Abstract
The nucleus is an ordered three-dimensional entity, and organization of the genome within the nuclear space might have implications for orchestrating gene expression. Recent technological developments have revealed that chromatin is folded into loops bringing distal regulatory elements into intimate contact with the genes that they regulate. Such intrachromosomal contacts appear to be a general mechanism of enhancer-promoter communication ‘in cis’. Tantalizing evidence is also emerging that regulatory elements might have the capacity to act ‘in trans’ to regulate genes on other chromosomes. However, unequivocal data required to prove that interchromosomal gene regulation truly represents another level of control within the nucleus is still lacking, and this concept remains highly contentious. Such controversy only emphasizes that our current understanding of the mechanisms that govern gene expression are far from complete.
Interchromosomal gene regulation in mammals: an emerging field
We are only just beginning to realize the extent of organization within the mammalian nucleus and how this might shape transcriptional regulation. Loci can interact with sequences elsewhere in the genome, leading to the hypothesis that genes can be subjected to regulation in trans by regulatory elements on other chromosomes. This concept is not without merit as a similar but well established phenomenon termed transvection takes place in Drosophila (Box 1). However, unlike classical transvection, interchromosomal gene regulation in mammals is still highly controversial. We will explore recent examples of interchromosomal associations (see Glossary) and discuss whether these represent a chance meeting of genes within the shared nuclear space or whether they provide evidence for functional regulation in trans.
Box1: Transvection in Drosophila melanogaster.
The term transvection was introduced in 1954 by Ed Lewis to describe the phenomenon that upon homologous association of two alleles, an element on one chromosome can affect gene expression on the homologous chromosome 98. In transvection both enhancers and silencers can act to influence gene expression in trans98. For example, at specific Drosophila loci the paring of alleles is required to achieve wild type levels of transcription, and deletion of regulatory elements on one chromosome can be rescued in trans by sequences on the homologous chromosome98. Similarly, the silencing effect of the polycomb response element (PRE) is enhanced by the pairing of two allelic copies of the PRE98. Interestingly, the degree of paring in transvection is highly locus-, tissue-, and fly line dependent, suggesting that, even though well established, the current model of transvection is incomplete and too simplistic99.
Nuclear organization
More than a century ago, studies by Rabl and then Boveri suggested that chromatin was not randomly organized within the nucleus but occupied distinct regions. However, only recent advances in technology have allowed the confirmation that metaphase chromosomes are indeed organized into discrete, non-overlapping ‘territories’1. Furthermore, chromosomes adopt non-random positions within the nucleus with gene-rich chromosomes being located preferentially towards the center of the nucleus, an arrangement that is not only retained in many different cell types but also appears to be conserved through evolution2-11. It is also well documented that heterochromatin and euchromatin segregate within the nucleus, forming chromatin ‘neighborhoods’ with similar properties12. Within the relatively fixed nuclear positions of chromosome territories, loci undergo constrained diffusion within a small (<1μm) corral13. However, gene activation and gene silencing events can be accompanied by dynamic chromatin movements (of up to 5μm) that potentially determine access to the transcriptional machinery 13-16. For example, on activation the major histocompatibility complex (MHC) genes form a large (megabase) chromatin loop, which extends out from its chromosome territory17. By contrast, during T cell development, silencing of the recombination activating gene 1 (Rag1) is accompanied by its relocation to pericentromeric heterochromatin18. Interestingly, when a gene is physically moved to a different genomic location (such as in transgenesis) it frequently becomes sensitive to ‘position effects’ resulting in aberrant expression. This might result from the influence of chromatin proximal to the site of integration and/or potentially from an altered position within the nucleus. Conversely, large BAC transgenes, transgenes containing locus control regions (LCR), or rearrangements caused by balanced translocations are frequently resistant to such position effects, even though they are introduced into both a novel genomic location and presumably an altered position within the nucleus 19, 20. This suggests that not all sequences are influenced by either their genomic or nuclear location. From a different perspective, artificially targeting loci to ectopic sites in the nucleus (such as the inner nuclear membrane) can also result in aberrant gene activation and/or silencing, providing evidence that gene positioning might actively regulate gene expression, rather than being a passive consequence of gene expression and/or silencing 21, 22. Thus, not only is the nucleus a highly structured organelle, the ordering of the genome within the nuclear space possibly represents an additional level of gene regulation.
Intrachromosomal associations: looping in cis
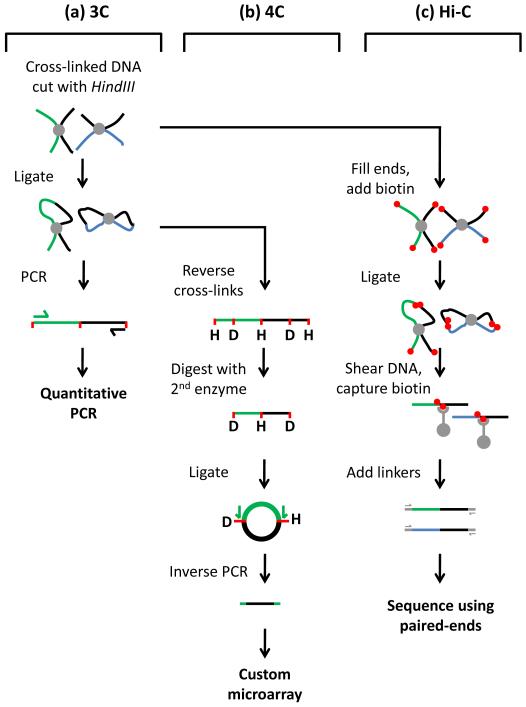
Although the nucleus is a well organized three-dimensional structure, transcriptional regulation in mammals was, until relatively recently, considered a linear process in which regulatory elements such as promoters and enhancers regulate proximal genes in cis. However, a strictly linear model is difficult to reconcile with the ability of enhancers to function when located over a megabase away from their target gene23. Development of technologies such as chromosome conformation capture (3C) has enabled detailed analysis of chromosome folding, revealing for the first time how promoters communicate with distal regulatory elements. A brief overview of 3C-based methodologies is provided in Figure 124-27. In mammalian cells, 3C was first used to investigate promoter-enhancer communication at the β-globin locus28. These studies revealed that, through DNA looping, hypersensitive sites within the LCR come into close physical proximity with the active globin genes situated 40-60kb away, forming a structure termed the ‘active chromatin hub’ (ACH). Additional distal hypersensitive sites were also found in close association with the LCR and active globin genes, whereas intervening sequences and olfactory receptor genes where looped out of this complex (Figure 2)28. Using a complimentary technique, termed ‘RNA TRAP’, a second group independently reported looping between the β-globin gene and its LCR29. It has been suggested that clustering of regulatory elements within the ACH increases the local concentration of transcription factors and maintains active chromatin domains facilitating high levels of transcription28, 30.
Figure 1.
3C based methodologies.
Development of chromosome conformation capture (3C) technologies has enabled detailed analysis of chromosome conformations for the first time. (a) 3C begins with the formaldehyde treatment of living cells, which cross-links DNA sequences in close proximity at the time of fixation. Cross-linked DNA is purified and subjected to restriction digest, in this case HindIII (H). Fragments are diluted and incubated with DNA ligase resulting in intramolecular-ligation of cross-linked fragments. Ligation of pre-selected genomic regions is quantified using locus specific PCR primers24, 28. 3C is thus limited to the analysis of pre-determined regions of interest. (b) 4C is a variation of 3C. Cross linked and ligated templates are generated as in 3C. Cross-links are then reversed and the template is digested with a second ‘frequent cutting’ restriction enzyme such as DpnII (D) to reduce fragment sizes before a second round of ligation. This generates small DNA circles that form the template for inverse PCR using primers designed within the bait region (green). The resulting PCR products are hybridized to custom arrays (or sequenced), allowing interrogation of genome wide associations for a single pre-selected bait sequence26. (c) Hi-C is the most recent adaptation of 3C and potentially allows analysis of all associations genome wide. In Hi-C cross-linked, digested 3C material is tagged with biotin and the ‘sticky ends’ generated by restriction digestion are filled in. The material is ligated and then sheared to generated small fragments, which are captured using streptavidin beads. Linkers are added and the material is subjected to paired end sequencing25.
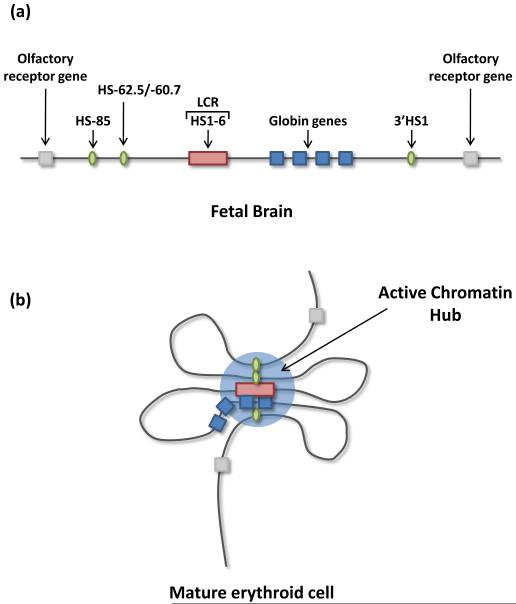
Figure 2.
Cis looping: the active chromatin hub (ACH). (a)In fetal brain, where the β-globin genes are not expressed, the locus adopts a linear conformation. (b) In mature erythroid cells, where the β-globin gene is expressed, the locus forms chromatin loops which bring the LCR and distal hypersensitive sites (upstream HS-85/HS-62.5/HS-60.7 and downstream HS1) into proximity with the expressing globin gene, to form an active chromatin hub. Intervening sequences and olfactory receptor genes are looped out of this complex28.
Similar findings have subsequently been noted for many other genes, in some instances with enhancers being reported to ‘loop’ into contact with regulated genes from over one megabase away31. Thus, intrachromosomal looping interactions appear to be a general phenomenon of long-range enhancer-promoter communication.
Interchromosomal associations
The ability of regulatory sequences to control transcription in cis through long-range looping intrachromosomal interactions combined with the observation that certain genes and/or sequences can adopt preferred locations within the nucleus raises the exciting possibility that regulatory elements, such as enhancers or LCRs, located on one chromosome could coordinately regulate genes on a different chromosome through interchromosomal associations. In potential support of this hypothesis, studies of promoter-enhancer communication have revealed that in living cells or nuclear extracts, transcription from plasmids containing promoter sequences can be activated in trans by enhancer sequences on separate plasmids32-34. Furthermore, in vitro experiments have shown that RNA polymerase II (Pol II) can be transferred from a plasmid containing the β-globin LCR to a second plasmid containing the β-globin gene, in a process that is facilitated by the erythroid transcription factor NFE235. Although highly artificial, these experiments reveal that there is no absolute prerequisite for enhancer and promoter sequences to be present on the same DNA strand. Subsequently, specific interchromosomal associations have been identified in the mammalian nucleus; the function of these associations is discussed below.
Alternative expression of cytokine genes
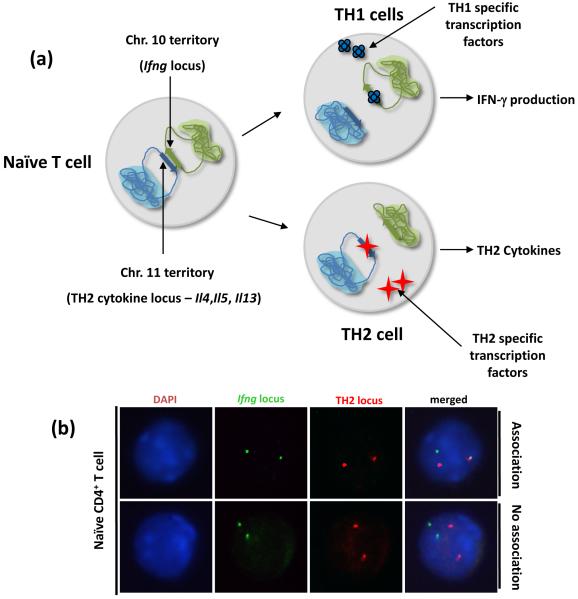
One of the first systems in which interchromosomal associations have been reported is the differentiation of naïve CD4+ T helper cells into TH1 and TH2 subsets. In mice, the TH2 cytokines interleukin 4 (Il4), interleukin 5 (Il5) and interleukin 13 (Il13) and their LCR are located in a single gene cluster on chromosome 11, whereas the TH1 cytokine interferon gamma (Ifng) is located on chromosome 10. In murine naïve CD4+ T cells, which do not express any of these genes, there is an interchromosomal association between the regulatory regions of the TH2 cytokine locus and the Ifng promoter region36. Polarization towards either the TH1 or TH2 fate results in initiation of appropriate cytokine gene expression, paralleled by a loss of interchromosomal associations. It was postulated that this intimate association is responsible for poising the two classes of cytokine genes for immediate expression (within 3-6 hours) upon T cell receptor (TCR) stimulation of naïve CD4+ cells36. In support of this hypothesis, mutations within the TH2 LCR not only affect TH2 cytokine expression but also influence expression of Ifng in stimulated naïve T cells and polarized effector TH1 cells (Figure 3)36. Since these studies, similar trans associations have been detected between the TH2 locus and the tumor necrosis factor alpha/lymphotoxin (Tnfα/Lt) locus on chromosome 17, and also between the TH2 locus and the interleukin 17 (Il17) locus on chromosome 1 (C.G.S, and Lark Kyun Kim, unpublished observations). [sc1]
Figure 3.
Interchromosomal associations of cytokine genes. (a) In murine naïve CD4+ T cells the TH2 locus (containing Il4, Il5, Il13 and Rad50) on chromosome 11 associates with the Ifng gene on chromosome 10. Differentiation into TH1 or TH2 effector cell results in loss of interchromosomal associations concomitant with induction of Ifng and TH2 cytokine expression respectively36. (b) Representative three-dimensional fluorescence in situ hybridization (3D-FISH) and confocal analysis of associations in naïve CD4+ T cells. Red color represents a TH2 BAC probe, and green represents an Ifng BAC probe. DNA staining with DAPI is shown in blue. Each spot represents one allele. The upper panel shows a representative nucleus displaying an interchromosomal between a single allele of Ifng and a single allele of the TH2 locus. The lower panel shows a representative example nucleus displaying no interchromosomal associations between the Ifng and TH2 loci.
Olfactory receptor choice
Although there are >1300 olfactory receptor genes dispersed over several mouse chromosomes, only a single olfactory receptor is ever expressed in any one neuron. It has been proposed that a single enhancer element on chromosome 14 (called the H enhancer) stochastically establishes an interchromosomal association with any one of the 1300 olfactory receptor genes, resulting in the exclusive expression of that receptor37. Subsequently, however, it was reported that deletion of the H enhancer only affected expression of a small number of proximal cis-linked olfactory receptor genes and did not affect those on other chromosomes38, 39. In addition, in heterozygous mice, expression of olfactory receptor genes proximal to the H− mutant allele was not rescued by the wild type H+ allele in trans39. Whereas the latter data convincingly show that the H enhancer is not required for olfactory receptor gene expression and choice in trans, it is possible that the additional compensatory enhancer elements exist in the genome. Further experimentation using genome-wide 3C based methods could be informative in determining whether such regulatory elements actually exist.
Imprinting
Genomic imprinting is a regulatory mechanism that establishes parent-of-origin-specific gene expression patterns 40. Imprinted genes are expressed from only one of the two alleles depending on the parental origin, leading to monoallelic expression40. One of the first indications that interchromosomal associations might be important in imprinting came from observations made by LaSalle et al in 1996. These authors noted that in human T lymphocytes, the imprinted 15q11-q13 region undergoes transient homologous association during late S phase41. Mutations within this region result in the genetic disorders Prader-Willi syndrome (PWS) and Angelman syndrome (AS)42. Interestingly cells from either PWS or AS patients did not show homologous associations, revealing that homolog pairing might play a role in establishing and/or maintaining imprinting41. Homologous associations were also shown to occur at the Beckwith-Wiedemann syndrome locus (BWS) of human chromosome 11p15, which contains which contains imprinted insulin-like growth factor 2 (IGF2) and H19 (a non-coding RNA) genes, suggesting that these associations might represent a general mechanism for regulating imprinting41. However, when, Teller et al revisited these findings they found no evidence for an increase of the fraction of nuclei with paired, oppositely imprinted AS/PWS or BWS loci at late S phase43. However, in agreement with the findings of LaSalle et al they did observe a significant homologous association between the centromeres of chromosome 11 (approximately 4 megabases away from the AS/PWS locus) during late S phase. Teller et al predicted that this is mediated by a nucleolar organizer region (NOR) (which are known to undergo paring events) linked to the centromere of chromosome 15 (and by default the AS/PWS region)43. Indeed analysis of lymphoblastoid cells from Gorilla gorilla, in which the AS/PWS region is not linked to a NOR they actually observed increased distances between AS/PWS loci during late S phase43. This led the authors to tentatively suggest that the associations reported by LaSalle et al result from a side effect of the conversion of NORs, and have nothing to do with an imprinting mechanism43.
Recently, using the 3C based ‘associated chromosome trap’ method, the lab of Andrew Hoffman identified an interchromosomal association between the murine Igf2/H19 imprinting control region (ICR) on the maternal chromosome 7 and an intergenic sequence between WD repeat and SOCS box-containing 1 (Wsbl) and neurofibromin 1 (Nf1) on the paternal chromosome 1144. Knockdown of the zinc-finger containing CCCTC-binding factor (CTCF) or deletion of the maternal ICR from chromosome 7 (but not the paternal ICR) not only abolished these associations but also reduced expression of Wsb1 and Nf1 from chromosome 11, suggesting that the ICR mediates interchromosomal gene regulation44. However, as CTCF is known to interact with, and recruit Pol II, these findings must be interpreted with care45. Indeed, CTCFs involvement in mediating associations might stem from its role in regulating transcription; associations could be lost simply as consequence of reduced transcription resulting from deletion of CTCF or loss of the maternal ICR CTCF sites.
Using 4C (Figure 1) the lab of Rolf Ohlsson has extended these studies and identified >100 chromosomal fragments associating with the maternal allele of the H19 ICR27. Moreover, within this panel of association partners imprinted regions were over-represented27. Mutation of CTCF sites within the maternal H19 ICR (but not the paternal allele) abolished these associations, resulting in dysregulated expression of normally associated genes and loss of asynchronous replication at numerous trans-associated imprinted loci27, 46. These observations led the authors to speculate that the H19 ICR is a hub for the “transvection of parent-of-origin-specific effects to non-allelic loci on other chromosomes”46. Taken together these data indicate the interchromosomal regulation is important in directing imprinting and that the H19 ICR could be the ‘master regulator’ of these events. Furthermore, although these studies highlight CTCF as a central player in this process its exact role is unclear as associated alleles do not show enrichment for the presence of CTCF sites 27, 46.
X chromosome inactivation
In mammals equivalent gene ‘dosage’ between XY males and XX female cells is achieved by chromosome wide transcriptional silencing of one of the two female X chromosomes47. This silencing event is exquisitely controlled by a small region of the X chromosome termed the X inactivation center (XIC). The XIC contains the non-coding RNA gene named X (inactive)-specific transcript (Xist), which is expressed specifically from the inactive X chromosome and triggers wide spread gene silencing47. Two other non-coding RNAs, XIST antisense RNA (Tsix) and X-inactivation intergenic transcription element (Xite), are expressed solely from the active X chromosome and are important for ‘choice’ (which chromosome to silence) and ‘counting’ (how many chromosomes to silence) events48. Recent evidence suggest that immediately before initiation of X inactivation the two X chromosomes undergo transient (<1 hour) homologous association mediated by the XIC49-51. Deletion of either Tsix or Xite from the XIC inhibits homologous association, concomitant with a failure in counting and choice, resulting in random X inactivation of 0, 1 or both X chromosomes48-51.
Interestingly, multi-copy transgene arrays comprised of short sequences containing Tsix or Xite are sufficient to initiate de novo ectopic paring between the autosomal site of integration and the endogenous X chromosome51. Furthermore, these new associations are formed at the expense of endogenous homologous associations resulting in a failure of X inactivation48, 51, 52. Knockdown of either CTCF or Oct4 (also known as POU class 5 homeobox 1) precludes homologous association; surprisingly, whereas depletion of CTCF results in a loss of X inactivation, depletion of Oct4 results in silencing of both X chromosomes52, 53. However, interpretation of these findings is complicated by the role of CTCF in mediating transcription and recruitment of Pol II45. This is especially relevant given that transcription itself is required for X chromosomes paring 52.
Together these findings provide considerable data to support the notion that although homologous interchromosomal association is not essential for X inactivation per se, it is likely to play a role in chromosome counting and choice. Furthermore, deletion and transgenic analysis imply that discrete sequences mediate these associations. The mechanism behind how the alleles locate each other and how homologous association regulates counting, choice and inactivation has yet to be resolved.
Estrogen responsive genes
Recent data suggest that interchromosomal associations can form rapidly in response to extracellular cues. For example, estrogen inducible genes are rapidly inducible on treatment with 17β-estradiol (E2). However, most estrogen receptor (ER-α) binding sites are intergenic and distal from E2 inducible genes, suggesting they form long-range looping associations54-56. To explore this possibility Hu et al, developed a novel variant of 3C technology termed ‘deconvolution of DNA interactions by DSL’ (3D)57. Using this method they identified a series of intrachromosomal associations between the E2-regulated trefoil factor 1 (TFF1) gene on human chromosome 21 and other ER-α bound loci on the same chromosome 57. In addition, they identified an interchromosomal association between TFF1 and an E2-regulated gene, named gene regulated by estrogen in breast cancer protein (GREB1), on chromosome 2. In untreated cells these associations were absent but formed rapidly (within 15 minutes) on treatment with E2. Remarkably, these associations were paralleled by relocalization of the entire nuclear territories of chromosome 21 and chromosome 2 which became intimately associated57. Formation of E2-inducible interchromosomal associations was dependent upon nuclear actin and nuclear myosin-I, suggesting coalescence might be mediated through active and directed large-scale nuclear reorganization events57. Although this represents an exciting possibility it must be noted that actin and myosin have previously been shown to be functional components of Pol II and chromatin remodeling complexes58, 59. Thus, if associations result from stochastic events driven by transcription and/or chromatin remodeling, then manipulating actin or myosin will not only affect transcription but will also indirectly influence these associations.
Finally, providing some evidence that these associations might have functional significance is the observation that expression of associated alleles was dramatically increased relative to non-associated alleles57. However, at this stage the evidence is correlative and does not prove that interchromosomal association between these two loci regulates their transcription.
Viral induction of IFN-β
The human antiviral response is initiated by transcriptional activation of type I interferons including interferon beta (IFN-β). Viral infection activates NF-kB (and other factors) resulting in enhanceosome assembly on the IFNB enhancer, inducing stochastic expression of IFNB from a single allele. Using 4C to study how this activation is rendered monoallelic, Apostolu et al identified three unique sequences on different chromosomes that rapidly (within 2 hours) associate with the expressing IFNB allele following viral infection60. These three sequences were each composed of Alu repeats that contained a functional NF-kB binding site. Transfected plasmids containing these sequences associated with the endogenous IFNB gene in an infection dependant manner, driving elevated levels of IFNB expression. Mutation of the NF-kB site suppressed this effect. The authors suggest a model in which viral infection induces NF-kB binding to these three sequences, which in turn, through intrachromosomal and interchromosomal association deliver NF-kB to a single IFNB allele initiating monoallelic expression60. Thus, the coordinated engagement of multiple transcription factor binding sites with a single allele, through intrachromosomal and interchromosomal associations, might represent a general mechanism in which gene expression is rendered monoallelic. However, such a mechanism is difficult to reconcile with the extremely short residence times of most transcription factors61.
Erythropoiesis
In mice and humans, multiple genes required for erythropoiesis have been reported to associate within RNA polymerase II transcription factories or at SC35[sc2]-enriched splicing speckles when expressed62-64. Of particular interest is the interchromosomal association between the human α-globin and β-globin genes. As the expression patterns of these two genes are similar during adult erythropoiesis and their gene products are required in equimolar amounts, it is tempting to speculate that their association is important in their coordinate regulation. However, a number of observations reveal that this is unlikely. For example, the murine globin genes do not show trans-association to the same degree as in humans62, 63. Furthermore, when the mouse α-globin locus is replaced with human the α-globin cluster this ‘humanized’ allele associates much less frequently with murine β-globin locus, but is still regulated appropriately62. Finally deletion of the human α-globin locus does not affect β-globin expression65. This implies that these associations likely represent sharing of common resources such as transcription factories or splicing speckles, rather than providing evidence of interchromosomal gene regulation. Recruitment of related genes to such specialized factories could help in coordinating gene expression and possibly increasing efficiency of transcription. Indeed, there is some precedent to suggest the existence of dedicated transcription factories which specialize in transcribing ‘similar’ genes66. However, work by Brown et al convincingly show that co-transcribed erythroid genes do not in fact cluster within transcription factories but rather associate around common SC35-enriched splicing speckles62, 63. Although clustering appeared to be largely stochastic it was influenced by constraints imposed by the chromatin neighborhood of the gene as well as its transcriptional status62, 63. Thus, in this case, interchromosomal association between co-transcribed genes appears to be a random byproduct resulting from active genes sharing common SC35 enriched splicing speckles.
Tackling the same questions, but using 4C, the de Laat group established that the active β-globin locus made intrachromosomal and interchromosomal contacts preferentially with transcribed regions of the genome26. And although their study confirmed the presence of previously described associations (between the β-globin locus and other erythropoietic genes), the bulk of associations involving the active β-globin locus were made with multiple active genes, but not necessarily tissue-specific genes, on the same chromosome, implying that specific clustering of similar genes does not occur26.
Unanswered questions and future perspectives
What is mediating associations?
It has been suggested that multiple genes share transcription factories and following activation, genes translocate to pre-existing factories64, 66, 67. Interestingly, RNA polymerases can generate ‘pulling’ forces substantially larger than cytoskeletal motor proteins such as kinesin and myosin, providing the possibility that polymerases might play a role in shaping the genome68. However, 4C experiments failed to reveal genome reorganization on inhibition of RNA polymerase, implying transcription is not essential to maintain (but not necessarily shape) global genome architecture69.
Currently the best candidate for mediating association is the multifunctional zinc-finger protein CTCF70. In a number of systems CTCF has been implicated in forming chromatin loops that bring distal regulatory elements into close proximity with promoter sequences71-74. CTCF is also important for interchromosomal associations seen in imprinting and X inactivation27, 44, 46, 52, 53. Importantly, CTCF can form dimers and maybe even oligomers, potentially providing a biophysical basis for intrachromosomal and interchromosomal associations75. As CTCF has 13000 to 36000 possible binding sites within the genome it is likely to play a major role in regulating global genome architecture76-79. However, this would imply that CTCF must collaborate with additional factors to provide specificity in intrachromosomal and interchromosomal associations. Interestingly, recent data show that CTCF is able to recruit cohesins to specific genomic locations80-88. Cohesins, better known for their role in mediating sister chromatid cohesion during mitosis, have established chromatin bridging potential and are thus an obvious candidate for orchestrating spatial organization of the genome. To date more than 15 proteins, including transcription factors, chromatin remodeling complexes and architectural proteins, have been shown to establish or maintain chromatin loops89. The concerted efforts of these factors (and likely unidentified members) could potentially maintain global genome architecture whilst providing sufficient flexibility to mediate specific intrachromosomal and interchromosomal associations. Interestingly, a number of loci involved in interchromosomal associations in mammals encode non-coding RNAs (for example the XIC in X inactivation). In such cases association might result simply from the process of transcribing these non-coding RNAs. Alternatively, it is tempting to speculate that non-coding RNAs themselves might serve a structural role or even function as trans-allelic messengers.
One final possibility that cannot be excluded is that nothing is directly mediating these associations. Instead associations might arise by stochastic meetings afforded by constraints imposed by chromatin context and/or transcriptional status62, 63, 90. Manipulation of any factor that could influence transcription or chromatin remodeling might indirectly disrupt these stochastic associations giving the impression that they are in fact directed, thereby complicating the analysis of interchromosomal associations. Indeed, as mentioned above CTCF is directly implicated in binding and recruitment of Pol II45. Thus, deletion of CTCF and/or its binding sites might indirectly influence associations by altering rates of transcription. Caution must therefore be used when interpreting such experiments.
How dynamic are associations?
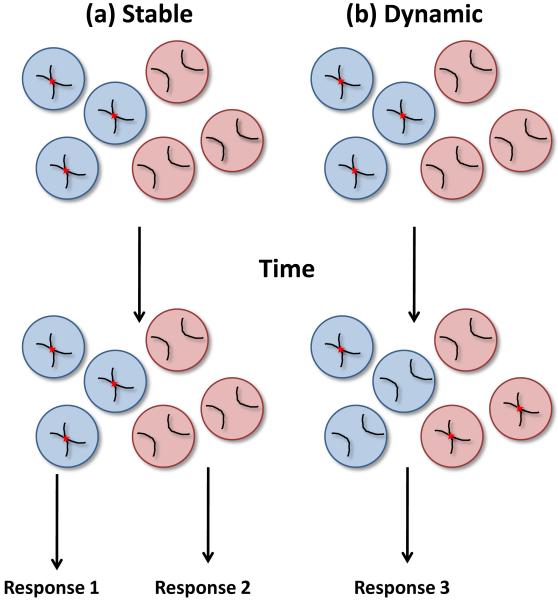
As 3C based methods require a very high number of PCR cycles to reveal interchromosomal associations it is difficult to quantitatively interpret such data. Furthermore, due to the nature of the technique, it is theoretically possible to generate a 3C product from any two regions of the genome by chance. This is most apparent in 4C experiments where specific interchromosomal associations appear to be embedded in a sea of thousands of low-level non-specific (presumably) background interchromosomal associations26. Thus, 3C based experiments must be extremely well controlled and interpreted with care. Importantly, these methods are routinely supported by microscopy-based techniques, such as fluorescent in situ hybridization (FISH), which although limited in resolution (approximately 250-500nm), allow quantification of association frequency at the single cell level. Interestingly, the most striking observation from microscopic analysis is that associations are detected in only limited proportion of cells at any one time; frequencies of association in the range 5-15% are generally reported, but frequencies as high as 30-60% have also been noted26, 27, 36, 37, 41, 43, 44, 46, 50, 51, 57, 60, 62-64, 67. Can associations present in a minority of cells within a population be physiologically relevant? Unfortunately, as current methodologies rely on the fixation of cells prior to analysis, creating a ‘snap-shot’ of a single moment in time, we know little regarding the dynamics of association. What do associations caught ‘in flagrante’ by 3C and FISH actually represent? Do they correspond to stable associations present in only a few cells or are they dynamic, transient events that occur stochastically in all cells? If assuming that associations are functional then these two extreme possibilities obviously have very different biological implications (Figure 4). For example, if an association is stable, but present in only a proportion of cells, these cells might generate a different response to cells in which the association is absent. In contrast, if an association is dynamic then most probably each cell would generate a similar response. Thus, it will be particularly informative to track associations by microscopy using transgenic arrays of Lac operator repeats tagged in vivo with GFP-Lac repressor fusion proteins91-94. Such methodologies have already been utilized to study transvection in Drosophila, revealing associations to be stable for hours rather than minutes95. The presence of such stable associations is more in line with our current understanding of chromosome dynamics; as mentioned previously, loci are constrained within a small (<1μm) diffusion corral 13. However, the possibility that associations can result from specific dynamic and large-scale chromatin movements has yet to be excluded.
Figure 4.
Dynamics of chromosomal association. (a) If associations are stable but present in only a proportion of cells, these cells might generate different responses (response 1 or 2). (b) If associations are dynamic all cells within a population have the potential to form associations. Most probably each cell would generate a similar response (response 3). However, it is also possible that cells with or without associations at the moment of stimulation might respond differently as in (a).
How general are functional interchromosomal associations?
Do co-transcribed genes generally coalesce? Although anecdotal evidence suggests that this could be the case, emerging 4C data suggest that this might be a rare event. Most likely, chromatin folding will be directed by self-organizing principles26. Thus, positioning of a locus might be probabilistic, determined by the sum of properties of neighboring sequences and the chromosome as a whole 96, 97. It is important to note that some sequences (such as nucleolar organizer regions) might be dominant in shaping the genome, and transgenes can sometimes override endogenous positioning of the site of integration49, 51.
Following early examples of regulatory interchromosomal associations there has only been a few identified cases. Does this imply that such associations are rare, or are we just looking in the wrong places? It is difficult to draw decisive conclusions as very few loci have been studied in detail. These are questions that will be addressed by the next generation of open-ended high throughput 3C-based methods (Figure 1). Indeed, a recent publication reported whole genome-wide associations at a 1 megabase resolution25. As the resolution increases and the cost decreases these immensely powerful methods will allow interrogation of genome-wide associations in different cell types and under different conditions. This might reveal a plethora of new associations and potentially provide an estimate of the global frequency of such events.
Do associations represent functional interactions?
Perhaps the most pressing question is whether associations are functionally significant? Although appealing, it is erroneous to suppose that coalescing of similar genes implies co-regulation and that association between genes and regulatory sequences on other chromosomes gives evidence for trans-regulation. It is more probable that the bulk of associations simply reflect stochastic encounters due to the constraints of sharing limited space and resources within the nucleus (Figure 5). Although unambiguously proving associations are functionally relevant is technically difficult, a number of criteria should be addressed, such as presented by Brown et al62. (1) Are associations conserved across species? If not, it is less likely that they represent functional interactions. (2) Does the mutation of alleles affect association and/or gene expression in trans? Potential effects from loss of gene products (protein or non-coding RNA) from modified alleles must be taken into account. Furthermore, mutation of candidate binding sites is preferable to ablation of protein mediators, which is often accompanied by pleiotropic effects. (3) Do transgenes form the same associations? If not are they regulated appropriately? Does their presence affect endogenous associations and/or gene regulation? (4) Are there other sequences (such as NORs) on the same chromosome, proximal to the region of interest, which might instead be responsible for driving association?
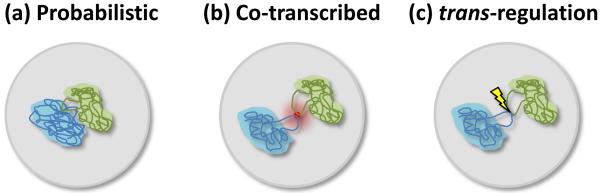
Figure 5.
Three types of Interchromosomal association. (a) Positioning of loci is probabilistic, determined by the sum of properties of neighboring sequences and the chromosome as a whole. Associations have no obvious functional significance. (b) Co-transcribed genes coalesce in and around nuclear bodies such as transcription factories and splicing speckles. As such bodies might be specialized in transcribing similar genes, these associations could help in coordinating gene expression and increase efficiency of transcription. Alternatively, these associations may be probabilistic as in (a) and have no functional significance. (c) Sequences regulate gene expression in trans through interchromosomal contacts.
Concluding remarks
Identification of interchromosomal associations in mammalian cells has initiated a new and exciting field in mammalian biology. Currently, the physiological significance of these associations has only been studied in a handful of cases, and although it is now well accepted that genomic regions can associate in trans, it is still controversial whether these associations represent the basis of a mammalian equivalent of Drosophila transvection. Fueled by recent technological developments we are beginning to build a detailed three-dimensional picture of genome organization. We must now focus on how such ordering is achieved and what the implications are for regulating gene expression. Importantly, a more thorough interrogation of how chromatin context and transcriptional status effects interchromosomal association is urgently required. It is likely that the nucleus is predominantly governed by self-organizing principles; however, within a sea of probabilistic intrachromosomal and interchromosomal associations, identification of specific and functional interactions should continue to reveal exciting aspects of nuclear biology and gene regulation. An understanding of the mechanisms governing these functional interactions might establish a more complete paradigm of gene regulation in mammals.
Acknowledgements
We would like to thank Stephanie C. Eisenbarth for critical reading of the manuscript and Fran Manzo for assistance with manuscript preparation. C.G.S. was, and A.W. is, a Howard Hughes Medical Institute Associate. R.A.F. is an Investigator of the Howard Hughes Medical Institute.
Glossary
- Active chromatin hub (ACH)
The clustering of active genes and cis-regulatory elements into a complex. It is thought that formation of an ACH increases the local concentration of transcription factors allowing high transcription rates.
- Chromosome territory
The discrete space or volume occupied by a single chromosome in the interphase nucleus. Chromosomal territories are essentially non-overlapping but their borders are not well defined and intermingling between chromosomes occurs at these junctions.
- Enhanceosome
A multi-protein complex that binds to the enhancer region of a gene and stimulates transcription.
- Homologous association
A meeting between identical sequences on homologous chromosomes.
- Interchromosomal association
A meeting between sequences and/or genes on different chromosomes. Homologous associations also fall into this class. These can also be described as trans-associations.
- Intrachromosomal association
A meeting between sequences and/or genes on the same chromosome. These can also be described as cis-associations.
- Locus control region
A class of powerful cis-regulatory elements with functional properties overlapping with classical enhancers, insulators and boundary elements. They are defined by their ability to confer copy-number dependent, position-independent expression in transgenesis.
- Nucleolar organizer region (NOR)
A chromosomal region containing several tandem copies of ribosomal RNA genes around which the nucleolus forms. In humans, the NOR contains genes for 5.8S, 18S, and 28S rRNA clustered on the short arms of chromosomes 13, 14, 15, 21 and 22.
- Transcription factories
Nucleoplasmic complexes containing multiple molecules of RNA polymerase II. Transcription factories have been shown to assemble at new sites of transcriptional activity. Alternatively, it has been suggested that on activation, genes translocate to a limited number of pre-formed factories, which they are obliged to share with other active genes.
- Transvection
A phenomenon where regulatory elements on one chromosome are able to regulate gene expression on another chromosome in trans (see Box 1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no financial conflict of interest.
References
- 1.Cremer T, et al. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum Genet. 1982;60:46–56. doi: 10.1007/BF00281263. [DOI] [PubMed] [Google Scholar]
- 2.Mayer R, et al. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005;6:44. doi: 10.1186/1471-2121-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanabe H, et al. Inter- and intra-specific gene-density-correlated radial chromosome territory arrangements are conserved in Old World monkeys. Cytogenet Genome Res. 2005;108:255–261. doi: 10.1159/000080824. [DOI] [PubMed] [Google Scholar]
- 4.Mora L, et al. Chromosome territory positioning of conserved homologous chromosomes in different primate species. Chromosoma. 2006;115:367–375. doi: 10.1007/s00412-006-0064-6. [DOI] [PubMed] [Google Scholar]
- 5.Tanabe H, et al. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci U S A. 2002;99:4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle S, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 7.Neusser M, et al. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma. 2007;116:307–320. doi: 10.1007/s00412-007-0099-3. [DOI] [PubMed] [Google Scholar]
- 8.Tanabe H, et al. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat Res. 2002;504:37–45. doi: 10.1016/s0027-5107(02)00077-5. [DOI] [PubMed] [Google Scholar]
- 9.Bolzer A, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cremer M, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- 11.Croft JA, et al. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon N. Heterochromatin structure and function. Biol Cell. 2004;96:631–637. doi: 10.1016/j.biolcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Chuang CH, et al. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 14.Dundr M, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnowski A, et al. Silencing and nuclear repositioning of the lambda5 gene locus at the pre-B cell stage requires Aiolos and OBF-1. PLoS One. 2008;3:e3568. doi: 10.1371/journal.pone.0003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams RR, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 17.Volpi EV, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 18.Brown KE, et al. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 19.Kioussis D, Festenstein R. Locus control regions: overcoming heterochromatin-induced gene inactivation in mammals. Curr Opin Genet Dev. 1997;7:614–619. doi: 10.1016/s0959-437x(97)80008-1. [DOI] [PubMed] [Google Scholar]
- 20.Kleinjan DJ, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 21.Finlan LE, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy KL, et al. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 23.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 24.Dekker J, et al. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 28.Tolhuis B, et al. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 29.Carter D, et al. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 30.Palstra RJ, et al. Beta-globin regulation and long-range interactions. Adv Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 31.Amano T, et al. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Dunaway M, Droge P. Transactivation of the Xenopus rRNA gene promoter by its enhancer. Nature. 1989;341:657–659. doi: 10.1038/341657a0. [DOI] [PubMed] [Google Scholar]
- 33.Mahmoudi T, et al. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller-Storm HP, et al. An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell. 1989;58:767–777. doi: 10.1016/0092-8674(89)90110-4. [DOI] [PubMed] [Google Scholar]
- 35.Vieira KF, et al. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J Biol Chem. 2004;279:50350–50357. doi: 10.1074/jbc.M408883200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spilianakis CG, et al. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 37.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 38.Fuss SH, et al. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Nishizumi H, et al. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proc Natl Acad Sci U S A. 2007;104:20067–20072. doi: 10.1073/pnas.0706544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.da Rocha ST, Ferguson-Smith AC. Genomic imprinting. Curr Biol. 2004;14:R646–649. doi: 10.1016/j.cub.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 41.LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- 42.Gurrieri F, Accadia M. Genetic imprinting: the paradigm of Prader-Willi and Angelman syndromes. Endocr Dev. 2009;14:20–28. doi: 10.1159/000207473. [DOI] [PubMed] [Google Scholar]
- 43.Teller K, et al. Maintenance of imprinting and nuclear architecture in cycling cells. Proc Natl Acad Sci U S A. 2007;104:14970–14975. doi: 10.1073/pnas.0704285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 45.Chernukhin I, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandhu KS, et al. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–2603. doi: 10.1101/gad.552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wutz A. Xist function: bridging chromatin and stem cells. Trends Genet. 2007;23:457–464. doi: 10.1016/j.tig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Lee JT. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 49.Augui S, et al. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 50.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 51.Xu N, et al. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 52.Xu N, et al. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 53.Donohoe ME, et al. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 55.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 56.Lin CY, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carmo-Fonseca M. How genes find their way inside the cell nucleus. J Cell Biol. 2007;179:1093–1094. doi: 10.1083/jcb.200711098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Lanerolle P, et al. Actin and myosin I in the nucleus: what next? Nat Struct Mol Biol. 2005;12:742–746. doi: 10.1038/nsmb983. [DOI] [PubMed] [Google Scholar]
- 60.Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 61.Mueller F, et al. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J. 2008;94:3323–3339. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown JM, et al. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 65.Weatherall DJ, Clegg JB. The thalassaemia syndromes. Blackwell Science; 2001. [Google Scholar]
- 66.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Osborne CS, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin H, et al. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 69.Palstra RJ, et al. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burke LJ, et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurukuti S, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Majumder P, et al. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yusufzai TM, et al. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 76.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 77.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 78.Jothi R, et al. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bowers SR, et al. A conserved insulator that recruits CTCF and cohesin exists between the closely related but divergently regulated interleukin-3 and granulocyte-macrophage colony-stimulating factor genes. Mol Cell Biol. 2009;29:1682–1693. doi: 10.1128/MCB.01411-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Degner SC, et al. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nativio R, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 85.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sekimata M, et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 89.Sexton T, et al. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 90.Lawrence JB, Clemson CM. Gene associations: true romance or chance meeting in a nuclear neighborhood? J Cell Biol. 2008;182:1035–1038. doi: 10.1083/jcb.200808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Belmont AS, Straight AF. In vivo visualization of chromosomes using lac operator-repressor binding. Trends Cell Biol. 1998;8:121–124. doi: 10.1016/s0962-8924(97)01211-7. [DOI] [PubMed] [Google Scholar]
- 92.Chubb JR, et al. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 93.Levi V, et al. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tumbar T, et al. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vazquez J, et al. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Laat W, Grosveld F. Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr Opin Genet Dev. 2007;17:456–464. doi: 10.1016/j.gde.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 97.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 99.Fedorova E, et al. The nuclear organization of Polycomb/Trithorax group response elements in larval tissues of Drosophila melanogaster. Chromosome Res. 2008;16:649–673. doi: 10.1007/s10577-008-1218-6. [DOI] [PubMed] [Google Scholar]