; Howe et al., 2008) and a monoclonal outgrowth in a patient with CGD (Ott et al., 2006). Two major reasons have been discussed for the adverse outcome: First, vector integration seems to play a role in the activation of adjacent proto-oncogenes. Second, expression of the therapeutic transgene itself has been shown to play at least a partial role in clonal outgrowth (reviewed in Baum et al., 2004).
These findings have raised the question whether other integrating viral gene delivery systems with improved safety characteristics, based on integration profiles, might be more appropriate. Foamy viruses (Rethwilm, 2007) and lentiviruses (Cockrell and Kafri, 2007) have been investigated as therapeutic gene transfer vehicles. We have studied gene transfer vectors in clinically relevant canine and nonhuman primate transplantation models (Trobridge et al., 2005). Beard and colleagues compared the integration profiles of lentiviral, gammaretroviral, and foamy viral vectors in long-term repopulating cells in the dog and found gammaretroviral vectors to be likely associated with the highest potential for adverse gene activation (Beard et al., 2007b) because they were more frequently inserted in and near proto-oncogenes transcription start sites (TSSs). Here, we analyzed in a long-term follow-up six dogs after transplantation with lentiviral vector-transduced CD34+ cells (Horn et al., 2004; Beagles et al., 2005). We did not observe any adverse event such as leukemia relating to the HSC gene therapy procedure and found no trend toward the development of monoclonality; the clonality of repopulating cells was heterogeneous at all times analyzed. We also verified the long-term multilineage potential of gene-modified repopulating cells by tracking repopulating clones in both myeloid and lymphoid lineages.
Materials and Methods
Animals, lentiviral vectors, transduction, and transplantation protocols
Animal husbandry, preparation of lentiviral vectors, gene transfer into canine CD34-enriched bone marrow and peripheral blood stem cells, as well as flow cytometric analysis have been described previously (Horn et al., 2004; Beagles et al., 2005) for all animals but G293. Briefly, this dog obtained 3.1 × 107 bone marrow-derived CD34-enriched cells transduced with a green fluorescent protein (GFP)-encoding lentiviral vector at a multiplicity of infection (MOI) of 100 plus 4.8 × 107 peripheral blood-derived CD34-enriched cells transduced with a yellow fluorescent protein (YFP)-encoding lentiviral vector at an MOI of 100 after total body irradiation of 920 cGy without cyclosporine. Transduction of both cell populations was carried out without prestimulation as described previously (Horn et al., 2004).
Linear amplification-mediated polymerase chain reaction and cloning of individual integration sites
Lentiviral integration site analysis was performed on canine DNA isolated from peripheral blood mononuclear cells (PBMCs). Linear amplification-mediated polymerase chain reaction (LAM-PCR) and sequence analysis of individual LAM fragments were carried out according to a published protocol (Beard et al., 2007b). Briefly, for each analyzed point in time, 100 ng of PBMC DNA was subjected to linear amplification three times independently. Reactions were pooled and further analyzed according to the previously mentioned protocol. LAM-PCR products were analyzed on 4 to 20% acrylamide–TBE (Tris–borate–EDTA) gradient gels (Invitrogen, Carlsbad, CA).
Subset sorting
PBMCs were collected after lysis of red blood cells. CD3-positive lymphocytes were labeled with an anti-CD3 antibody (clone 16.6B3, kindly provided by P. Moore, University of California, Davis, CA). Granulocytes were selected with a DM5 antibody (a gift from B. Sandmaier, Fred Hutchinson Cancer Research Center, Seattle, WA). Both subsets were stained with a secondary phycoerythrin-labeled polyclonal goat-anti-mouse antibody (Dako, Carpinteria, CA). Sorting was carried out with an ARIA cell sorter (BD Biosciences, San Jose, CA). DNA from cell subsets with purity greater than 98% based on back gating was prepared with a QIAamp mini spin column kit (Qiagen, Valencia, CA). To determine vector-specific DNA, PCR was carried out with Platinum Taq polymerase (Invitrogen) and primers Lenti 2F (5′-AGAGA-TGGGTGCGAGAGCGTCA-3′) and Lenti 2R (5′-TGCCTTG-GTGGGTGCTACTCCTAA-3′) to detect a 471-bp fragment. The integrant within the RAP1GDS1 proto-oncogene was amplified by nested PCR using primers RAP1-1 (5′-CGA-CCTCTTGCTCTGTC-3′) and LVLTRII (Beard et al., 2007b) in the first PCR and RAP1-1 in combination with LVLTRIII (Beard et al., 2007b) in the nested PCR to detect a 524-bp fragment. Accordingly, the PCR for the integrant in the proto-oncogene CEP1 (Beard et al., 2007b) was carried out with primer CEP1-4 (5′-AGACAGAACCAGGCATTAC-3′) to detect a 318-bp fragment. Detection of integrants 29.15 and 10.17 with distances greater than 50 kb from the nearest known proto-oncogene TSS (K.A. Keyser, unpublished data) were amplified by PCR, using primers 29.15a (5′-GTGGTAAAG-GTGAGTTGACTG-3′) and 10.17c (5′-CCCTGCTGGACTA-AATGTAC-3′), respectively. PCR products correspond to 244 bp for integrant 29.15 and 322 bp for integrant 10.17.
Results
Stable long-term persistence of lentivirus-transduced hematopoietic repopulating cells in dogs
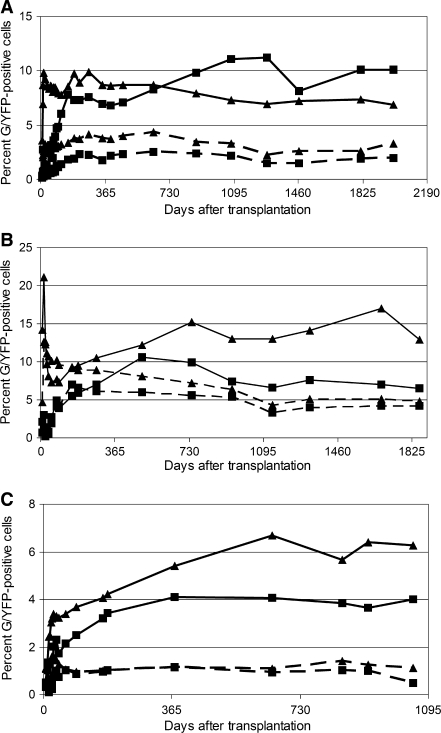
Efficient and safe long-term repopulation of genetically modified cells is a critical aspect of HSC gene therapy to make this approach appropriate for a variety of clinical applications. Here, we report on the long-term follow-up of six dogs that were transplanted with lentivirus-transduced CD34-enriched cells, using an overnight or 2-day transduction protocol with either GFP- or YFP-expressing lentiviral vectors. Figure 1 shows stable long-term gene marking in granulocytes and lymphocytes for more than 5 years in dog G206 (2002 days; Fig. 1A) and dog G236 (1862 days; Fig. 1B). Dog G293 showed a comparable engraftment pattern over a period of almost 3 years after transplantation (1233 days; Fig. 1C). We observed persistent marking in all six dogs without evidence of hematologic abnormalities (data not shown). Marking was also detectable and stable in red blood cells and in platelets in all six dogs (data not shown).
FIG. 1.
Stable gene expression levels in peripheral blood cell subsets of dogs. Animals were treated with lentivector-transduced GFP/YFP-expressing CD34-enriched cells in an autologous transplantation setting. (A) G206; (B) G236; (C) G293. x axis, days posttransplantation (DPT); y axis, relative percentage of G/YFP-expressing cells. Solid lines, GFP marking; dashed lines, YFP marking; squares, lymphocytes; triangles, granulocytes.
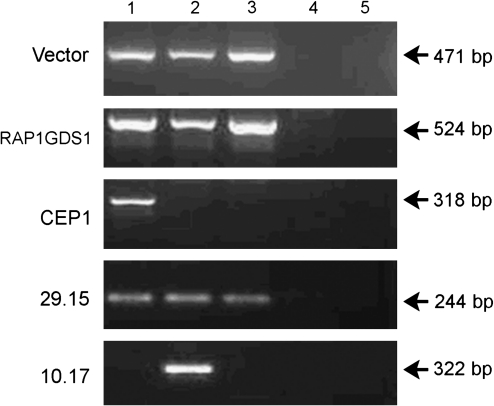
To confirm multilineage long-term repopulation by gene-modified cells, we analyzed four integration sites identified from whole white blood cell DNA preparations in sorted myeloid and lymphoid cells from dog G206. We were able to detect two integrants, one within RAP1GDS1 (Beard et al., 2007b), and the insertion 29.15 (K.A. Keyser, unpublished) in FACS-sorted granulocyte and CD3+ lymphocyte subsets 2188 days posttransplantation (Fig. 2, RAP1GDS1 and 29.15). The presence of these integrants in both lymphocyte and myeloid subsets of G206 supports the interpretation that lentiviral vectors are able to transduce long-term multilineage HSCs. To assess whether these findings are due to suboptimal purity of the cell populations, we mixed PBMC DNA from a nontransplanted animal with increasing percentages of DNA from the G206 lymphocyte or granulocyte cell population to determine the detection limit of the PCR. We found the detection limit of one specific integrant to be 10% of specific subset DNA or higher (data not shown). Thus, given the purity of our sorting results (>95%), this suggests that the clone tracking data correspond to multipotential gene-modified clones and not to cellular contamination from another hematopoietic subset.
FIG. 2.
Detection of two common integrants in FACS-sorted granulocyte and CD3+ lymphocyte subsets. Granulocyte and CD3+ lymphocyte subsets were sorted from PBMCs from dog G206, using subset-specific antibodies. DNA was prepared from the collected cells and subjected to PCR analysis (see Materials and Methods) using either two vector-specific primers (Vector) or one vector-specific primer in combination with integrant-specific primers (see Materials and Methods, Subset Sorting). Lane 1, unsorted PBMCs from G206; lane 2, G206 granulocyte subset; lane 3, G206 CD3 lymphocyte subset; lane 4, PBMCs from a nontransplanted animal; lane 5, water control.
No evidence of monoclonality after long-term repopulation of lentiviral vector-transduced canine CD34-enriched cells
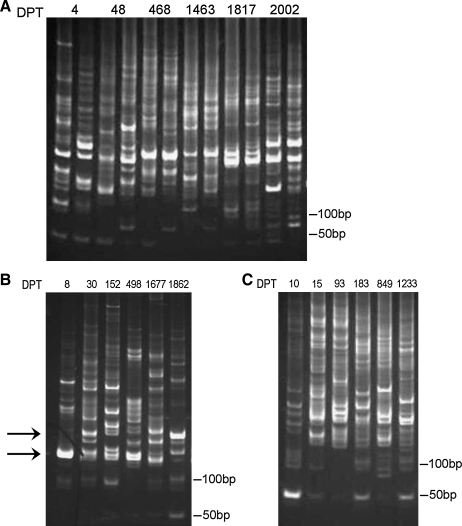
Long-term surviving animals were monitored for heterogeneous repopulation of the hematopoietic system. To determine trends toward mono- or oligoclonality, we performed LAM-PCR at various time points after transplantation for G206, G236, and G293. Mono- or oligoclonality would have been evidenced by the occurrence of either a single or only a few distinguishable LAM-PCR fragments (Schmidt et al., 2007). As seen in Fig. 3, all dogs appeared to have multiple clones contributing to hematopoiesis because we were able to detect multiple LTR-specific amplified PCR products on a polyacrylamide gel at all time points analyzed. There was no obvious difference in the number of PCR products at early or later time points after transplantation. Also, after analysis of LAM-PCR products on acrylamide gels, it appeared that several bands became progressively more intense over time. In an attempt to determine whether this was due to progressive outgrowth of a dominant clone or simply coincidental migration of LAM-PCR products with similar molecular weights, we directly cloned and sequenced several products. As examples, we analyzed 155-bp PCR fragments that appeared in the LAM-PCR analysis 8, 30, and 498 days posttransplantation, and 250-bp fragments present 30 and 1862 days posttransplantation in G236 (Fig. 3B, arrows). After isolation and cloning, we analyzed their chromosomal location. The canine sequence isolated from the 155-bp LAM-PCR fragment 8 days posttransplantation was different from the sequences we obtained on days 30 and 498 posttransplantation, respectively. We found one specific integrant on day 30 posttransplantation, which was also detectable as one of two canine sequences 498 days posttransplantation (data not shown). The canine sequences isolated from the 250-bp LAM-PCR fragment 30 and 1862 days posttransplantation differed from each other. Thus, in the two cases here, the presence of LAM-PCR fragments at similar molecular weights corresponded to coincidental migration of distinct fragments and not the progression of a single dominant clone.
FIG. 3.
Polyclonal hematopoietic repopulation after transplantation with lentivirus-transduced CD34-enriched cells. Peripheral blood samples from (A) dog G206, (B) dog G236, and (C) dog G293 at various time points after transplantation were analyzed by LAM-PCR, showing that multiple different clones contribute to the repopulation. The number above each lane indicates how many days after transplantation (DPT) the DNA was prepared from the PBMCs of each dog. For G206, LAM-PCRs were carried out in duplicate. The arrows in (B) indicate the 155- and 250-bp fragments cloned from G236 DNA (see Results).
Hematopoietic repopulation also was investigated by analyzing the PBMC DNA of G115, G136, and G221 by LAM-PCR. We found multiple LAM-PCR-amplified bands at the most recent time points for these three dogs, suggesting that transplantation of lentiviral vector-transduced cells did not result in monoclonality in these animals (data not shown).
In addition, to verify provirus integration sites, we carried out sequence analysis of LAM-PCR-amplified products by shotgun cloning and sequencing at the most recent time points after transplantation (Beard et al., 2007b). We analyzed almost 700 sequence reads and found 52 novel unique integrants, applying stringent criteria for patterns defining a true vector integration event (Beard et al., 2007a). The numbers of sequences found in each dog are given in Table 1 (column IS). This further suggests that no mono- or oligoclonal repopulation of lentiviral vector-transduced HSCs developed even after periods longer than 5 years after transplantation. Information about the gene closest to the lentivirus integration sites described here can be found in Supplementary Table 1 (see Supplementary Table 1 at www.liebertonline.com/hum).
Table 1.
Overview of Dogs Transplanted with Lentivector-GFP/YFP-Transduced CD34-Positive G/YFP-Expressing Repopulating Cellsa
| Dog no. | Current state | Median % G/YFP-positive PBMCs | IS | DPT |
|---|---|---|---|---|
| G115 | Euthanized because of seizures | 0.95% YFP (BM), 2.31% GFP (PB) | 5 | 659 |
| G136 | Euthanized because of growth behind eye | 3.47% YFP (PB), 5.34% GFP (BM) | 7 (+ 7) | 801 |
| G206 | Alive | 2.58% YFP (BM), 7.31% GFP (PB) | 6 | 2002 |
| G221 | Euthanized after episode related to Addison's disease | 7.11% YFP (PB), 15.25% GFP (BM) | 6 | 363 |
| G236 | Alive | 4.6% YFP (BM), 11.5% GFP (PB) | 8 | 1862 |
| G293 | Euthanized | 0.76% YFP (PB), 4.92% GFP (BM) | 14 | 1233 |
Abbreviations: BM, bone marrow; DPT, days post transplantation; GFP, green fluorescent protein; IS, insertion site; PB, peripheral blood; PBMCs, peripheral blood mononuclear cells; YFP, yellow fluorescent protein.
The IS column shows the number of true unique lentiviral integration sites determined for each dog at the indicated time after transplantation. The Median % G/YFP-positive PBMCs column indicates the origin of donor cells transduced with either YFP or GFP lentiviral vectors. The “+ 7” in column IS, for dog G136, refers to the number of sequences retrieved from the tumor.
Analysis of euthanized animals
Several dogs were killed after transplantation because of nonhematopoietic issues. G115 was killed 2 years after transplantation because of a seizure disorder. Animal G136 had developed a growth behind its eye, and dog G221 developed Addison's disease 1 year after transplantation.
It is unlikely that the diseases in G115, G136 and G221 were caused by a dominant repopulating clone in the hematopoietic system of these animals because we found multiple integrants in their peripheral blood cell DNA. Pathologic examination of the tumor behind the left eye of G136 showed an invasive carcinoma with features of moderately differentiated squamous cell carcinoma characterized by nests and sheets of tumor cells with occasional keratin pearls, necrosis, and keratinocyte differentiation. To rule out that these cells originated from a clonally expanded gene-marked progenitor, we analyzed DNA extracted from the tumor for trends toward monoclonality. There were multiple bands detectable by LAM-PCR, likely from blood contamination after resection and incomplete washing before DNA isolation. Shotgun cloning of the LAM-PCR fragments followed by sequence analysis of 96 randomly picked clones revealed 7 new previously undescribed integrants (see Supplementary Table 1). Furthermore, if the tumor arose from a malignant clone, gene marking of the tumor DNA by real-time PCR would indicate high levels of provirus. Therefore, we performed quantitative PCR with lentivirus-specific primers. As would be expected from a tumor or any organ contaminated with gene-marked blood, gene marking in the tumor DNA was similar to marking in G136 peripheral blood (data not shown). Altogether, these data suggest that the tumor behind the eye was not caused by gene-modified cells.
Clone tracking of lentiviral vector integrants
We also investigated the time-dependent contribution to hematopoiesis of four individual clones to the overall marking in G221 and, as a control, in G206. Independent of whether the integrant was within a proto-oncogene or not (Beard et al., 2007b), all four integrants in each dog showed a similar time-dependent fluctuation, suggesting that lentiviral vector integration within or near a proto-oncogene did not provide a strong proliferative advantage (data not shown).
The presence of the previously published insertions within proto-oncogenes RAP1GDS1, CEP1, ZNF198, DST, and WHSC1L1 (Beard et al., 2007b) was analyzed at the most recent time point (2188 days posttransplantation) in dog G206. RAP1GDS and CEP1 had already been detected during the subset sorting analysis (see Fig. 2, RAP1GDS1 and CEP1). The ZNF198 integrant was not detectable at this time whereas the DST and WHSC1L1 integrants were amplified by PCR out of the PBMC DNA of G206 (data not shown). In G236, the presence of integrant USP38 (Beard et al., 2007b) was detected 1994 days posttransplantation (data not shown). From Addison's disease-affected animal G221, PBMC DNA was harvested at the time of euthanasia (363 days posttransplantation). Both the integrants within proto-oncogenes HIVEP1 and ANXA1 (Beard et al., 2007b) were detectable at this time (data not shown).
Discussion
Here, we report the long-term follow-up of six dogs transplanted with lentivirus-transduced CD34-enriched cells. We found that gene marking and expression of the transgenes encoding GFP and YFP were stable for the entire observation period (up to more than 5 years after transplantation). We also found long-term marking and expression in all hematopoietic subpopulations analyzed. Importantly, we did not observe any overt side effects associated with gene-marked cells, including no evidence of monoclonality or leukemia.
Side effects in two of the three major clinical gene therapy trials to treat X-SCID and X-CGD have occurred 31–68 months (X-SCID) and 3 months (X-CGD) after transplantation (Ott et al., 2006; Hacein-Bey-Abina et al., 2008). All these trials have used gammaretroviral vectors. More recently, lentiviral vectors have been developed and explored for gene therapy. However, not only gammaretroviral but also lentiviral vectors induced insertional gene dysregulation (Maruggi et al., 2009) and clonal dominance (Kustikova et al., 2009). Thus, the safety assessment of lentiviral vector systems needs further investigation. We have reported early engraftment data with lentivirus-transduced cells in baboons and dogs with a follow-up median of 36 weeks (baboons; Horn et al., 2002) and 51 weeks (dogs; Horn et al., 2004), respectively. An and colleagues have studied lentiviral vectors in rhesus macaques with a reported follow-up of 28 months (An et al., 2001), which, so far to our knowledge, is the longest follow-up of lentivirus-mediated gene transfer into hematopoietic stem cells in a large animal study. To further assess safety issues concerning the use of lentiviruses to transduce HSCs, we monitored our previously reported dogs for up to more than 5 years. To our knowledge this is the longest follow-up of HSCs gene-modified with HIV-derived lentiviral vectors in large animal models. We found stable long-term contribution to the repopulation of the hematopoietic system by the gene-marked cells without treatment-related side effects.
In the clinical trial for X-SCID and CGD, patients developed monoclonality with corrected cells, which either led to leukemic outgrowth or a myelodysplastic syndrome (MDS)-like state. We monitored the animals in the present report for up to more than 5 years and did not observe any trend toward oligo- or monoclonality when analyzing vector integrations by LAM-PCR. In addition, vector integrants close to or within proto-oncogenes did not differ from other integrants with respect to their contribution to repopulation. No gross increase in gene-modified cells resulting in an increase in gene marking (Zhang et al., 2008) was observed in granulocyte or lymphocyte subpopulations, further indicating that neither malignant nor benign outgrowth of gene-modified cells had occurred in the hematopoietic system of the animals.
One of the putative reasons for clonal outgrowth caused by individual insertions is the presence of viral promoter/enhancer elements in the long terminal repeats (LTRs) of the gene transfer vector (Baum et al., 2004). One of the major differences between the lentiviral vectors in our studies and the gammaretroviral vectors used in the clinical studies is the presence of self-inactivating (SIN) LTRs. As we did not observe malignant events in our experimental animals, it is possible that the use of such SIN vectors in our experimental setting provides safer alternatives for future gene therapy trials. Of note, SIN gammaretroviral vectors are currently being developed for CGD therapy (Moreno-Carranza et al., 2009).
Some of our animals developed health problems during the observation time. However, when we assessed the stability of repopulation by LAM-PCR and sequence analysis of individual integrants in these dogs, we could rule out that the observed adverse effects were caused by monoclonal evolution of gene-marked cells. Thus, it is likely that the diseases observed in the animals were due to either natural circumstances or other treatment-related parameters. Radiation- and drug-induced secondary tumors have been a long-standing problem in transplantation regimens (Lowe et al., 2007); all of the dogs monitored in this report received 920 cGy total body irradiation, and all but dog G293 received immunosuppressive agents (see Materials and Methods).
One integrant within a proto-oncogene and one integrant not near any proto-oncogene could be detected in both granulocyte and a lymphocyte subpopulations in dog G206 almost 6 years after transplantation, respectively, indicating the ability of lentiviral vectors to transduce true repopulating stem cells. (This is particularly important when the genetic disease affects certain hematopoietic subpopulations, suggesting that lentiviral vectors might be versatile tools for the treatment of any kind of genetic disease affecting particular subsets of a patient's hematopoietic system.)
On review of all the previously published insertion sites in proto-oncogenes (Beard et al., 2007b), all but one could be detected at the most recent time points (data not shown). The animals harboring these integrants are either still alive and do not show any trend to monoclonal repopulation (G206 and G236) or had to be killed for reasons unrelated to their hematologic parameters (G221, Addison's disease). Thus, it is likely that the lentiviral integration sites near proto-oncogenes did not influence oncogene expression leading to malignant outgrowth.
In conclusion, our data show polyclonal long-term hematopoiesis with lentivirus-transduced repopulating cells without any evidence of monoclonality or leukemia. These data suggest that the use of HIV-derived SIN lentiviral vectors should allow for relatively safe gene correction of hematopoietic diseases, and improved safety characteristics of lentiviral vectors are preferred over currently used gammaretroviral vectors.
Supplementary Material
Acknowledgments
The authors thank the technicians in the canine facilities of the Fred Hutchinson Cancer Research Center and at San Diego. The authors also thank Helen Crawford and Bonnie Larson for assisting with the preparation of this manuscript. This work has been supported in part by the NIH (Bethesda, MD; grants HL36444, HL200010, KD56465, and DK47754). H.P.K. is a Markey Molecular Medicine Investigator and the recipient of the Jose Carreras/E.E Thomas Endowed Chair for Cancer Research.
Author Disclosure Statement
The authors declare no competing financial interests.
References
- Aiuti A. Slavin S. Aker M. Ficara F. Deola S. Mortellaro A. Morecki S. Andolfi G. Tabucchi A. Carlucci F. Marinello E. Cattaneo F. Vai S. Servida P. Miniero R. Roncarolo M.G. Bordignon C. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- An D.S. Kung S.K. Bonifacino A. Wersto R.P. Metzger M.E. Agricola B.A. Mao S.H. Chen I.S. Donahue R.E. Lentivirus vector-mediated hematopoietic stem cell gene transfer of common γ-chain cytokine receptor in rhesus macaques. J. Virol. 2001;75:3547–3555. doi: 10.1128/JVI.75.8.3547-3555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum C. Von Kalle C. Staal F.J. Li Z. Fehse B. Schmidt M. Weerkamp F. Karlsson S. Wagemaker G. Williams D.A. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences [review] Mol. Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Beagles K.E. Peterson L. Zhang X. Morris J. Kiem H.-P. Cyclosporine inhibits the development of green fluorescent protein (GFP)-specific immune responses after transplantation of GFP-expressing hematopoietic repopulating cells in dogs. Hum. Gene Ther. 2005;16:725–733. doi: 10.1089/hum.2005.16.725. [DOI] [PubMed] [Google Scholar]
- Beard B.C. Dickerson D. Beebe K. Gooch C. Fletcher J. Okbinoglu T. Miller D.G. Jacobs M.A. Kaul R. Kiem H.-P. Trobridge G.D. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol. Ther. 2007a;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- Beard B.C. Keyser K.A. Trobridge G.D. Peterson L.J. Miller D.G. Jacobs M. Kaul R. Kiem H.-P. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, and foamy virus. Hum. Gene Ther. 2007b;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- Cockrell A.S. Kafri T. Gene delivery by lentivirus vectors [review] Mol. Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- Edelstein M.L. Abedi M.R. Wixon J. Gene therapy clinical trials worldwide to 2007: An update [review] J. Gene Med. 2007;9:833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Le Deist F. Carlier F. Bouneaud C. Hue C. De Villartay J.P. Thrasher A.J. Wulffraat N. Sorensen R. Dupuis-Girod S. Fischer A. Davies E.G. Kuis W. Leiva L. Cavazzana-Calvo M. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M. McCormack M.P. Wulffraat N. Leboulch P. Lim A. Osborne C.S. Pawliuk R. Morillon E. Sorensen R. Forster A. Fraser P. Cohen J.I. De Saint Basile G. Alexander I. Wintergerst U. Frebourg T. Aurias A. Stoppa-Lyonnet D. Romana S. Radford-Weiss I. Gross F. Valensi F. Delabesse E. Macintyre E. Sigaux F. Soulier J. Leiva L.E. Wissler M. Prinz C. Rabbitts T.H. Le Deist F. Fischer A. Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 [erratum appears in Science 2003;302:568] Science. 2003a;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M. Le Deist F. Wulffraat N. McIntyre E. Radford I. Villeval J.L. Fraser C.C. Cavazzana-Calvo M. Fischer A. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003b;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Garrigue A. Wang G.P. Soulier J. Lim A. Morillon E. Clappier E. Caccavelli L. Delabesse E. Beldjord K. Asnafi V. Macintyre E. Dal Cortivo L. Radford I. Brousse N. Sigaux F. Moshous D. Hauer J. Borkhardt A. Belohradsky B.H. Wintergerst U. Velez M.C. Leiva L. Sorensen R. Wulffraat N. Blanche S. Bushman F.D. Fischer A. Cavazzana-Calvo M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn P.A. Morris J.C. Bukovsky A.A. Andrews R.G. Naldini L. Kurre P. Kiem H.-P. Lentivirus-mediated gene transfer into hematopoietic repopulating cells in baboons. Gene Ther. 2002;9:1464–1471. doi: 10.1038/sj.gt.3301820. [DOI] [PubMed] [Google Scholar]
- Horn P.A. Keyser K.A. Peterson L.J. Neff T. Thomasson B.M. Thompson J. Kiem H.-P. Efficient lentiviral gene transfer to canine repopulating cells using an overnight transduction protocol. Blood. 2004;103:3710–3716. doi: 10.1182/blood-2003-07-2414. [DOI] [PubMed] [Google Scholar]
- Howe S.J. Mansour M.R. Schwarzwaelder K. Bartholomae C. Hubank M. Kempski H. Brugman M.H. Pike-Overzet K. Chatters S.J. De Ridder D. Gilmour K.C. Adams S. Thornhill S.I. Parsley K.L. Staal F.J. Gale R.E. Linch D.C. Bayford J. Brown L. Quaye M. Kinnon C. Ancliff P. Webb D.K. Schmidt M. Von Kalle C. Gaspar H.B. Thrasher A.J. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustikova O.S. Schiedlmeier B. Brugman M.H. Stahlhut M. Bartels S. Li Z. Baum C. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Mol. Ther. 2009;17:1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. Bhatia S. Somlo G. Second malignancies after allogeneic hematopoietic cell transplantation [review] Biol. Blood Marrow Transplant. 2007;13:1121–1134. doi: 10.1016/j.bbmt.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Maruggi G. Porcellini S. Facchini G. Perna S.K. Cattoglio C. Sartori D. Ambrosi A. Schambach A. Baum C. Bonini C. Bovolenta C. Mavilio F. Recchia A. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol. Ther. 2009;17:851–856. doi: 10.1038/mt.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Carranza B. Gentsch M. Stein S. Schambach A. Santilli G. Rudolf E. Ryser M.F. Haria S. Thrasher A.J. Baum C. Brenner S. Grez M. Transgene optimization significantly improves SIN vector titers, gp91phox expression and reconstitution of superoxide production in X-CGD cells. Gene Ther. 2009;16:111–118. doi: 10.1038/gt.2008.143. [DOI] [PubMed] [Google Scholar]
- Ott M.G. Schmidt M. Schwarzwaelder K. Stein S. Siler U. Koehl U. Glimm H. Kuhlcke K. Schilz A. Kunkel H. Naundorf S. Brinkmann A. Deichmann A. Fischer M. Ball C. Pilz I. Dunbar C. Du Y. Jenkins N.A. Copeland N.G. Luthi U. Hassan M. Thrasher A.J. Hoelzer D. Von Kalle C. Seger R. Grez M. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Rethwilm A. Foamy virus vectors: An awaited alternative to gammaretro- and lentiviral vectors [review] Curr. Gene Ther. 2007;7:261–271. doi: 10.2174/156652307781369092. [DOI] [PubMed] [Google Scholar]
- Schmidt M. Schwarzwaelder K. Bartholomae C. Zaoui K. Ball C. Pilz I. Braun S. Glimm H. Von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- Stocking C. Baum C. Gene transfer into haemopoietic cells [review] Baillieres Clin. Haematol. 1997;10:445–465. doi: 10.1016/s0950-3536(97)80020-0. [DOI] [PubMed] [Google Scholar]
- Trobridge G. Beard B.C. Kiem H.-P. Hematopoietic stem cell transduction and amplification in large animal models. Hum. Gene Ther. 2005;16:1355–1366. doi: 10.1089/hum.2005.16.1355. [DOI] [PubMed] [Google Scholar]
- Zhang X.-B. Beard B.C. Trobridge G.D. Wood B.L. Sale G.E. Sud R. Humphries R.K. Kiem H.-P. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J. Clin. Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.