Abstract
HIV-1-based lentiviral vectors are a promising tool for gene therapy. However, integration of a lentiviral vector into host cell genes may lead to the development of cancer. Therefore, control of integration site selection is critical to the successful outcome of gene therapy approaches that use these vectors. The discovery that integration site selection by HIV-1 and HIV-1-based vectors is controlled by the LEDGF/p75 protein has presented new opportunities to control integration site selection. In this study, we tested the hypothesis that a fusion protein containing the C-terminal HIV integrase-binding portion of LEDGF/p75, and the N-terminal chromodomain of heterochromatin protein-1α (HP1α), can target HIV-1 vector DNA outside of genes. We show that this fusion protein, termed TIHPLE, associates with the heterochromatin hallmark trimethylated Lys-9 of histone H3 (H3K9me3). Transient overexpression of TIHPLE alters integration site selection by an HIV-1-based vector and decreases the number of integration events that occur in genes. This change in integration site selection was achieved without a reduction in overall integration efficiency. Furthermore, we show that TIHPLE increases integration in the vicinity of H3K9me3 and in repetitive DNA sequences. These data provide a novel approach to address the problem of the tendency of retroviral vectors to integrate at undesirable sites of the human genome.
Introduction
Gene therapy depends on the successful transfer of a desired gene into a patient's cells. A vast number of gene therapy trials employed to this purpose employ retroviral vectors. These are versatile tools that can successfully transduce a gene into a target cell's genome, in a process called retroviral DNA integration. Two major problems are associated with the use of retroviral vectors. First, efficiency of retroviral transduction is a rate-limiting step of gene therapy and is, in many cases, quite low and insufficient to achieve the therapeutic objectives (Iwase et al., 2007). Second, vector DNA integration may occur at an undesirable site in the host cell genome. This has been unfortunately demonstrated in a gene therapy trial, where insertion of a murine leukemia virus-based vector into the LMO2 oncogene led to T cell leukemia (Hacein-Bey-Abina et al., 2003). It therefore became critical to develop technologies that target integration away from genes, and thus increase the safety of retroviral vectors.
Integration of viral DNA into host DNA is an essential step in the replication cycle of retroviruses, retroviral vectors, and retrotransposons (Coffin et al., 1997; Skalka, 1999; Flint et al., 2000). The first two steps in integration, denoted processing and joining, are catalyzed by the retroviral protein integrase and require specific sequences at the ends of viral DNA. In the first step, nucleotides (usually two) are removed from the 3′ ends of the viral DNA, and in the second step, these newly created ends are joined to staggered phosphates in the complementary strands of the host cell DNA. The host cell DNA essentially suffers a double-strand break whose ends are held together by single-stranded links to viral DNA sequences. Opposing short gaps in the complementary strands are generated by the staggered joining reaction. Complete, stable integration of the viral DNA depends on repair of these gaps, in a process called postintegration repair (Daniel, 2006).
Retroviral vectors that are used for gene transfer can be divided into three classes. These are murine leukemia virus (MLV)-based, human immunodeficiency virus (HIV)-based, and avian sarcoma virus (ASV)-based vectors. The MLV-based vectors are the most widely used retroviral vectors in gene therapy trials. The MLV genome is relatively simple and the MLV-based vectors are consequently well characterized. The HIV-based, or lentiviral, vectors are rapidly gaining in popularity and may soon replace the MLV-based vectors in widespread use. The initial disadvantage of these vectors, that is, a large, complex, and not fully characterized genome, was overcome by a detailed analysis of the genome and deletion of accessory genes that were not needed for gene transfer (Zufferey et al., 1997). In addition, these (HIV-based) vectors possess a crucial advantage over the MLV-based vectors: Unlike the latter, they can efficiently integrate into nondividing cells (Weinberg et al., 1991; Lewis et al., 1992). This is due to their ability to enter the nucleus through the nuclear pores (Bukrinsky et al., 1992, 1993; Coffin et al., 1997). The MLV-based vectors cannot do so and must wait for mitosis, when the nuclear envelope is dissolved (Roe et al., 1993). The HIV-based vectors are thus more versatile than MLV-based vectors. Finally, some laboratories use the ASV-based vectors (Barsov and Hughes, 1996; Daniel et al., 1999). However, these are not yet fully characterized and also integrate into nondividing cells less efficiently than do the HIV-based vectors (Katz et al., 2002).
It has been long observed that retroviral vectors of all classes can integrate at virtually any site of the human genome (Coffin et al., 1997; Skalka, 1999; Flint et al., 2000). This phenomenon was associated with unspecific binding of cellular DNA by retroviral integrases (Coffin et al., 1997; Skalka, 1999; Flint et al., 2000). Thus, it had been long thought that the distribution of integration sites in DNA of infected cells is essentially random (Coffin et al., 1997; Skalka, 1999; Flint et al., 2000). However, more recent analyses, which involved large-scale cloning and sequencing of integration sites, demonstrated that the MLV-based vectors preferentially integrate in promoter regions (Wu et al., 2003). This unfortunate fact is also responsible for the disastrous outcome of the previously mentioned gene therapy trial, which employed MLV-based vectors (Hacein-Bey-Abina et al., 2003). Similarly, HIV-based vectors show preferences for genes, although they do not specifically prefer the promoter region (Schroder et al., 2002). The ASV-based vectors possibly integrate randomly into the human genome (Mitchell et al., 2004). However, some results suggest that they show MLV-like preferences (Narezkina et al., 2004).
The initial attempts to target retroviral DNA integration date to the mid-1990s. Because integration is catalyzed by integrase, this protein became the focus of these early efforts. Three laboratories (headed by A.M. Skalka, S.A. Chow, and F.D. Bushman) used a strikingly similar approach to the problem. All three laboratories constructed fusion proteins, which consisted of the integrase protein, either from HIV-1 or ASV, and a DNA-binding sequence from a cellular or bacterial protein. In two cases, the DNA-binding domain (DBD) was from the Escherichia coli LexA repressor (Katz et al., 1996; Holmes-Son and Chow, 2000); in other work it was from the zinc finger protein E2C (Bushman and Miller, 1997) and finally from the zinc finger zif268 (Tan et al., 2004). DBDs were fused to either the N terminus or C terminus of the integrase protein. In all cases, the fusion protein(s) was shown to target integration to a predetermined sequence in the test tube (Katz et al., 1996; Holmes-Son and Chow, 2000; Tan et al., 2004). The Chow laboratory successfully extended this work to in vivo experiments and demonstrated that integrase–E2C fusion proteins do increase integration in vivo into predetermined chromosomal regions (about 10-fold; Tan et al., 2006). However, most of the integration events still occurred outside of the target region (Tan et al., 2006).
These issues induced the development of innovative approaches to targeting retroviral vectors. One example of such an approach has been described. This method uses integrase-deficient lentiviral vectors (IDLVs) to target gene addition to specific regions (Lombardo et al., 2007). The function of the integrase protein in this case was replaced by zinc finger nucleases (ZFNs). In this study, we test a different approach that is based on modifications of a cellular protein that plays a key role in integration.
In 2003, the laboratory headed by Z. Debyser used the yeast two-hybrid system to isolate a new HIV-1 integrase-binding protein, termed LEDGF/p75 (lens epithelium-derived growth factor; Cherepanov et al., 2003). Ironically, knockout experiments have demonstrated that LEDGF/p75 is not a lens growth factor (Sutherland et al., 2006). Instead, animals lacking the mouse LEDGF/p75 homolog, Psip1, have skeletal abnormalities indicating that this protein is involved in bone development (Sutherland et al., 2006). However, suppression of LEDGF/p75 with small interfering RNA (siRNA), as well as experiments with primary LEDGF/p75 null cells from the LEDGF/p75 null transgenic animals, showed that integration of HIV-1-based vectors is reduced 95% (20-fold) in the absence of LEDGF/p75 (Llano et al., 2006a; Shun et al., 2007). Therefore, LEDGF/p75 is required for efficient integration of HIV-1 DNA. Interestingly, LEDGF/p75 does not bind to the MLV integrase (Busschots et al., 2005). Further analysis showed that LEDGF/p75 is a transcription factor and has a C-terminal integrase-binding domain and an N-terminal chromatin-binding domain (Vanegas et al., 2005; Meehan et al., 2009). An analysis of the integration sites in LEDGF/p75 null cells showed that the residual integration, which can still be found in these cells, no longer occurs preferentially in genes, but is distributed seemingly randomly around the genome (Shun et al., 2007). LEDGF/p75 is thus essential for efficient integration and targets integration into active genes by tethering the integrase protein to chromatin. These data led us to form the following hypothesis: If the N-terminal domain is replaced by a domain that binds to a specific chromatin structure, or a DNA sequence, the resulting fusion protein may target integration to a predetermined chromosomal region. In eukaryotes, there are two major types of chromatin: euchromatin and heterochromatin. Euchromatin contains most of the genes. In contrast, heterochromatin contains many repetitive sequences and few genes (Dimitri et al., 2005). The hallmark of heterochromatin is the trimethylated Lys-9 of histone H3 (H3K9me3), which serves as a binding site for heterochromatin protein-1α (HP1α). We have constructed a fusion protein that consists of the HP1α chromodomain and the C terminus of LEDGF/p75, which contain the integrase-binding domain. In this study, we demonstrate that transient expression of this fusion protein retargets integration of an HIV-1-based vector away from genes, without any significant drop in overall integration efficiency. These results constitute a proof-of-principle that it is possible to retarget integration by manipulating its cellular cofactors, and should stimulate the development of new methods to control integration targeting by retroviral vectors.
Materials and Methods
Cells
293T/17 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics.
Plasmids
The TIHPLE (targeting integration heterochromatin protein LEDGF)-encoding gene was designed to contain amino acids 1–73 of HP1α (RefSeq numbers NP_036249, NP_001120793, and NP_001120794), which comprise the HP1α chromodomain (Jones et al., 2000), and the C terminus of LEDGF/p75 (GenBank accession number AAF25870.1). The synthetic gene was constructed by GENEART (Regensburg, Germany), codon-optimized for expression in human cells and cloned into the pcDNA3.1 vector (Invitrogen, Carlsbad, CA) between the HindIII and XhoI sites, and expressed via the cytomegalovirus (CMV) promoter (for the TIHPLE amino acid sequence, see Supplementary Fig. 1 at www.liebertonline.com/hum). The pcDNA3.1 plasmid was purchased from Invitrogen. The deletion mutant HP1δTIHPLE was constructed by polymerase chain reaction (PCR)-mediated cloning, by which the DNA sequence encoding amino acids 2–71 was removed and the deletion mutant was subcloned between the HindII and XhoI sites of pcDNA3.1.
HIV-1-based vectors
All vesicular stomatitis virus (VSV) G-pseudotyped HIV-1-based vectors were prepared as described previously (Naldini et al., 1996; Daniel et al., 2004), and carry the enhanced green fluorescent protein (EGFP)-encoding reporter gene (Daniel et al., 2004).
Transfections
Regularly maintained 293T/17 (referred to as 293T subsequently) cells were plated onto 60-mm cell culture dishes at a density of 2 × 106 cells per dish. DMEM was used, supplemented with 10% inactivated fetal bovine serum. The next day the plates were transfected, using Lipofectamine 2000 transfection reagent (cat. no. 11668027; Invitrogen) with a TIHPLE plasmid, control pcDNA3.1 plasmid, or mock control (transfection reagent only). Transfections were carried out with 8 μg of DNA and 20 μl of transfection reagent. The next morning, the DNA and transfection reagent were removed, cells were washed with phosphate-buffered saline (PBS), and fresh DMEM was added. The cells were allowed to grow until the next day (48 hr posttransfection), when they were infected with a VSV G-pseudotyped HIV-based vector containing the EGFP gene run by the CMV promoter.
Infections
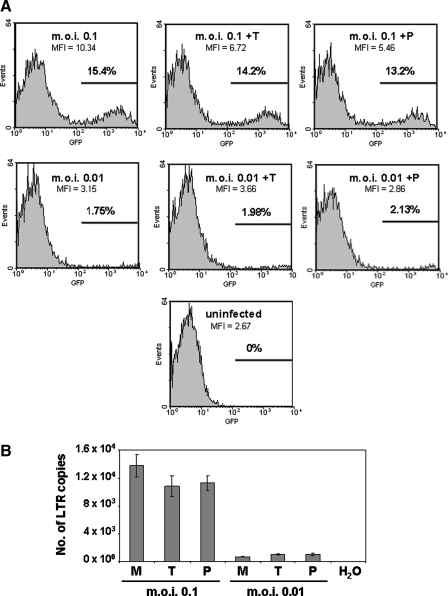
For analysis of integration sites, viral particles with a titer of 5 × 106 particles per milliliter were applied to each 60-mm culture dish at a multiplicity of infection (MOI) of 4. The viral particles were supplemented with dextran to a final concentration of 10 μg/ml to enhance infection. The virus was removed and the medium was changed after a 16-hr incubation period. Four days postinfection the 60-mm dishes were trypsinized and genomic DNA extraction was performed on the cells with a DNeasy blood and tissue kit (cat. no. 69506; Qiagen, Hilden, Germany). The genomic DNA was eluted in 200 μl of AE buffer provided with the kit. Concentration of the genomic DNA sample was determined by ultraviolet spectroscopy. Approximately 1 μg of sample was electrophoresed on an 0.6% agarose gel to determine the overall size of the DNA before proceeding. Infections for analyses of integration efficiency and EGFP expression by fluorescence-activated cell sorting (FACS) were performed under the same conditions, only at lower MOI (0.01–0.1, as shown in Fig. 4A), and once at an MOI of 1, with a virtually identical result with respect to the effect of TIHPLE on EGFP expression (data not shown).
FIG. 4.
Effect of TIHPLE on vector transduction and expression of the EGFP reporter. 293T cells were transfected with TIHPLE and control plasmids and infected with the HIV-1-based vector as described previously (see Materials and Methods and Fig. 2). Cells were analyzed 15 days later. (A) FACS analysis of expression of the EGFP reporter, which is carried by the HIV-1-based vector. Left panels: 293T cells infected with the HIV-1-based vector; right panels: +P, 293T cells transfected with pcDNA3.1 and infected with the HIV-1-based vector; middle panels: +T, 293T cells transfected with the TIHPLE plasmid and infected with the HIV-1-based vector. Horizontal bars and numbers indicate the percentage of EGFP-positive cells. MOIs and mean fluorescence intensity (MFI) are indicated. (B) Number of vector DNA copies per sample (300 ng of DNA), determined by real-time PCR. M, mock-transfected 293T cells infected with the HIV-1-based vector; P, 293T cells transfected with pcDNA3.1 and infected with the HIV-1-based vector; T, 293T cells transfected with the TIHPLE plasmid and infected with the HIV-1-based vector.
Real-time PCR analysis of vector DNA copies in infected cells
Real-time PCR was performed with a Stratagene Mx3005P with MxPro software (Stratagene/Agilent Technologies, La Jolla, CA). Reactions contained 12.5 μl of 2 × QuantiFast probe PCR master mix (Qiagen), 100 μM probe, and 200 μM primers. Cycling conditions were as follows: 95°C for 3 min followed by 50 cycles at 95°C for 3 sec and 60°C for 30 sec. Samples were run in triplicate. Primers and probe sequences were as follows: LTR forward, 5′-TGTGTGCCCGTCTGTTGTGT-3′; Gag reverse, 5′-CCTGCGTCGAGAGAGCTC-3′; probe, 5′-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3′ (Integrated DNA Technologies, Coralville, IA).
Construction of GenomeWalker libraries
The following protocol is adapted from the GenomeWalker universal kit (cat. no. 638904; Clontech/Takara Bio, Mountain View, CA). Approximately 3 μg of genomic DNA was digested with a mixture of blunt-cutting restriction enzymes that have at least a 6-base recognition site. Each digestion included at least three of the following enzymes: DraI, StuI, MscI, and MslI. These enzymes were chosen because of several characteristics. First, they create blunt-ended cuts, allowing the GenomeWalker adaptor to be ligated in a later step. Second, they do not cut within the viral long terminal repeat (LTR), which is essential for the experimental design to successfully clone intact integration sites. Restriction digests were made up of the following: 3 μg of genomic DNA, 80 units of combined restriction enzymes, 10 μl of 10 × restriction enzyme buffer (a final concentration of 1×), and nuclease-free water up to 100 μl. The reactions were mixed gently and put at 37°C overnight (approximately 16 hr).
The next day the digested genomic DNA was purified and concentrated by phenol chloroform extraction and ethanol precipitation. The dried pellets were dissolved in 20 μl of nuclease-free water. To confirm digestion, 1 μl of each sample was electrophoresed on an 0.6% agarose gel. Adaptors were then ligated to digested DNA as follows.
The ligation reaction was composed of 4 μl of digested, purified DNA, 1.9 μl of GenomeWalker adaptor (25 μM), 1.6 μl of 10 × ligation buffer, and 0.7 μl of T4 DNA ligase enzyme (6 units/μl). These were combined and mixed in a 0.2-ml PCR tube and incubated overnight at 16°C in a PCR thermal cycler. The reactions were stopped by adjusting the thermal cycler to 70°C for 5 min. Last, 72 μl of TE buffer (10 mM Tris, 1 mM EDTA; pH 7.5) was added to each reaction and vortexed at slow speed for 15 sec.
Ligation products were then used to clone integration sites by nested PCR. The first-round PCR was run with an outer adaptor primer (AP1, from the GenomeWalker kit) and a custom primer termed GagR2. AP1 is a forward primer located in the adaptor, whereas GagR2 is a reverse primer located immediately downstream of the 3′ end of the U5 LTR of the virus. The second-round PCR was run with an inner adaptor primer (AP2, also from the GenomeWalker kit) and another custom primer termed U3RU2. The U3RU2 primer is also a reverse primer, this time located in the viral LTR immediately downstream of the 5′ end of the U5 LTR. Primer sequences used in these experiments were as follows: HIV-1 GagR2, 5′-TTTTGGCGTACTCACCAGTCG-3′; AP1, 5′-GTAATACGACTCACTATAGGGC-3′; HIV-1 U3RU2, 5′-TGAGGGATCTCTAGTTACCAGAGT-3′; AP2, 5′-ACTATAGGGCACGCGTGGT-3′; M13 forward primer, 5′-GTAAAACGACGGCCAG-3′.
PCRs were carried out with TaKaRa Ex Taq polymerase (cat. no. RR001A; TakaRa Bio Company, Madison, WI). Each primary PCR contained the following: 36.8 μl of water, 5 μl of 10 × PCR buffer, 4 μl of dNTPs (10 mM each), 0.2 μl of Ex Taq DNA polymerase, 1 μl of AP1 and GagR2 (10 μM each), and 1 μl of DNA template. The parameters were set up with a predenaturation step of 5 min at 94°C, followed by 30 cycles of 94°C for 1 min, 55°C for 45 sec, and 72°C for 3 min. A final 5-min 72°C extension step was added at the end of the reaction.
Second-round PCR was performed with 1 μl of the primary PCR product as DNA template. The primers were exchanged for AP2 and U3RU2. The cycling parameters and reaction contents remained the same.
The nested PCR products were cloned into the TOPO vector, using a TOPO TA Cloning kit (cat. no. K452001; Invitrogen). Individual plasmid-containing colonies were then expanded for plasmid DNA extraction.
Sequencing
Samples that contained a sufficiently high DNA concentration were sent to the Sidney Kimmel Nucleic Acid Facility at the Kimmel Cancer Center at Thomas Jefferson University (Philadelphia, PA) for sequencing. They were prepared by combining 0.4 μg of plasmid DNA, 3.2 pmol of M13 forward primer, and water to 12 μl. The facility uses a 3730 DNA analyzer and BigDye terminator cycle sequencing kits (both from Applied Biosystems, Foster City, CA).
Integration site analysis and statistics
Raw sequences were analyzed with the BLAT program (University of California, Santa Cruz, Santa Cruz, CA; Human Genome Project Working Draft February 2009 Freeze; http://www.genome.ucsc.edu/cgi-bin/hgBlat). Integration sites were considered authentic if the genomic portion was positioned between a recognizable adaptor sequence and the correct viral LTR end (either … CA or TG … , depending on the orientation). Also, the genomic sequence must match with: 98% identity and be unequivocally placed at a single site in the genome to be considered in the statistical analysis. Integration was determined to occur in a gene only if it was located within the boundaries of a RefSeq gene (National Center for Biotechnology Information Reference Sequence Project). Other sequence features were analyzed as described (Smit, 1999; Schroder et al., 2002).
Alu-ChIP
293T cells were transfected with TIHPLE and control plasmids as described previously. One day posttransfection, they were replated onto 100-mm dishes and the next day infected at an MOI of 1. One day postinfection, cells were harvested and a chromatin immunoprecipitation (ChIP) assay was performed as described previously (Smith et al., 2008) with the H3K9me3 antibody (1 μg per sample; cat. no. 07-523; Upstate Cell Signaling Solutions/Millipore), H3K4me3 antibody (cat. no. 07-473; Upstate Cell Signaling Solutions/Millipore), or phosphatidylinositol-3-kinase antibody (sc-55589; Santa Cruz Biotechnology, CA). One microliter of purified DNA was then subjected to Alu-PCR. The first round of Alu-PCR employed a primer targeting the cellular Alu sequence 5′-GCC TCC CAA AGT GCT GGG ATT ACA G-3′ as well as the primer targeting the HIV GagR2 region (see previously). Samples were subjected to 30 PCR cycles of 95°C for 30 sec, 60°C for 45 sec, and 72°C for 5 min, and after the final round samples were kept at 72°C for 10 min. One microliter of the first-round product was diluted and used in the 30-cycle second round (nested) with viral LTR primer 5′-GGA TTG TGC TAC AAG CTA GTA CC-3′ and the U3RU2 primer (see previously). The second-round PCR was cycled as follows: 95°C for 5 min; 35 cycles of 95°C for 40 sec, 55°C for 45 sec, and 72°C for 60 sec; this was followed by 72°C for 10 min. PCR products were then analyzed on a 2% agarose gel.
Western blot analyses
To detect TIHPLE and LEDGF proteins, cell lysates (106 cells per sample) were subjected to Western blot analysis with the anti-LEDGF/p75 antibody (cat. no. A300-848A; Bethyl Laboratories, Montgomery, TX), which recognizes the C terminus of the LEDGF protein. To detect the HP1 portion of TIHPLE, we used the HP1α antibody (ab9057; Abcam, Cambridge, MA).
Coimmunoprecipitations
To detect association of the TIHPLE protein with methylated histones, TIHPLE-transfected, HP1δTIHPLE-transfected, and control cells were harvested 2 days after transfection and lysed. Lysates of 106 cells per sample were briefly sonicated to break genomic DNA, centrifuged, and the supernatants were immunoprecipitated with H3K4me3 and H3K9me3 antibodies (see previously; 1 μg of antibody per sample). As a negative control, lysates were immunoprecipitated with rabbit IgG (ab37415; Abcam). Lysates were resolved by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis (SDS–PAGE) and filters were blotted with the LEDGF/p75 antibody (see previously). To detect association of the TIHPLE protein with integrase, cells were transfected as described previously and infected with the HIV-1-based vector at an MOI of 20 in the presence of dextran. Eight hours after the addition of virus, cells were harvested and lysed. Whole cell lysates of 106 cells per sample were immunoprecipitated with the integrase antibody (10 μl per sample; AIDS Research and Reference Reagent Program, antiserum, cat. no. 756, from D.P. Grandgenett [Grandgenett and Goodarzi, 1994]). Filters were blotted with the LEDGF/p75 antibody, as described previously.
Cell proliferation/toxicity assay
Cells were transfected with TIHPLE and control plasmids as described previously. Two days posttransfection, cells were replated onto a 96-well plate (1000 or 3000 cells per well, three wells per point). Four days later, cell proliferation was measured with an XTT assay kit (cat. no. 11 465 015 001; Roche Diagnostics, Mannheim, Germany), according to the manufacturer's instructions.
Results
TIHPLE construction and expression in human cells
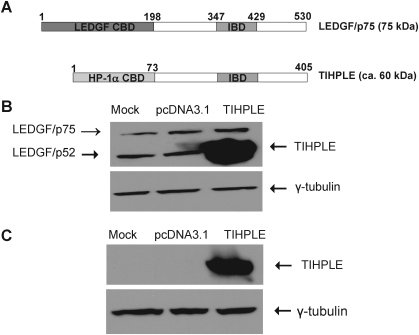
The cellular LEDGF protein has two isoforms, p75 and p52, which arise as a result of alternative splicing (Singh et al., 2000). The p75 isoform (referred to as LEDGF/p75), but not the p52 isoform, has an integrase-binding domain in its C terminus (residues 340–417; the whole p75 protein is 530 amino acid residues in length) (Singh et al., 2000). The N terminus of LEDGF/p75 contains a chromatin-binding domain (residues 1–198; Llano et al., 2006b). It is not currently known whether this domain binds to a specific chromatin structure (Llano et al., 2006b). Nevertheless, the LEDGF/p75 protein targets the HIV-1 preintegration complex to transcriptionally active chromatin regions (Shun et al., 2007). In contrast to LEDGF/p75, the chromatin-binding domain of the cellular heterochromatin protein-1α (HP1α CBD) is well characterized and it has been shown that it binds to the trimethylated Lys-9 residue of histone H3 (H3K9me3; Nielsen et al., 2001). H3K9me3 is a hallmark of heterochromatin (Grewal and Moazed, 2003). HP1α thus localizes to heterochromatin regions (Kellum, 2003; Maison and Almouzni, 2004). The TIHPLE protein consists of residues 1–73 of HP1α, which contain the HP1α chromatin-binding domain, and residues 198–530 of the LEDGF/p75 protein, which contain the integrase-binding domain (Fig. 1A). The synthetic TIHPLE gene was assembled from synthetic oligonucleotides and PCR products, as described in Materials and Methods. TIHPLE DNA was subcloned into a pcDNA3.1 plasmid vector (Invitrogen) under the control of the CMV promoter. To evaluate TIHPLE expression in human cells, the TIHPLE-encoding plasmid and the empty pcDNA3.1 vector were transfected into 293T cells. Two days later, transfected and control cells were harvested and subjected to Western blotting analysis with LEDGF/p75 and HP1α antibodies. We observed a strong band of the predicted size (about 60 kDa) in TIHPLE-transfected cells, but not in control cells. The band was detected with both an LEDGF/p75 antibody that recognizes the LEDGF/p75 C terminus, and an HP1α antibody (Fig. 1B and C). The LEDGF/p75 antibody also recognized the expected LEDGF/p75 and p52 bands (Fig. 1B). As shown, TIHPLE is expressed at levels that exceed those of endogenous LEDGF/p75 and p52. We noted that the TIHPLE gene was codon-optimized for expression in human cells (see Materials and Methods). This fact, together with the strong CMV promoter, likely accounts for the high expression of TIHPLE in 293T cells (Fig. 1B).
FIG. 1.
Structure and expression of TIHPLE. (A) Structures of LEDGF/p75 and TIHPLE proteins. IBD, integrase-binding domain; CBD, chromatin-binding domain. (B) TIHPLE expression in 293T cells as detected with an LEDGF antibody (see Materials and Methods). 293T cells were transfected with the TIHPLE-encoding and control plasmids and LEDGF expression was analyzed 2 days posttransfection, as described in Materials and Methods. Mock, mock-transfected 293T cells; pcDNA3.1, 293T cells transduced with the empty vector pcDNA3.1; TIHPLE, 293T cells transduced with the TIHPLE plasmid. (C) TIHPLE expression in 293T cells as detected with an HP1α antibody (see Materials and Methods). Cell lysates from (B) were analyzed by Western blotting to detect HP1α amino acid sequences in TIHPLE. Terminology as in (B).
Association of TIHPLE with H3K9me3 and integrase
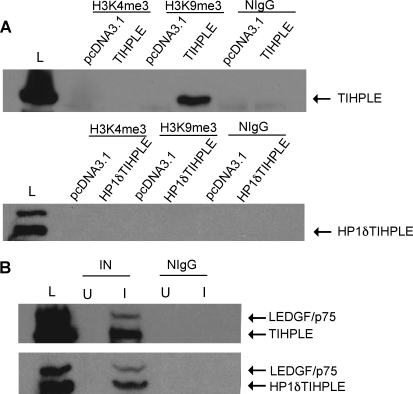
To determine whether TIHPLE associates with methylated histones, 293T cells were transfected with TIHPLE-encoding and control pcDNA3.1 plasmids. Two days posttransfection, cells were lysed and lysates were immunoprecipitated with antibodies recognizing either the trimethylated Lys-4 of histone H3 (H3K4me3) or the trimethylated Lys-9 of histone H3 (H3K9me3). Lysates were resolved by SDS–PAGE and the presence of TIHPLE protein was detected with the LEDGF/p75 antibody. TIHPLE was found in the H3K9me3 immunoprecipitates, but not in the H3K4me3 immunoprecipitates or control samples that were precipitated with normal rabbit IgG (Fig. 2A, top). We conclude that the TIHPLE protein associates with H3K9me3, which is a hallmark of heterochromatin. To test the hypothesis that the HP1α CBD mediates TIHPLE binding to H3K9me3, we created a TIHPLE deletion mutant (HP1δTIHPLE), which is missing the HP1α CBD. This mutant failed to associate with H3K9me3. The HP1α CBD is thus necessary for TIHPLE association with H3K9me3 (Fig. 2A, bottom).
FIG. 2.
Coimmunoprecipitation of TIHPLE protein with H3K9me3 and integrase. (A) 293T cells were transfected with TIHPLE (top), HP1δTIHPLE (bottom), and pcDNA3.1 plasmids as described in Materials and Methods. Two days posttransfection, cells were harvested and lysed, and lysates were sonicated to break down genomic DNA. Lysates were then cleared by centrifugation and H3K9me3 and H3K4me3 were immunoprecipitated by addition of the corresponding antibodies (see Materials and Methods). As a control, immunoprecipitations were performed with normal rabbit IgG. Immunoprecipitates were then resolved by SDS–PAGE and TIHPLE was detected with the LEDGF antibody (see Materials and Methods). pcDNA3.1, 293T cells transfected with the pcDNA3.1 plasmid; TIHPLE, cells transfected with the TIHPLE plasmid; H3K4me3, lysates immunoprecipitated with the H3K4me3 antibody; H3K9me3, lysates immunoprecipitated with the H3K9me3 antibody; NIgG, normal rabbit IgG (see Materials and Methods); L, whole cell lysate of TIHPLE-transfected cells (top) or HP1δTIHPLE-transfected cells (bottom). Arrows indicate the TIHPLE and HP1δTIHPLE bands. (B) Cells were transfected as in (A), and infected 2 days posttransfection with the HIV-1-based vector at an MOI of 20. Eight hours after infection, cells were harvested and lysed, and lysates were immunoprecipitated with the integrase or control antibodies. Top: TIHPLE-transfected cells. Bottom: HP1δTIHPLE-transfected cells. U, uninfected cells; I, infected cells; IN, lysates immunoprecipitated with the integrase. Arrows indicate TIHPLE, HP1δTIHPLE, and LEDGF/p75 bands. Other terminology as in (A).
To determine whether TIHPLE associates with the integrase protein, we infected TIHPLE-expressing and control cells with the HIV-1-based vector and harvested cells 8 hr postinfection. Cell lysates were then immunoprecipitated with the HIV-1 integrase antibody. As shown in Fig. 2B (top), TIHPLE was present in integrase immunoprecipitates from infected, but not uninfected, cells. Similarly, HP1δTIHPLE, which contains the LEDGF/p75 integrase-binding domain, associates with integrase (Fig. 2B, bottom). We conclude that TIHPLE associates with both HIV-1-integrase and H3K9me3.
Effects of TIHPLE on cell proliferation and viability
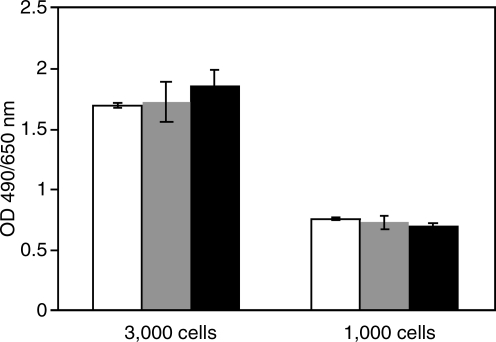
To determine whether TIHPLE affects cellular processes in uninfected cells, TIHPLE and the control pcDNA3.1 plasmid were transfected into 293T cells and the number of viable cells was determined 5 days posttransfection, using the XTT assay. As shown in Fig. 3, TIHPLE-expressing cells proliferated at a rate that was virtually indistinguishable from that of control cells. We conclude that TIHPLE does not appear to affect the proliferation or viability of 293T cells.
FIG. 3.
Effect of TIHPLE on the growth of 293T cells. 293T cells were transfected with TIHPLE and control plasmids as described in Materials and Methods. One day posttransfection, cells were replated onto a 96-well plate, at a density of 1000 or 3000 cells per well. Four days after plating, cell density was measured by XTT assay, as described in Materials and Methods. Open columns, mock-transfected 293T cells; shaded columns, 293T cells transduced with the empty vector pcDNA3.1; solid columns, 293T cells transduced with the TIHPLE plasmid. Error bars indicate standard deviations.
Effect of TIHPLE on HIV-1 transduction and expression of vector-transduced marker
Gene therapy approaches depend on high-efficiency transduction and expression of a vector-transduced gene. The HIV-1-based vector, which we used in this study, carries an enhanced green fluorescent protein (EGFP) marker. To determine whether TIHPLE affects transduction and/or expression of the vector-transduced EGFP, we analyzed the number of vector DNA copies and EGFP expression 15 days after transduction of TIHPLE-expressing cells. At this time point, cells do not contain any unintegrated vector DNA. We observed that neither the number of vector DNA copies, nor the number of EGFP-expressing cells, nor the EGFP intensity was significantly affected in TIHPLE-expressing cells when compared with control cells, which were transduced with an empty plasmid (Fig. 4A and B). Likewise, we attempted to determine whether TIHPLE affects EGFP expression from stably integrated proviral DNA. We transfected TIHPLE-expressing and control plasmids into cells, which were previously transduced with the HIV-1-based vector. We then analyzed EGFP expression in these cells by flow cytometry. We observed that TIHPLE did not affect EGFP expression under these conditions (data not shown). We conclude that TIHPLE does not affect HIV-1 transduction, or the expression of a reporter gene that was carried by the HIV-1-based vector.
Association of vector DNA with methylated Lys-4 and Lys-9 of histone H3
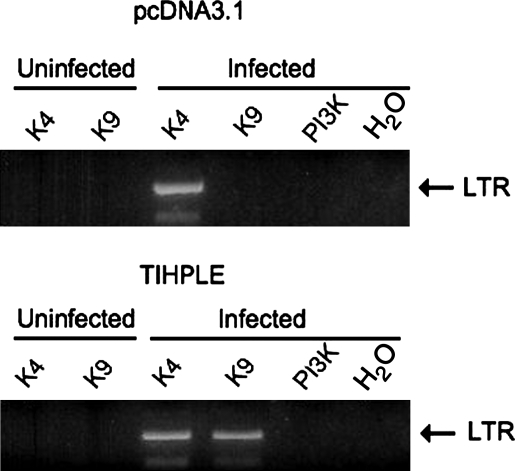
As a quick method to evaluate the TIHPLE effect on integration site selection, we developed a chromatin immunoprecipitation-based assay that we termed Alu-ChIP. To establish whether integration occurs in the vicinity of certain chromatin hallmarks, we immunoprecipitated chromatin from nuclear extracts with antibodies detecting H3K4me3 (mark of transcriptionally active chromatin) or H3K9me3 (mark of heterochromatin) and screened for the presence of integrated viral DNA, using Alu-PCR. This method was termed Alu-ChIP, because it combines ChIP and Alu-PCR. As shown in Fig. 5, in control cells, viral DNA was found to be associated with H3K4me3, but not H3K9me3. However, in the TIHPLE-expressing cells, viral DNA was also found to be associated with the TIHPLE target, H3K9me3. We conclude that TIHPLE expression may bias integration site selection toward integration in the vicinity of the heterochromatin hallmark, H3K9me3. However, our ultimate objective is to use TIHPLE to target integration away from genes, and the Alu-CHIP method, although fast, does not currently permit us to quantitatively determine the frequency of integration in genes and outside of genes. Thus, we have isolated and analyzed integration sites from TIHPLE-expressing and control cells, as described subsequently.
FIG. 5.
Presence of integrated viral DNA in the vicinity of H3K4me3 and H3K9me3 in control and TIHPLE-expressing cells. 293T cells were transfected with pcDNA3.1 or TIHPLE. All cells were then infected with the HIV-1-based vector at an MOI of 1. ChIP (see Materials and Methods for details) was performed with antibodies recognizing the H3K4me3 or H3K9me3 residue, or with control PI-3K antibody, or no antibody (H2O). Alu-PCR was performed with Alu and HIV gag primers for 30 cycles in the first round, and LTR primers for 35 cycles in the second round. pcDNA3.1, cells transfected with pcDNA3.1 (top); TIHPLE, TIHPLE-expressing cells (bottom). Products were analyzed on a 2% agarose gel. The LTR band is indicated and antibodies are indicated. Cells were harvested 1 week postinfection.
Analysis of integration sites in TIHPLE-expressing and control cells
To elucidate the effects of TIHPLE on the distribution of integration sites in the genome of human 293T cells, we transfected these cells with TIHPLE-expressing and control plasmids. We then infected these and untransfected cells with an HIV-1-based vector. We isolated genomic DNA 4 days postinfection and the virus–host DNA junctions were then amplified by linker-mediated PCR and subcloned into a cloning vector for expansion in bacterial cells and sequencing, as described in Materials and Methods. We identified a total of 308 integration sites. Of these, 116 were in the mock-transfected cells, 121 were in cells that were transfected with the control plasmid, and 129 were in TIHPLE-expressing cells. All sequences were then analyzed with the BLAT program (see Materials and Methods).
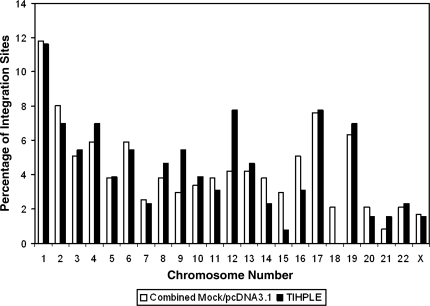
We initially mapped the integration sites to human chromosomes. As shown in Fig. 6, vector integration sites were distributed broadly in the 293T cell genome, and we did not detect any particular bias toward a chromosomal location in any of the samples, except for a high number of integration sites in smaller, gene-rich chromosomes, which was reported by others (Schroder et al., 2002). Next, we determined the frequency of integration into genes. HIV-1 and HIV-1-based vectors are known to prefer integration into genes. The first genome-wide analysis of HIV-1 integration site selection reported that 69% of HIV-1 (and HIV-1-based vector) integrations occurred in genes (Schroder et al., 2002). In contrast, random integrations should result in approximately 33% of integration events occurring in genes given that the human genome is approximately 33% genes (Schroder et al., 2002). Our data showed that 72.4% of HIV-1-based vector integration events in mock-transfected (transfection reagent only) 293T cells occurred in genes, and 76.9% of integration events occurred in genes of 293T cells that were transfected with the control plasmid (Table 1). There is no statistically significant difference between these control samples (p = 0.8255). These data are also consistent with the reported distribution of integration sites of HIV-1-based vectors (Schroder et al., 2002; and see Table 3). In contrast, only 59.7% of integration sites were found in genes of TIHPLE-expressing cells (Table 1). This is still a higher frequency of integration in genes than would be expected for random integration. However, the difference in integration frequency between TIHPLE-expressing and control cells (72.4% for untransfected cells and 76.9% for pcDNA3.1.-transfected cells) was statistically significant (p = 0.025 and p = 0.003, respectively) and highly significant when we compared integration frequency in genes of TIHPLE-expressing cells with a pool of integration sites in genes of mock- and pcDNA3.1-transfected cells (p = 0.002). Thus, we conclude that TIHPLE reduces the integration in genes.
FIG. 6.
Comparison of chromosomal distribution of integration sites between control (mock- and pcDNA3.1-transfected) and TIHPLE-expressing cells. Integration sites were analyzed as described in Results and in Materials and Methods and integration sites were then assigned to individual chromosomes (x axis). y axis, percentage of integration sites per chromosome.
Table 1.
Frequency of Integration Within Genes
| Percentage in human genome | Percentage in mock integration sites | Percentage in pcDNA3.1 integration sites | Percentage in TIHPLE integration sitesa | |
|---|---|---|---|---|
| In gene | ∼33% | 72.4% | 76.9% | 59.7% |
| p = 0.025b | ||||
| p = 0.003c | ||||
| p = 0.002d | ||||
| Not in gene | ∼67% | 27.6% | 23.1% | 40.3% |
| Total cloned integration sites | N/A | 116 | 121 | 129 |
Abbreviations: N/A, not applicable; TIHPLE, targeting integration heterochromatin protein LEDGF.
p Values are from Fisher's exact one-sided test.
p Value a comparison between TIHPLE and mock.
p Value a comparison between TIHPLE and pcDNA3.1.
p Value a comparison between TIHPLE and combined mock/pcDNA3.1.
Table 3.
| Chromosomal feature | Schroder et al. (2002) | Percentage in mock integration sites | Percentage in pcDNA3.1 integration sites | Percentage in TIHPLE integration sites |
|---|---|---|---|---|
| Genes | 69% | 72.4% (p = 0.792) | 76.9% (p = 0.966) | 59.7% (p = 0.028) |
| LINE | 17% | 7.8% (p = 0.998) | 11.6% (p = 0.950) | 16.3% (p = 0.621) |
| Alu (SINE) | 15.9% | 21.6% (p = 0.091) | 25.6% (p = 0.010) | 19.4% (p = 0.200) |
| MIR (SINE) | 0.7% | 0% (p = 1.000) | 0.8% (p = 0.647) | 1.6% (p = 0.339) |
| DNA elements | 2.2% | 5.2% (p = 0.088) | 4.1% (p = 0.198) | 2.3% (p = 0.595) |
| LTR elements (HERV) | 3.7% | 5.2% (p = 0.291) | 0% (p = 1.000) | 12.4% (p = 0.000) |
| Satellite |
0.4% |
0% (p = 0.670) |
0% (p = 0.660) |
0% (p = 0.644) |
| Total cloned integration sites | 524 | 116 | 121 | 129 |
p Values are a comparison of each population from this study against data from Schroder et al. (2002), using Fisher's exact one-sided test.
We then examined the distribution of integration sites with respect to sequence features of human DNA (Table 2). Because heterochromatin is associated with repetitive sequences, we first determined the frequency of integration events in highly repetitive sequences found in the human genome. Next, we examined integrations in short interspersed nuclear elements (SINEs), long interspersed nuclear elements (LINEs), and LTR repeat elements (human endogenous retroviruses, HERVs). We also compared the frequency of integration in these sequences with the results reported by Schroder and colleagues, who first reported the preferences of HIV-1 and HIV-1-based vectors for integration into genes and other elements of the genome. Our data indicate that the fraction of integration sites in SINEs and LINEs of control cell populations largely corresponded to the frequency of integration into these elements as reported by others (Table 3). We also noted a high number of integration sites in Alu elements (a type of SINE), which occur frequently in genes, even somewhat higher than published (Schroder et al., 2002). We believe this difference could be attributable to the different cell type we used in our experiments (293T cells as opposed to SupT1 cells). Similarly, integration into the LTR elements, which comprise endogenous retrotransposons and retroviruses, occurred in control cells at a frequency lower than that which would be expected from the size of the fraction of genome they occupy, but consistent with the reported frequency for HIV-1 and HIV-1-based vectors (Schroder et al., 2002). However, we observed that the frequency of integration into LTR elements of TIHPLE-expressing cells was much higher than that of control cells (12.4 vs. 0–5.2%). The difference between TIHPLE and combined control cells is statistically significant (p = 0.000; Table 2). LTR elements were reported to be associated with heterochromatin (Huang et al., 2004). The high frequency of integration into the LTR elements of TIHPLE-expressing cells is thus consistent with the hypothesis that TIHPLE diverts integration of the HIV-1-based vector into heterochromatin. We also observed increased integration into LINEs, when TIHPLE-expressing cells are compared with control cells (Table 2). This finding is consistent with the lower frequency of integration into genes of TIHPLE-expressing cells, because LINEs, unlike SINEs, are usually found in gene-poor regions (Smit, 1999). We again note that LINEs are associated with heterochromatic regions (Smit, 1999). We have also examined the frequency of integration into the vicinity of other sequence features. These include CpG islands and upstream and downstream of genes (Table 4). We found that 3.4% of integration events occurred within 5 kb upstream of genes in combined mock-transfected cells and pcDNA3.1-transfected cells and 5.4% in TIHPLE-expressing cells, but this difference does not appear to be statistically significant (p = 0.248). Similarly, we have not found statistically significant differences between TIHPLE-expressing and control cells when we compared integration sites in the vicinity of CpG islands or downstream of genes (Table 4).
Table 2.
Integration in Repetitive Sequences
| Chromosomal feature | Percentage in human genomea | Percentage in mock integration sites | Percentage in pcDNA3.1 integration sites | Percentage in TIHPLE integration sites |
|---|---|---|---|---|
| LINE | 20% | 7.8% | 11.6% | 16.3% (p = 0.048)b |
| Alu (SINE) | 10.6% | 21.6% | 25.6% | 19.4% (p = 0.857)b |
| MIR (SINE) | 2.2% | 0% | 0.8% | 1.6% (p = 0.285)b |
| DNA elements | 2.8% | 5.2% | 4.1% | 2.3% (p = 0.923)b |
| LTR elements (HERV) | 8.3% | 5.2% | 0% | 12.4% (p = 0.000)b |
| Satellite | UNc | 0% | 0% | 0% |
| Total cloned integration sites | 116 | 121 | 129 |
Abbreviations: HERV, human endogenous retrovirus; LINE, long interspersed nuclear element; LTR, long terminal repeat; MIR, mammalias interspersed repeat; SINE, short interspersed nuclear element.
Data from Schroder and colleagues (2002).
p Values are a comparison of combined mock/pcDNA3.1 data and TIHPLE, using Fisher's exact one-sided test.
Unknown, from Schroder and colleagues (2002).
Table 4.
Integration into Various Chromosomal Features
| Percentage in mock integration sites | Percentage in pcDNA3.1 integration sites | Percentage in combined control integration sites | Percentage in TIHPLE integration sites | |
|---|---|---|---|---|
| Within ± 1 kb of CpG islands | 1.7% | 1.7% | 1.7% | 1.6% (p = 0.643)a |
| Within 5 kb upstream of genes | 6.0% | 0.83% | 3.4% | 5.4% (p = 0.248)a |
| Within 5 kb downstream of genes |
3.4% |
0.83% |
2.1% |
3.1% (p = 0.397)a |
| Total cloned integration sites | 116 | 121 | 237 | 129 |
p Values are a comparison of combined mock/pcDNA3.1 data and TIHPLE, using Fisher's exact one-sided test.
We conclude that TIHPLE primarily reduces the frequency of integration into genes and increases integration into repetitive elements that are associated with heterochromatin.
Discussion
In this study, we examined the effect of a fusion protein consisting of the HP1α chromodomain and the LEDGF/p75 C terminus, termed TIHPLE, on integration site selection by an HIV-1-based vector. We present evidence that TIHPLE can be efficiently expressed in human cells and associates with the H3K9me3 modification in chromatin, and does not affect the transduction or expression of the EGFP reporter of the HIV-1-based vector, nor does it affect the viability or proliferation of 293T cells. To determine whether TIHPLE affects integration site selection, we first used a chromatin immunoprecipitation-based assay. Our results suggest that a higher fraction of integration sites in TIHPLE-expressing cells (when compared with control cells) can be found in the vicinity of H3K9me3, which is the binding site for the HP1α chromodomain and a hallmark of heterochromatin (Nielsen et al., 2001; Grewal and Moazed, 2003). We analyzed 366 integration sites in control and TIHPLE-expressing cells to determine whether TIHPLE diverts integration away from genes, as its binding to heterochromatin would suggest. We found that 72.4–76.9% of integration sites in control cells can be found in genes. However, only 59.7% of integration sites in TIHPLE-expressing cells were found in genes. The difference between control and TIHPLE-expressing cells is statistically significant. In addition, we found a higher frequency of integration into LTR elements and LINEs in TIHPLE-expressing cells. Because these elements were found to be associated with heterochromatin, these data are again consistent with the hypothesis that TIHPLE drives integration away from genes and into heterochromatin.
What is the mechanism underlying the observed effect of TIHPLE? The TIHPLE C terminus contains the LEDGF/p75 integrase-binding domain, and the N terminus binds to the heterochromatin hallmark, H3K9me3. Thus, it appears likely that TIHPLE targets integration by an LEDGF/p75-like mechanism, that is, it tethers the preintegration complexes to its target chromatin structure (heterochromatin). One possible alternative explanation is that TIHPLE acts as a dominant negative LEDGF/p75 mutant and blocks the LEDGF/p75-mediated targeting of integration into genes. However, inhibition of LEDGF/p75 leads to a dramatic drop in integration efficiency (Shun et al., 2007). In contrast, the efficiency of transduction of the EGFP reporter into TIHPLE-expressing cells is about the same as that of normal cells (Fig. 4). Moreover, TIHPLE does not simply reduce integration in genes, but also appears to significantly increase integration events that occur in LTR elements and in the vicinity of H3K9me3 (Fig. 5 and Tables 2 and 3). Finally, we note that the E.M. Poeschla laboratory demonstrated that a fusion protein, in which the LEDGF/p75 chromatin-binding domain was replaced with the sequence of histone H1, can functionally replace LEDGF/p75 and restore integration efficiency in LEDGF/p75-deficient cells (Meehan et al., 2009). Thus, LEDGF/p75-based proteins that contain a different chromatin-binding domain are functional. We conclude that the effect of TIHPLE is not likely due to simple interference with LEDGF/p75 function.
Another alternative explanation is that TIHPLE somehow binds to and affects expression of cellular genes and thus blocks integration into them. This explanation appears to be inconsistent with the coimmunoprecipitation results (Fig. 2), where we show that TIHPLE associates with H3K9me3 (which is found predominantly outside of genes), and does not associate with H3K4me3, which is associated with transcriptionally active chromatin (Schubeler et al., 2004). Moreover, TIHPLE does not seem to have any detrimental effects on cellular growth (Fig. 3). Thus, we conclude that our results appear to be most consistent with the original hypothesis, that is, TIHPLE tethers preintegration complexes to H3K9me3. In our future experiments, we plan to perform a large-scale analysis, which is important for full understanding of interactions of integrating vector DNA in TIHPLE-expressing cells with heterochromatin hallmarks and heterochromatin-containing regions, as demonstrated by Wang and colleagues (2007) for association of integration with hallmarks of euchromatin.
There are two caveats associated with targeting into heterochromatin. First, HP1α protein, along with other HP1 proteins, was shown to participate in silencing of some euchromatic genes, and associates in this instance with promoter-proximal regions (Smallwood et al., 2007). Thus, hypothetically, TIHPLE could target a fraction of integration events toward silent genes. This could, again hypothetically, lead to activation of these genes. However, the majority of HP1α is associated with gene-poor constitutive heterochromatin regions (Maison and Almouzni, 2004) and few integration events occurred in our analysis within 5 kb upstream of genes, suggesting that this type of targeting occurs only rarely, if ever. Second, heterochromatin regions are poorly characterized and it is possible that integration in these regions may disrupt an unknown, but important, cellular function. We will investigate these possibilities in future experiments. However, TIHPLE does reduce integration into genes and thus reduces the genotoxicity that is associated with retroviral vectors.
Of all integration events in TIHPLE-expressing cells, 59.7% can still be found in transcription units. This is a significant reduction, but it is not yet as strong a reduction as can be achieved with a vector in which the integrase function is replaced by transposase (Staunstrup et al., 2009). What are the possible reasons for the lower efficiency of TIHPLE? We delivered TIHPLE into cells by transfection. Although our transfection efficiency is quite high (80%; data not shown), a significant fraction of cells did not receive TIHPLE. In these cells, integration is still targeted to genes by LEDGF/p75. One way around this issue would be to develop cell lines that stably express TIHPLE. However, stable TIHPLE expression would be undesirable in a gene therapy protocol, and therefore we believe transient expression is a better approach. Nevertheless, if TIHPLE or TIHPLE-related proteins are to be used in any gene therapy trials, it will be necessary to achieve delivery of TIHPLE into all cells, preferably by means of an unintegrating, hit-and-run vector.
Even TIHPLE-expressing cells still contain the wild-type LEDGF/p75. Although the TIHPLE expression level is much higher than that of LEDGF/p75, the LEDGF protein may still target some integration into genes. Thus, to enhance the TIHPLE effect, it would most likely be necessary to transiently inhibit the wild-type LEDGF/p75, possibly by RNA interference treatment. We plan to test these approaches in our laboratory to determine their effect on integration site selection. Nevertheless, the results presented in this study indicate a new possible approach to targeting retroviral integration, and suggest a new way to increase the safety of retroviral vectors by targeting them to predetermined chromosomal regions.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA125272 and CA135214 and by a W.W. Smith Foundation AIDS Research Award to R.D.
Author Disclosure Statement
No competing financial interests exist.
References
- Barsov E.V. Hughes S.H. Gene transfer into mammalian cells by a Rous sarcoma virus-based retroviral vector with the host range of the amphotropic murine leukemia virus. J. Virol. 1996;70:3922–3929. doi: 10.1128/jvi.70.6.3922-3929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M.I. Sharova N. Dempsey M.P. Stanwick T.L. Bukrinskaya A.G. Haggerty S. Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M.I. Haggerty S. Dempsey M.P. Sharova N. Adzhubel A. Spitz L. Lewis P. Goldfarb D. Emerman M. Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F.D. Miller M.D. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J. Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busschots K. Vercammen J. Emiliani S. Benarous R. Engelborghs Y. Christ F. Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P. Maertens G. Proost P. Devreese B. Van Beeumen J. Engelborghs Y. De Clercq E. Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Coffin J.M. Hughes S.H. Varmus H. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- Daniel R. DNA repair in HIV-1 infection: A case for inhibitors of cellular co-factors? Curr. HIV Res. 2006;4:411–421. doi: 10.2174/157016206778560027. [DOI] [PubMed] [Google Scholar]
- Daniel R. Katz R.A. Skalka A.M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- Daniel R. Myers C.B. Kulkosky J. Taganov K. Greger J.G. Merkel G. Weber I.T. Harrison R.W. Skalka A.M. Characterization of a naphthalene derivative inhibitor of retroviral integrases. AIDS Res. Hum. Retroviruses. 2004;20:135–144. doi: 10.1089/088922204773004842. [DOI] [PubMed] [Google Scholar]
- Dimitri P. Corradini N. Rossi F. Verni F. The paradox of functional heterochromatin. Bioessays. 2005;27:29–41. doi: 10.1002/bies.20158. [DOI] [PubMed] [Google Scholar]
- Flint S.J. Enquist L.W. Krug A.M. Racaniello V.R. Skalka A.M. Molecular Biology, Pathogenesis, and Control. ASM Press; New York: 2000. Principles of virology. [Google Scholar]
- Grandgenett D.P. Goodarzi G. Folding of the multidomain human immunodeficiency virus type-I integrase. Protein Sci. 1994;3:888–897. doi: 10.1002/pro.5560030604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I. Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M. McCormack M.P. Wulffraat N. Leboulch P. Lim A. Osborne C.S. Pawliuk R. Morillon E. Sorensen R. Forster A. Fraser P. Cohen J.I. De Saint Basile G. Alexander I. Wintergerst U. Frebourg T. Aurias A. Stoppa-Lyonnet D. Romana S. Radford-Weiss I. Gross F. Valensi F. Delabesse E. Macintyre E. Sigaux F. Soulier J. Leiva L.E. Wissler M. Prinz C. Rabbitts T.H. Le Deist F. Fischer A. Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Holmes-Son M.L. Chow S.A. Integrase–LexA fusion proteins incorporated into human immunodeficiency virus type 1 that contains a catalytically inactive integrase gene are functional to mediate integration. J. Virol. 2000;74:11548–11556. doi: 10.1128/jvi.74.24.11548-11556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Fan T. Yan Q. Zhu H. Fox S. Issaq H.J. Best L. Gangi L. Munroe D. Muegge K. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32:5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S. Lan F. Bayliss P. De La Torre-Ubieta L. Huarte M. Qi H.H. Whetstine J.R. Bonni A. Roberts T.M. Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jones D.O. Cowell I.G. Singh P.B. Mammalian chromodomain proteins: Their role in genome organisation and expression. Bioessays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Katz R.A. Merkel G. Skalka A.M. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: In vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- Katz R.A. Greger J.G. Darby K. Boimel P. Rall G.F. Skalka A.M. Transduction of interphase cells by avian sarcoma virus. J. Virol. 2002;76:5422–5434. doi: 10.1128/JVI.76.11.5422-5434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R. HP1 complexes and heterochromatin assembly. Curr. Top. Microbiol. Immunol. 2003;274:53–77. doi: 10.1007/978-3-642-55747-7_3. [DOI] [PubMed] [Google Scholar]
- Lewis P. Hensel M. Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M. Saenz D.T. Meehan A. Wongthida P. Peretz M. Walker W.H. Teo W. Poeschla E.M. An essential role for LEDGF/p75 in HIV integration. Science. 2006a;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Llano M. Vanegas M. Hutchins N. Thompson D. Delgado S. Poeschla E.M. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006b;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- Lombardo A. Genovese P. Beausejour C.M. Colleoni S. Lee Y.L. Kim K.A. Ando D. Urnov F.D. Galli C. Gregory P.D. Holmes M.C. Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Maison C. Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell. Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Meehan A.M. Saenz D.T. Morrison J.H. Garcia-Rivera J.A. Peretz M. Llano M. Poeschla E.M. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009;5:e1000522. doi: 10.1371/journal.ppat.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R.S. Beitzel B.F. Schroder A.R. Shinn P. Chen H. Berry C.C. Ecker J.R. Bushman F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L. Blomer U. Gallay P. Ory D. Mulligan R. Gage F.H. Verma I.M. Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Narezkina A. Taganov K.D. Litwin S. Stoyanova R. Hayashi J. Seeger C. Skalka A.M. Katz R.A. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L. Oulad-Abdelghani M. Ortiz J.A. Remboutsika E. Chambon P. Losson R. Heterochromatin formation in mammalian cells: Interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- Roe T. Reynolds T.C. Yu G. Brown P.O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder A.R. Shinn P. Chen H. Berry C. Ecker J.R. Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Schubeler D. Macalpine D.M. Scalzo D. Wirbelauer C. Kooperberg C. Van Leeuwen F. Gottschling D.E. O'Neill L.P. Turner B.M. Delrow J. Bell S.P. Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun M.C. Raghavendra N.K. Vandegraaff N. Daigle J.E. Hughes S. Kellam P. Cherepanov P. Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.P. Kimura A. Chylack L.T., Jr. Shinohara T. Lens epithelium-derived growth factor (LEDGF/p75) and p52 are derived from a single gene by alternative splicing. Gene. 2000;242:265–273. doi: 10.1016/s0378-1119(99)00506-5. [DOI] [PubMed] [Google Scholar]
- Skalka A.M. Retroviral DNA Integration. Academic Press; New York: 1999. [Google Scholar]
- Smallwood A. Esteve P.O. Pradhan S. Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smith J.A. Wang F.X. Zhang H. Wu K.J. Williams K.J. Daniel R. Evidence that the Nijmegen breakage syndrome protein, an early sensor of double-strand DNA breaks (DSB), is involved in HIV-1 post-integration repair by recruiting the ataxia telangiectasia-mutated kinase in a process similar to, but distinct from, cellular DSB repair. Virol. J. 2008;5:11. doi: 10.1186/1743-422X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunstrup N.H. Moldt B. Mates L. Villesen P. Jakobsen M. Ivics Z. Izsvak Z. Mikkelsen J.G. Hybrid lentivirustransposon vectors with a random integration profile in human cells. Mol. Ther. 2009;17:1205–1214. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H.G. Newton K. Brownstein D.G. Holmes M.C. Kress C. Semple C.A. Bickmore W.A. Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations. Mol. Cell. Biol. 2006;26:7201–7210. doi: 10.1128/MCB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. Zhu K. Segal D.J. Barbas C.F., III Chow S.A. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W. Dong Z. Wilkinson T.A. Barbas C.F., III Chow S.A. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas M. Llano M. Delgado S. Thompson D. Peretz M. Poeschla E. Identification of the LEDGF/p75 HIV-1 integraseinteraction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- Wang G.P. Ciuffi A. Leipzig J. Berry C.C. Bushman F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J.B. Matthews T.J. Cullen B.R. Malim M.H. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 1991;174:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Li Y. Crise B. Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Zufferey R. Nagy D. Mandel R.J. Naldini L. Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.