Abstract
Myristoylation is the attachment of the 14-carbon fatty acid myristate to the N-terminal glycine residue of proteins. Typically a cotranslational modification, myristoylation of proapoptotic cysteinyl-aspartyl proteases (caspase)-cleaved Bid and PAK2 was also shown to occur post-translationally and is essential for their proper localization and proapoptotic function. Progress in the identification and characterization of myristoylated proteins has been impeded by the long exposure times required to monitor incorporation of radioactive myristate into proteins (typically 1–3 months). Consequently, we developed a nonradioactive detection methodology in which a bio-orthogonal azidomyristate analog is specifically incorporated co- or post-translationally into proteins at N-terminal glycines, chemoselectively ligated to tagged triarylphosphines and detected by Western blotting with short exposure times (seconds to minutes). This represents over a millionfold signal amplification in comparison to using radioactive labeling methods. Using rational prediction analysis to recognize putative internal myristoylation sites in caspase-cleaved proteins combined with our nonradioactive chemical detection method, we identify 5 new post-translationally myristoylatable proteins (PKCε, CD-IC2, Bap31, MST3, and the catalytic sub-unit of glutamate cysteine ligase). We also demonstrate that 15 proteins undergo post-translational myristoylation in apoptotic Jurkat T cells. This suggests that post-translational myristoylation of caspase-cleaved proteins represents a novel mechanism widely used to regulate cell death.—Martin, D. D. O., Vilas, G. L., Prescher, J. A., Rajaiah, G., Falck, J. R., Bertozzi, C. R., Berthiaume, L. G. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 22, 000–000 (2008)
Keywords: fatty acylation, myristoylation, Staudinger ligation, cell death, triaryphosphine
Protein Fatty acylation is the covalent addition of fatty acids to proteins. Although known mainly as an efficient means to direct proteins to various membranes, protein fatty acylation is also important in regulating protein function, protein-protein interactions, protein stability, and cell metabolism (1–4). Typically, the saturated fatty acids palmitate (16 carbon) or myristate (14 carbon) are covalently added onto proteins via processes known as S-acylation and N-myristoylation, respectively (5, 6).
In N-myristoylation, myristate is covalently linked in an irreversible fashion to an exposed N-terminal Gly residue by either of two functionally distinct N-myristoyl transferases (NMTs) (5). The myristoyl moiety is essential for promoting weak reversible protein-membrane interactions and often is found adjacent or distant to palmitoylated cysteine residue(s) or a polybasic domain (2, 6, 7). The general consensus sequence recognized by NMT is Gly2-X3-X4-X5-(Ser/Thr/Cys)6, where X represents most amino acids, with the exception of Pro, aromatic, or charged residues at X2. An exposed N-terminal Gly is absolutely essential for myristoylation to occur, and substitution of this residue to any other amino acid abrogates myristoylation (2, 5). In cotranslational myristoylation, the N-terminal glycine residue is exposed on removal of the initiator methionine by methionine aminopeptidase (6, 8). However, during apoptosis, 4 proteins (Bid, p21-activated protein kinase [PAK2], actin, and gelsolin) were shown to undergo post-translational myristoylation on cysteinyl-aspartyl proteases (caspase) cleavage and the resulting exposure of an internal Gly residue within a myristoylation consensus sequence (9–11).
Apoptosis, or programmed cell death, is critical for the maintenance of organisms and tissue homeostasis. Dysregulation of this process has detrimental consequences that can result in developmental abnormalities, neurodegeneration, or cancer (12, 13). A variety of processes can activate the two main cellular apoptotic pathways, which ultimately rely on the regulated and specific cleavage of hundreds of proteins by caspases (12, 13).
Post-translational myristoylation of caspase-cleaved proteins is rapidly emerging as a novel means to regulate programmed cell death (9–11). The programmed cleavage of hundreds of proteins during apoptosis warrants the search for additional post-translationally myristoylated proteins and the characterization of their involvement in the regulated demise of the cell. However, the identification and characterization of myristoylated proteins is a tedious process that relies on the metabolic labeling of cells with radioactive myristate (3H- or 14C-), which requires lengthy fluorographic or autoradiographic exposure time to detect radioactive myristate incorporation into proteins, typically weeks to months. Although [125I]iodo-fatty acid analogs have been developed to reduce exposure time needed to detect myristoylated or palmitoylated proteins (14), this technique requires the handling of large quantities (mCi) of the hazardous high energy 125I radioisotope. Therefore, the need for a nonradioactive methodology able to reliably detect myristoylated proteins is obvious. Recently, a new method involving the specific chemoselective ligation between an alkyl-azide and a triarylphosphine has been effectively adapted to study glycosylation and farnesylation using a variety of bio-orthogonal azido analogs (15, 16). We have adapted the principle of this methodology to efficiently detect myristoylated proteins using a bio-orthogonal azidomyristate analog and a variety of tagged triarylphosphines. We demonstrate that our new detection method is greater than 106× faster than fluorography when using [3H]myristate as a label.
Herein, we used our new detection method in conjunction with rational database analysis to identify 5 new candidates for post-translational myristoylation: mammalian STE20-like protein kinase 3 (MST3), B-cell receptor-associated protein 31 (BCR-associated protein Bap31), cytoplasmic dynein intermediate chain 2 (CD-IC2), glutamate-cysteine ligase catalytic subunit (GCLC), and novel protein kinase Cε (PKCε). Furthermore, we demonstrate the existence of up to 15 post-translationally myristoylated proteins in apoptotic Jurkat cell lysates. Although the involvement of these post-translationally myristoylated proteins in the apoptotic process is unknown, these findings strongly suggest that post-translational myristoylation of caspase-cleaved proteins plays a broad role in the regulation of apoptosis. Because our method tags azidomyristoylated proteins with biotin or FLAG epitopes, it will eventually be amenable to large-scale proteomic analyses of myristoylated proteins in cells. Overall, our new method will greatly simplify and accelerate the study of co- and post-translationally myristoylated proteins in various cellular processes.
MATERIALS AND METHODS
Materials
COS-7 and Jurkat cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cell culture media was purchased from Invitrogen (Carlsbad, CA, USA). [3H]myristate was acquired from Amersham (Arlington Heights, IL, USA). All GFP antibodies were provided by Eusera (Edmonton, AB, Canada) and/or Abcam (Cambridge, UK). Neutravidin-horseradish peroxidase (HRP) and anti-FLAG antibodies were obtained from Pierce Biotechnology (Rockford, IL, USA) and Sigma-Aldrich Corp. (St. Louis, MO, USA), respectively. Unless indicated otherwise, all chemicals were of the highest purity available (Sigma-Aldrich).
Nonradioactive metabolic labeling of cells with 12-azidododecanoic acid to detect exogenously expressed myristoylated chimeric enhanced green fluorescent proteins using a biotinylated phosphine
The ctPAK2-N15-EGFP and Yes-N11-EGFP constructs were from previous work (9). COS-7 cells were transiently transfected with the pEGFP-N1 (Invitrogen) vectors expressing the various chimeric enhanced green fluorescent proteins (EGFPs) using Fugene 6 (Roche, Indianapolis, IN, USA) in Dulbecco modified Eagle medium in the absence of FBS or antibiotics but supplemented with 1% fatty acid-free BSA (Sigma-Aldrich). At 10 h post-transfection, 40 µM 12-azidodo-decanoate (azidomyristate) in dimethyl sulfoxide (DMSO) or vehicle alone were added to the media and incubated for 12–14 h. The cells were washed with cold PBS, harvested, and lysed with 0.8 ml of 1% sodium dodecyl sulfate (SDS) -radio-immuno-precipitation assay (RIPA) buffer (50 mM Tris-hydrogen chloride [Tris-HCl] pH 8.0, 150 mM sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 2 mM magnesium chloride, 2 mM EDTA, and complete protease inhibitor [Roche]), followed by 2 rounds of sonication at an output of ~5.5. The supernatants of a 16,000 g (10 min at 4°C) centrifugation were collected and precipitated with 3 volumes of ice-cold acetone overnight at −20°C. The precipitates were washed once with 1 volume of ice-cold acetone, dried, and resuspended in phosphine reaction buffer (0.1 M sodium phosphate pH 7.2, 1% SDS). Typically, 200–300 µg of protein was conjugated with 250 µM phosphine-biotin or phosphine-FLAG (from a 5 mM stock in 30% dimethylform-amide in reaction buffer kept under argon gas) for 10 h at room temperature. Following acetone precipitation, the dried pellet was solubilized in 1% SDS-RIPA, diluted to 0.1% SDS-RIPA, and precleared with protein-G-sepharose (Amersham) by rocking for 45 min at 4°C. Chimeric EGFPs were immunoprecipitated with 0.5 µg affinity purified goat anti-green fluorescent protein (anti-GFP) per 100 µg of lysate for 16 h at 4°C, followed by addition of protein-G-sepharose for 2 h at 4°C. Following extensive washing with 0.1% SDS-RIPA, the immune complexes were boiled for 5 min in SDS-sample buffer, separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to polyvinylidene difluo-ride (PVDF) membranes. Where indicated, the membranes were treated with either 0.2 M Tris-HCl pH 7.0 or 0.2 M potassium hydroxide (KOH) as described before (9), and biotinylated azidomyristoylated proteins were detected using neutravidin-HRP or the appropriate anti-FLAG antibodies followed by electrochemiluminescence (ECL) detection (Amersham). EGFP was detected on membranes using rabbit anti-GFP serum.
Metabolic labeling of Jurkat cells with [3H]myristate or azidomyristate to detect endogenous post-translationally myristoylated proteins
A modified version for the metabolic labeling of Jurkat cells with [3H]myristate was used for azidomyristate (9). Approximately 1.0 × 107 Jurkat cells were incubated for 1 h at 37°C in the presence or absence of 1 mM 2-hydroxymyristic acid (HMA), an NMT inhibitor that does not inhibit protein palmitoylation (17–19), in RPMI without FBS or antibiotics but supplemented with 1.0% fatty acid-free BSA. Then, 40 µM azidomyristate or DMSO vehicle was added to the cells and incubated for 3 h prior to induction of apoptosis by the addition of 2.5 µM staurosporine (STS) and 5 µg/mL cyclo-heximide (CHX) (ICN Biochemicals, Costa Mesa, CA, USA) (9). After 4 h, cells were subjected to hypotonic lysis, and the postnuclear supernatant was separated into cytosolic and membrane fractions (9). Typically, 100–200 µg of protein was precipitated with acetone, dissolved in reaction buffer, conjugated with 250 µM phosphine-biotin, and analyzed by Western blotting as described above.
Endogenous caspase-cleaved [3H]myr- or azidomyr-ctPAK2 was immunoprecipitated from whole Jurkat cell lysates as described previously (9) (Supplemental Methods). Immune complexes were recovered with protein-G-sepharose as above, eluted with 100 µl phosphine reaction buffer at 80°C for 20 min, conjugated to 250 µM phosphine-biotin, and the reaction products were analyzed by Western blotting as described above. Where indicated, the membranes were treated with either 0.2 M Tris-HCl pH 7.0 or 0.2 M KOH as described above.
Organic syntheses: synthesis of 12-azidododecanoic and 14-azidotetradecanoic acids
The syntheses of the azidomyristate and azidopalmitate analogs were accomplished using a four-step protocol adapted from previously published procedures as described in detail in Supplemental Methods.
Syntheses of phosphine-biotin and phosphine-FLAG
The synthesis of biotinylated- or FLAG-tagged triarylphosphines was performed essentially as described earlier (15, 20, 21).
Identification of candidates for putative post-translational myristoylation, site-directed mutagenesis, and cloning
Putative internal myristoylation sequences adjacent to known caspase cleavage sites were identified from a published list of caspase substrates (22). Myristoylation candidate sequences were recognized using two prediction algorithms provided on http://www.expasy.org/tools/myristoylator/ (23) and http://mendel.imp.ac.at/myristate/SUPLpredictor.htm (24). The DNA encoding the first 10 amino acid sequences of 9 proteins predicted to be myristoylatable candidates were subcloned into pEGFP-N1 as described in Supplemental Methods.
RESULTS
Phosphine-biotin/FLAG-mediated detection of azidomyristoylated exogenous and endogenous proteins via Western blot analysis
To reduce exposure time required to detect and characterize myristoylated proteins using radioactive fatty acids, we sought to develop an alternative nonradioactive chemical detection protocol in which a bio-orthogonal azido analog of myristate could be incorporated into proteins upon metabolic labeling and detected by chemoselective ligation to a tagged triarylphosphine (Fig. 1). We found that 12-azidododecanoate was the optimal isosteric analog for amide-linked incorporation into proteins by NMT (data not shown). To establish our proof of concept, we sought to demonstrate the incorporation of 12-azidododecanoate (azidomyristate) into a variety of known myristoylated proteins. In the first series of experiments (Fig. 2A), the 34 kDa C-terminal portion of the known post-translationally myristoylated protein caspase-cleaved PAK2 (ctPAK2) and its nonmyristoylatable G2A (substitution of Gly in position 2 to Ala) counterpart were fused to the N terminus of EGFP, yielding an ~59 kDa fusion protein (WT-FL-ctPAK2-EGFP and G2A-FL-ctPAK2-EGFP, respectively). In the second and third series (Fig. 2B ), the first 11 or 15 amino acids of the known myristoylated proteins c-Yes non-receptor tyrosine kinase and ctPAK2, respectively, and their nonmyristoylatable G2A versions were appended at the N terminus of EGFP after an initiator methionine (N11 or N15 constructs), respectively (2, 9).
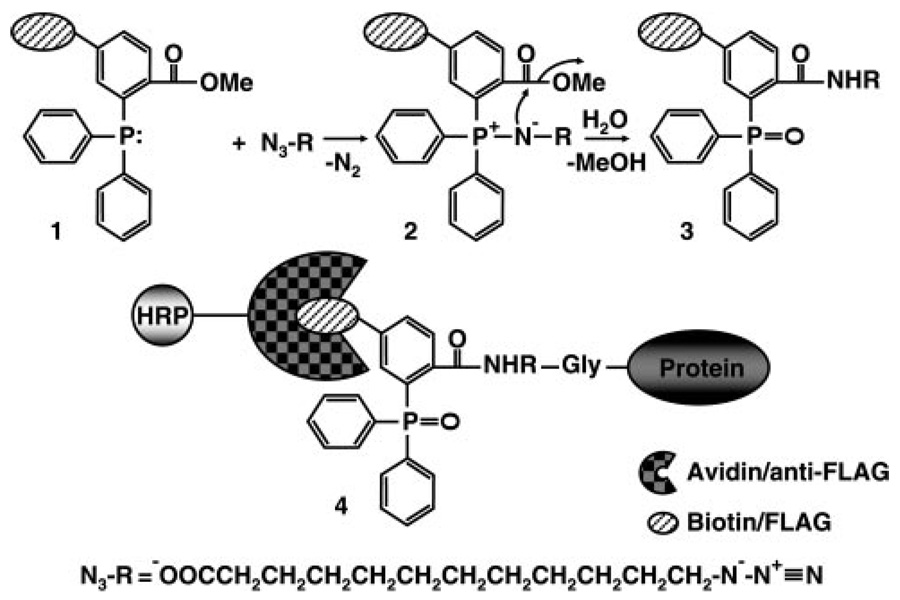
Figure 1.
Schematic of the Staudinger ligation. The triarylphosphine (1) specifically reacts with the azide moiety of azidomyristate to generate an aza-ylide intermediate (2), which cyclizes and is subsequently hydrolyzed to form the final product (3) that contains the triarylphosphine covalenty linked to the azidomyristate. In cells, the azidomyristate is added to proteins at an N-terminally exposed Gly by NMT. On solubilization of cells, the azidomyristoylated proteins are ligated to the tagged (biotin or FLAG epitope) triarylphosphine (4) via the Staudinger ligation.
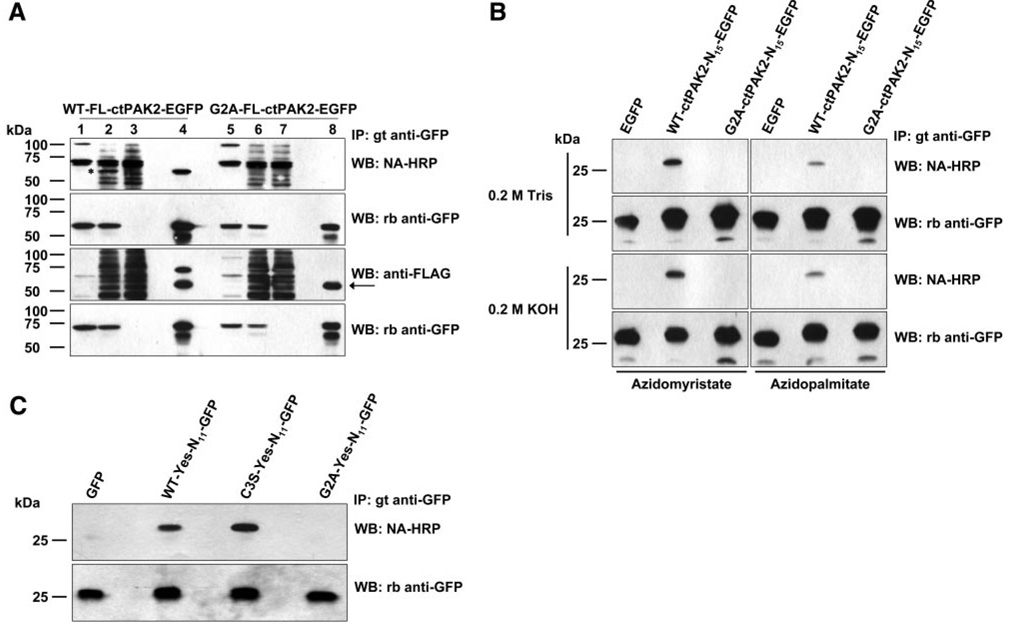
Figure 2.
Incorporation of azidomyristate into proteins exogenously expressed in COS-7 cells as detected by chemoselective ligation with phosphine-biotin and phosphine-FLAG. A) The lysates of COS-7 cells expressing wild-type full-length ctPAK2 with a C-terminal EGFP tag (WT-FL-ctPAK2-EGFP) or the non-myristoylatable G2A-FL-ctPAK2-EGFP labeled with azidomyristate were left untreated (lanes 1, 5) or conjugated with phosphine-biotin or phosphine-FLAG (lanes 2–4, 6–8). The biotin and FLAG covalently attached to the azidomyristate analog were detected in the lysates (lanes 2, 3, 6, 7) and immunoprecipitated proteins (lanes 4, 8) by Western blotting using neutravidin-HRP (NA-HRP) or anti-FLAG antibodies and ECL. Lanes 3 and 7 represent the postimmunoprecipitation supernatants. Anti-GFP immunoblots demonstrate the presence of WT-FL-ctPAK2-EGFPs and G2A-FL-ctPAK2-EGFPs in similar amounts. The asterisk marks the detection of the azidomyristoylated WT-FL-ctPAK2-EGFP in the crude cell lysate (lane 2) by NA-HRP but not in the corresponding postimmunoprecipitated supernatant (lane 3); the arrow indicates a nonspecific band. B) COS-7 cells transiently expressing truncated (first 15 amino acids only) WT-ctPAK2-N15-EGFP, G2A-ctPAK2-N15-EGFP (9), or EGFP were labeled with azidomyristate or azidopalmitate and conjugated to phosphine-biotin. Where indicated, the PVDF membranes were incubated in 0.2 M Tris pH 7.0 or 0.2 M KOH solutions. C) Azidomyristate labeling of COS-7 cells expressing truncated (first 11 amino acids) WT-Yes-N11-EGFP, C3S-Yes-N11-EGFP, G2A-Yes-N11-EGFP (2), or EGFP were handled as in A).
In Fig. 2, we show that metabolic labeling of COS-7 cells transiently transfected with the above-described EGFP constructs with the azidomyristate analog lead to the incorporation of the analog as judged by its subsequent detection with the phosphine-biotin and phosphine-FLAG as assessed by Western blotting. Only the WT-FL-ctPAK2-EGFP and WT-ctPAK2-N15-EGFP (Fig. 2) chimeras incorporated azidomyristate, while the G2A-FL-ctPAK2-EGFP and G2A-ctPAK2-N15-EGFP mutants did not. Of note, azidomyristoylated proteins could typically be detected in under-a-minute exposure to film, often only seconds. The fact that an ~59 kDa band (indicated by an asterisk) can be readily detected in lysates of cells transfected with vector expressing WT-FL-ctPAK2-EGFP (Fig. 2A ), but not in the lysates of cells expressing G2A-FL-ctPAK2-EGFP, further illustrates the sensitivity of the new method. In Fig. 2C , we show that our method allows the detection of azidomyristate incorporation in the truncated “N11” constructs WT-Yes- and C3S-Yes-N11-GFP in a matter of seconds, while their detection in McCabe et al. (2) required a 1-month exposure using [3H]myristate as a label. Anti-GFP blots attest to the similar presence of the EGFP chimeras in the various lanes. Interestingly, when transfected cells were labeled metabolically with azidopalmitate, acylation of WT-ctPAK2-N15-EGFP also was detected, albeit to a much lower extent (Fig. 2B ). Metabolic interconversion of palmitate into myristate (especially using a 10 h labeling period) and its subsequent incorporation into proteins is well documented and could explain this result (2). Alternatively, despite NMT’s high selectivity for myrstoyl-CoA, which normally excludes transfer of palmitate from palmitoyl-CoA, NMT may be able to use azidopalmitoyl-CoA more efficiently than palmitoyl-CoA, but less efficiently than azidomyristoyl-CoA. To investigate whether the azido-fatty-acyl moieties were incorporated into the EGFP chimeras via a thioester or amide bond, PVDF membranes were soaked in 0.2 M KOH for a total of 16 h, a treatment known to hydrolyze radiolabeled fatty acids incorporated into proteins via a thioester bond (9, 25, 26). Regardless of the analog used, the label incorporated into WT-ctPAK2-N15-EGFP, but not G2A-ctPAK2-N15-EGFP, was apparently totally resistant to alkali treatment, indicating that the azido moieties were attached to the chimeras via an amide bond and not an alkali-sensitive thioester bond. This result further supports the possibility that incorporation of azidopalmitate label into WT-ctPAK2-N15-EGFP was due to its likely conversion into azidomyristate. In Fig. 2A , we demonstrate that phosphine-FLAG can be an effective substitute for phosphine-biotin and yields highly similar results and conclusions.
To investigate whether our method could detect endogenously myristoylated proteins and perhaps facilitate the identification of new post-translationally myristoylated proteins in cells undergoing apoptosis, we sought to compare the detection of ctPAK2 myristoylation using azidomyristate or [3H]myristate as labels. We found that, like [3H]myristate, the incorporation of azidomyristate into immunoprecipitated ctPAK2 was abolished in the presence of the NMT inhibitor HMA (19) (Fig. 3A), suggesting that the azidomyristate is recognized as a substrate by the highly selective NMT (5). Of note, when incorporated into cells, HMA is metabolically activated to 2-hydroxymyristoyl-CoA, a specific inhibitor of NMT that does not inhibit protein palmitoylation (17–19). While preventing the incorporation of [3H]myristate or azidomyristate into ctPAK2 in cells induced to undergo apoptosis, HMA did not impair caspase 3-mediated PAK2 cleavage on STS treatment (data not shown). The myristoylation of endogenous ctPAK2 myristoylation was detected after a 5 s exposure on Western blot processing with neutravidin-HRP/ECL, while the detection of the incorporation of [3H]myristate into ctPAK2 required a 65-day exposure using a conventional fluorographic method (Fig. 3A ).
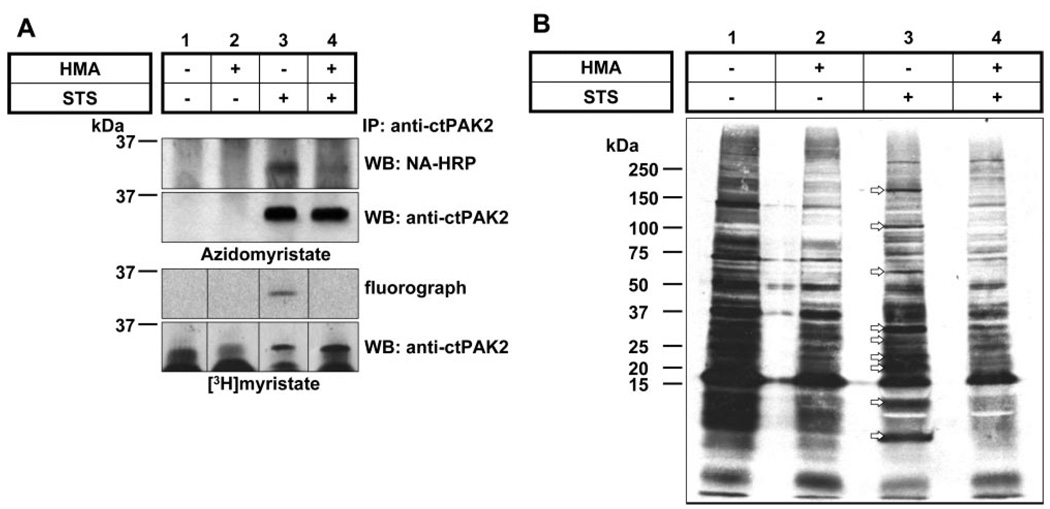
Figure 3.
Myristoylation of endogenous ctPAK2 in Jurkat cells. Jurkat cells metabolically labeled with A) azidomyristate or [3H]myristate were induced to undergo apoptosis with STS/CHX (lanes 3, 4) in the presence or absence of HMA (lanes 2, 4) (9). Endogenous ctPAK2 was immunoprecipitated and conjugated to phosphine-biotin for the nonradioactive detection, then separated by SDS-PAGE, and incorporation of azidomyristate was assessed by Western blot analysis with neutravidin-HRP using a 5-s exposure and incorporation of [3H]myristate via a 2-month fluorographic exposure (composite gel). The postimmuno-precipitation supernatants from the [3H]myristate samples in A were separated by SDS-PAGE and subjected to fluorography for 1 month. B) The arrows highlight the presence of at least 9 putative post-translationally myristoylated proteins in apoptotic Jurkat cells.
Overall, our results indicate that a bio-orthogonal azidomyristate analog can be incorporated into proteins via an amide bond and used to detect exogenously or endogenously expressed myristoylated proteins in a matter of seconds using phosphine-biotin instead of the typical 1–3 month fluorographic exposure to film required for the detection of [3H]myristoylated proteins (2, 9). This difference represents over a millionfold improvement in detection speed.
Azidomyristate incorporation into proteins and its detection with phosphine-biotin demonstrates the existence of several endogenous post-translationally myristoylated proteins in Jurkat T cells
To investigate if post-translational myristoylation is a widely used modification process during the onset of apoptosis, metabolically radiolabeled Jurkat T cell proteins contained in the post-PAK2-immunoprecipitation supernatants from the previous experiment were analyzed by SDS-PAGE followed by fluorography. The fluorogram (1-month exposure) in Fig. 3B demonstrates that on induction of apoptosis with STS and CHX, at least 9 proteins appear to exhibit post-translational myristoylation (Fig. 3B , lane 3). Because HMA inhibits NMT but not palmitoyl transferases, proteins corresponding to bands whose incorporation of [3H]myristate was drastically reduced in the presence of the NMT inhibitor HMA on induction of apoptosis with STS/CHX (Fig. 3B , lane 4) were considered as post-translationally myristoylated proteins.
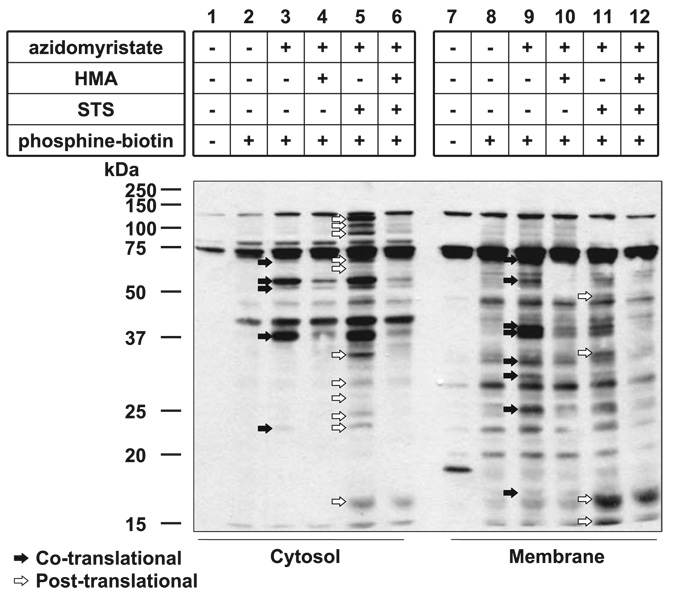
In comparison, we metabolically labeled Jurkat T cells undergoing STS/CHX-mediated apoptosis with the azidomyristate analog in the presence or absence of HMA. To reduce the complexity of the samples, post-nuclear supernatants of cell lysates were fractionated into membrane and cytosolic fractions following metabolic labeling and hypotonic lysis. Figure 4 shows that at least 15 and 13 proteins appear to be post- and cotranslationally myristoylated, respectively, as judged by the appearance of prominent bands on a short exposure (1 min) of the membrane to film. Interestingly, the vast majority of post-translationally azidomyristoylated proteins (11 of 15) were found in the cytosolic fraction while most cotranslationally azidomyristoylated proteins (8 of 13) were found in the membrane fraction. Untreated cells (no azidomyristate, no phosphine-biotin; Fig. 4, lanes 1, 7) showed 3 endogenous bands in the cytosolic fraction, and up to 8 proteins were detected by the neutravidin-HRP in the membrane fraction; these likely correspond to endogenous biotinylated carboxylases or their proteolytic fragments. We found that the membranous fractions presented a consistently higher background than the cytosolic ones. This might be a consequence of the hydrophobic nature of the 3 aryl groups of the phosphine moiety of the phosphine-biotin or the result of indirect detection of azido-labeled phospholipids nonspecifically bound to membrane proteins. Regardless, these results indicate that there are additional proteins that are post-translationally myristoylated during apoptosis and that this phenomenon might play an important role in the regulation of apoptosis.
Figure 4.
Detection of co- and post-translational azidomyristoylation of endogenous proteins in Jurkat cells using phosphine-biotin. Jurkat cells were metabolically labeled with azidomyristate and induced to undergo apoptosis with STS/ CHX (lanes 5, 6, 11, 12) in the presence or absence of HMA (lanes 4, 6, 10, 12), followed by hypotonic lysis and fractionation. The cytosolic (lanes 1–6) and membrane (lanes 7–12) fractions of the postnuclear supernatant were conjugated with the phosphine-biotin (lanes 2–6, 8–12) and subjected to Western blot analysis. Biotinylated, azidomyristoylated proteins were detected with neutravidin-HRP, using a 1-min exposure. Co- (lanes 3, 8) and post-translationally (lanes 5, 11) myristoylated proteins are indicated by black and white arrows, respectively.
The fact that numerous proteins are post-translationally myristoylated during apoptosis illustrates the need for their identification and the characterization of their roles during this process. Toward this end, we used our azidomyristate analog to label COS-7 cells transiently transfected with vectors expressing various N-terminal EGFP chimeras containing the first 10 amino acids of caspase-cleaved proteins containing putative myristoylation sites directly adjacent to their cleavage site (Supplemental Tables 1 and 2).
Development of a strategy for the rapid identification of potentially post-translationally myristoylated proteins
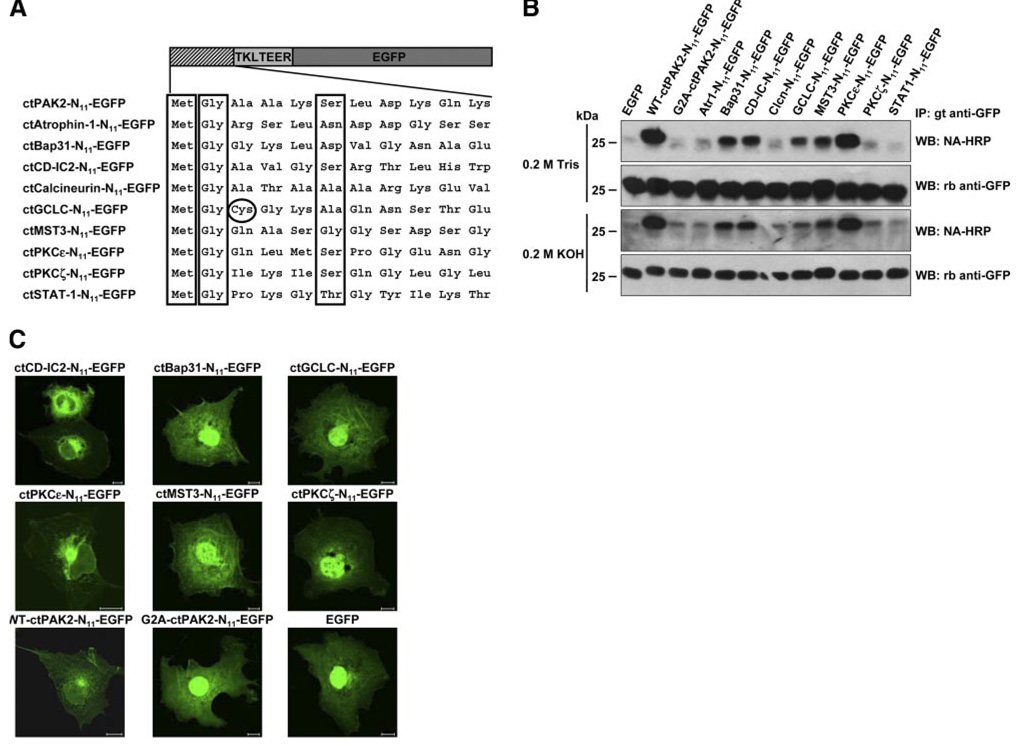
We identified potential substrates for post-translational myristoylation by locating internal myristoylation consensus sequences directly downstream of identified caspase cleavage sites (22). To do so, the complete carboxyl terminal sequences downstream of the caspase cleavage site were subjected to computational prediction analysis based on myristoylator and NMT–the Myr Predictor algorithms (23, 24). Of more than 280 proteins identified as caspase substrates (22), 48 occurred immediately upstream of a glycine residue, and 9 of these proteins received favorable scores for myristoylation and were identified as putative candidates for post-translational myristoylation (Supplemental Table 2). The first 10 amino acids downstream of the caspase cleavage sites of these 9 proteins were appended after an initiator methionine to the N terminus of EGFP at the cDNA level, as we reported earlier (2) (Fig. 5A). When we metabolically labeled COS-7 cells transiently transfected with vectors expressing the indicated EGFP chimeras with the azidomyristate analog, we show 5 of the 9 immunoprecipitated EGFP chimeras tested are myristoylatable substrates. Figure 5B shows that ctBap31-, ctCD-IC2-, ctGCLC-, ctMST3-, and ctPKCε-N11-EGFPs, as well as the positive control WT-ctPAK2-N11-EGFP, all incorporated significant, albeit different, amounts of the azidomyristate as judged by the positive detection of phosphine-biotin with neutravidin-HRP on the membrane. The signals were the strongest in ctPKCε-N11-EGFP and WT-ctPAK2-N11-EGFP. In contrast, ctAtr1-, ctCalcineurin-, ctPKCζ-, and ctSTAT1-N11-EGFP, as well as our negative control, the non-myristoylatable G2A-ctPAK2-N11-EGFP, did not appear to incorporate the azidomyristate analog as no reactive band was found on development of the film. The incorporation of the azidomyristate analog was resistant to alkali hydrolysis and therefore believed to occur via an amide bond. All chimeric EGFPs were expressed at similar levels (Fig. 5B ).
Figure 5.
Azidomyristoylation of putative post-translationally myristoylatable proteins. A) Representation of the N11-EGFP chimeras bearing the first 10 amino acids of the predicted putative substrates of post-translational myristoylation with an initiator methionine and a TKLTEER hydrophilic linker. The initiator methionine, essential Gly in position 2, and preferred amino acids at position 6 (Ser, Thr, Cys) are boxed. B) EGFP chimeras were transiently expressed in COS-7 cells and labeled with azidomyristate. The immunoprecipitated EGFP chimeras were reacted with phosphine-biotin and detected by Western blotting with neutravidin-HRP after the membranes were treated with 0.2 M Tris pH 7.0 or 0.2 M KOH. Chimeric ctPAK2-, ctCD-IC2-, ctPKCε-, ctMST3-, ctBap31-, and ctGCLC-N11-EGFPs all gave significant ECL signals on films, although of varying intensity. C) COS-7 cells transiently expressing the significantly azidomyristoylated chimeric EGFPs identified in B, the nonmyristoylatable ctPKCζ- or G2A-ctPAK2-N11-EGFP, and EGFP were fixed and photographed by fluorescence confocal microscopy. Scale bars = 10 µm.
To further assess the extent of myristoylation of these 5 sequences of amino acids appended to EGFP, we used confocal microscopy to look at transiently transfected COS-7 cells (Fig. 5C ). We previously have shown that myristoylation of short chimeric GFP constructs is sufficient to confer endomembrane binding and nuclear exclusion to the GFP reporter (2). We show that ctCD-IC2-N11-EGFP and ctPKCε-N11-EGFP as well as WT-ctPAK2-N11-EGFP presented significant perinuclear membrane association and nuclear exclusion while the extent of apparent membrane association and nuclear exclusion is significantly lower for ctBap31-, ctGCLC-, and ctMST3-N11-EGFPs. We believe the extent of nuclear exclusion and endomembrane localization to be highly indicative of the extent of myristoylation. It appears that the localization of ctPKCζ-N11-EGFP (Fig. 5C ), which did not incorporate the azidomyristate (Fig. 5B ), was like that of G2A-ctPAK2-N11-EGFP, which was indistinguishable from that of EGFP (found largely in the nucleus) (2).
When appended to the hydrophilic N terminus of a reporter protein, the first 10 amino acids of a known myristoylated protein usually are sufficient to confer myristoylation by NMT and are excellent indicators of protein N-myristoylation (2, 11). Altogether, ctCD-IC2 and ctPKCε therefore appear to be our most likely candidates as novel post-translationally myristoylatable substrates for NMT inside cells while ctGCLC, ctBap31, and ctMST3 may be only partially myristoylated. The significance of the post-translational myristoylation of these proteins is currently being investigated.
DISCUSSION
We developed an alternative and highly sensitive detection method to study the myristoylation of proteins to replace the time consuming, expensive, hazardous, and laborious labeling of cells with radioactive fatty acids. This alternative method takes advantage of the Staudinger ligation’s unique chemoselective reactivity that can covalently link alkyl azides, such as azidomyristate, to a tagged triarylphosphine via an amide bond, thereby allowing specific probing of azidomyristoylated proteins within the cell (15). Indeed, the fact that neither alkyl azides nor phosphines are found in the biological milieu and that they do not react with cellular nucleophiles at ambient temperatures makes them ideal for in vivo labeling and subsequent tagging of post-translationally modified proteins (15, 16, 27, 28). Important to the biological study of myristoylation, 12-azidododecanoate is an alkyl azide analog of myristate that, like most alkyl azides, has been shown previously to be nontoxic to cells and animals (29). Furthermore, 12-azidododecanoate also is efficiently converted to its azidomyristoyl-CoA derivative by fatty acyl CoA synthetase (29), which is a prerequisite to its use by NMT.
Herein, we demonstrate that the isosteric analog of myristate (29), azidomyristate (12-azidododecanoate) can be specifically incorporated in a co- or post-translational fashion via an alkali-resistant amide bond at the N-terminal glycine of exogenously or endogenously expressed proteins and readily detected with the phosphine-biotin using neutravidin/ECL. Recently, Hang et al. (30) also demonstrated that a variety of azido-fatty acid analogs can be used as chemical probes to monitor protein fatty acylation using the Staudinger ligation, therefore establishing the proof-of-principle for such a methodology. Of relevance to our study, they also showed that 12-azidododecanoate (azidomyristate) is preferentially incorporated into proteins via an amide bond (30). In our study, we established the proof-of-principle in greater detail and exploited this new technique to demonstrate the existence of several post-translationally myristoylated proteins in Jurkat T cells undergoing apoptosis. Furthermore, we designed a strategy that allowed us to successfully identify new post-translationally myristoylated proteins.
Our strategy is based on several studies that showed that the first 10–15 N-terminal amino acids of a known myristoylated protein are sufficient to confer myristoylation when appended to reporter proteins such as tumor necrosis factor or GFP (2, 3, 11, 31). Since we showed that the myristoylation of short chimeric WT-ctPAK2-N15-EGFP and WT-Yes-N11-GFP can readily be detected using azidomyristate cell labeling and reaction with phosphine-biotin (Fig. 2), we first identified internal myristoylation sites adjacent to caspase cleavage sites using computational prediction analysis (23, 24) and, second, incorporated these predicted NMT substrate sequences at the N terminus of EGFP at the cDNA level (Supplemental Table 2). Finally, using our nonradioactive azidomyristate labeling/phosphine-biotin-based detection method, we assessed the myristoylation status of the chimeric EGFP proteins transiently expressed in COS-7 cells. In doing so, we identified ctBap31-, ctCD-IC2-, ctGCLC-, ctMST3-, and ctPKCε-N11-EGFP as potential new substrates for NMT and reconfirmed the myristoylation of ctPAK2-N11-EGFP. In our assay, ctPKCζ-N11-EGFP was not a substrate for NMT, confirming the results of Utsumi et al. (11) using tumor necrosis factor as a reporter protein and [3H]myristate as the label.
ctCD-IC2-N11-EGFP and ctPKCε-N11-EGFP provided the strongest signal via Western blot analysis and were almost completely excluded from the nucleus like ctPAK2-N11-EGFP as visualized by confocal microscopy (Fig. 5B, C) and, therefore, are likely very efficiently myristoylated and represent strong candidates for post-translational myristoylation of their respective full-length proteins in apoptotic cells. In contrast, despite the significant incorporation of azidomyristate into ctBap31-, ctGCLC-, and ctMST3-N11-EGFP as assessed by Western blot analysis, these chimeras did not show significant nuclear exclusion, suggesting that they are only partially myristoylated, although this may still be of significance inside cells. It is possible that their endogenous counterparts may act as more desirable substrates for NMT or that there may be some unknown interferences in our system because we were using reporter constructs, which required cotranslational myristoylation of chimeric reporters as a means to assess the myristoylation status of naturally post-translationally myristoylated proteins. Furthermore, there are two NMT isoforms (NMT-1 and NMT-2) expressed in mammalian cells, which differ primarily at their N termini. While the NMT-1 N terminus is thought to be responsible for ribosome interactions required for cotranslational myristoylation, NMT-2 is more cytosolic (5, 32). The two isoforms have both overlapping and nonover-lapping substrate specificities (33). Presently, we do not know which isoform is required for post-translational myristoylation, but based on localization studies and recent evidence that NMT-2 interacts with caspase 3 and Bcl-2, we hypothesize that NMT-2 may be the primary enzyme involved in post-translational myristoylation (32, 34).
Like other previously identified post-translationally myristoylated proteins, the proteins we identified are kinases, proapoptotic proteins, or regulators of the cytoskeleton structure (9, 10, 31). Like ctPAK2, caspase cleavage of PKCε results in the loss of the N-terminal regulatory domain to generate a constitutively active C-terminal kinase domain (35). PKCε contains two caspase cleavage sites at Asp383 and Asp451. The former appears to be the primary site of cleavage and is located within the hinge domain, between the regulatory and kinase domains, whereas cleavage at the second site is delayed and is found within the catalytic domain (35). PKCε is a calcium-independent and diacylglycerol (DAG)-dependent kinase and post-translational myristoylation of ctPKCε at the primary caspase cleavage site (Asp383) may substitute the DAG-binding domain (found in the N terminus) to relocate the kinase domain to a new site within the cell where it could phosphorylate and regulate the activity of specific proteins during apoptosis.
Interestingly and unlike PAK2 and PKCε, the C-terminal cleavage product of MST3 contains the regulatory domain of the protein (36). Therefore, myristoylation of ctMST3 could relocalize its regulatory domain away from the catalytic domain, resulting in a constitutively active enzyme that could diffuse in the cytosol and translocate to the nucleus via its nuclear localization sequence (36), unlike constitutively active myr-ctPAK2, which was found primarily at the plasma membrane and at the surface of endosomes (9). It is also possible that the two fragments remain associated, like cleaved Bid, and that the myristate provides a novel localization site for cleaved MST3 (10). Similar to ctPAK2, MST3 regulates cell morphology but also plays a role in cell cycle progression (37). In addition, overexpression of full-length MST3 or its N-terminal cleavage product is proapoptotic (36). Although the myristoylation of ct-MST3-N11-EGFP appeared to be partial, it may still be relevant and of significance inside cells.
CD-IC2 is required for the interaction between cytoplasmic dynein and dynactin through binding of p150Glued via its N terminus. Dynein and dynactin regulate the secretory and endocytic pathways and microtubule organization at interphase, and the ER serves as cargo for cytoplasmic dynein in Xenopus egg extracts. During apoptosis, CD-IC2 is cleaved within the p150Glued binding domain but remains associated with the heavy and intermediate light chains of dynein, probably via its overlapping WD-40 repeats (38). Very interestingly, light chain 8 of dynein associates with the proapoptotic protein Bcl-2 interacting mediator of cell death in normal cells, and during apoptosis this complex translocates to membranes of various organelles, including mitochondria, where it binds and inhibits BCL-2 or functional homologs (39). It is possible that myr-ctCD-IC2 may be involved in this complex or required for a similar pathway to direct the progression of apoptosis.
Bap31 is an integral membrane protein of the ER and is thought to regulate the export of cellubrevin from the ER. During apoptosis, Bap31 forms a complex with caspase-8, BCL-2 or BCL-XL, and what is thought to be a CED-4 homologue (40, 41). The ~20 kDa N-terminal fragment is a potent activator of apoptosis when overexpressed in cells. Interestingly, the C-terminal cleavage product, which has not been studied to the extent of the N-terminal fragment, contains overlapping leucine zippers and a weak homology death-effector domain that has been shown to interact with exogenously expressed CED-4 in mammalian cells, which, in turn, interacts with caspase-8 (41). Again, these protein-binding domains may be required for complete and proper localization of the myr-ctBap31 protein. Like myr-ctBid and myr-ctPAK2, myristoylation of ctBap31 may ameliorate the proapoptotic effect of Bap31 by translocating caspase-8 to new substrates.
GCLC is the catalytic subunit of the heterodimeric enzyme glutamate-l-cysteine ligase, which regulates the rate-limiting step in the synthesis of the antioxidant glutathione. A common feature during apoptosis is the depletion of glutathione in the cell. Although overex-pression of full-length GCLC is antiapoptotic, the effect of the caspase cleavage products of GCLC and the subsequent myristoylation of ctGCLC remains unclear at this time (42).
Using this alternative detection methodology, we also demonstrate the existence of at least 15 proteins that undergo post-translational myristoylation in apoptotic Jurkat T cells (Fig. 4). This result suggests that post-translational myristoylation of caspase-cleaved proteins represents a mechanism widely used to regulate cell death. While the majority of cotranslationally myristoylated proteins were found in the membrane fraction (Fig. 4) as assessed by our methodology, the majority of the post-translationally myristoylated proteins were found in the cytosolic fraction, perhaps suggesting alternative roles for myristoylation other than membrane tethering.
Because the azide moiety bound to myristate can be ligated with high chemoselectivity with a tagged phosphine via a highly stable amide bond, one of the most exciting future applications of this methodology resides in the proteomic identification of the complete set of myristoylated proteins in living and dying cells via affinity chromatography and identification by mass spectrometry. Although this methodology is designed as a chemical tool to label, discover, and identify myristoylated proteins, it is important to note that the biophysical properties of azidomyristate likely differ from that of myristate, and this dissimilarity obviously potentially restricts the use of azidomyristate in subcellular localization studies.
The ease, high sensitivity, and power of our new methodology to detect and identify new myristoylated proteins is undeniable and will greatly facilitate the study of the biology of co- and post-translational myristoylation of proteins in cells and eventually animals with the potential of unraveling new roles for this type of post-translational protein modification.
Supplementary Material
Acknowledgments
We thank G. Lambkin and G. Plummer for technical assistance. This work was supported by Canada Institutes of Health Research (CIHR) grant MOP-81248 and a bridge funding grant from the Alberta Cancer Board to L.G.B., who is a senior scholar of the Alberta Heritage Foundation for Medical Research. D.D.O.M. held a Province of Alberta Graduate Scholarship and holds a Canada Graduate Scholarships Doctoral Award from CIHR. Both D.D.O.M. and G.L.V. held Faculty of Medicine and Dentistry 75th Anniversary Graduate studentships. Additional financial support was provided to J.R.F. by the Robert A. Welch Foundation and the U.S. National Institutes of Health (GM31278). J.A.P. was supported by a predoctoral fellowship from the HHMI.
REFERENCES
- 1.Berthiaume L, Deichaite I, Peseckis S, Resh MD. Regulation of enzymatic activity by active site fatty acylation. A new role for long chain fatty acid acylation of proteins. J. Biol. Chem. 1994;269:6498–6505. [PubMed] [Google Scholar]
- 2.McCabe JB, Berthiaume LG. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol. Biol. Cell. 1999;10:3771–3786. doi: 10.1091/mbc.10.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCabe JB, Berthiaume LG. N-terminal protein acylation confers localization to cholesterol, sphingolipid-enriched membranes but not to lipid rafts/caveolae. Mol. Biol. Cell. 2001;12:3601–3617. doi: 10.1091/mbc.12.11.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE. 2006;2006 doi: 10.1126/stke.3592006re14. re14. [DOI] [PubMed] [Google Scholar]
- 5.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 6.Resh MD. Membrane targeting of lipid modified signal transduction proteins. Subcell. Biochem. 2004;37:217–232. doi: 10.1007/978-1-4757-5806-1_6. [DOI] [PubMed] [Google Scholar]
- 7.Maurer-Stroh S, Gouda M, Novatchkova M, Schleiffer A, Schneider G, Sirota FL, Wildpaner M, Hayashi N, Eisenhaber F. MYRbase: analysis of genome-wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins. Genome Biol. 2004;5:R21. doi: 10.1186/gb-2004-5-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Towler DA, Eubanks SR, Towery DS, Adams SP, Glaser L. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J. Biol. Chem. 1987;262:1030–1036. [PubMed] [Google Scholar]
- 9.Vilas GL, Corvi MM, Plummer GJ, Seime AM, Lambkin GR, Berthiaume LG. Posttranslational myristoylation of caspase-activated p21-activated protein kinase 2 (PAK2) potentiates late apoptotic events. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6542–6547. doi: 10.1073/pnas.0600824103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 11.Utsumi T, Sakurai N, Nakano K, Ishisaka R. C-terminal 15 kDa fragment of cytoskeletal actin is posttranslationally N-myristoylated upon caspase-mediated cleavage and targeted to mitochondria. FEBS Lett. 2003;539:37–44. doi: 10.1016/s0014-5793(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 12.Wyllie AH. Apoptosis: an overview. Br. Med. Bull. 1997;53:451–465. doi: 10.1093/oxfordjournals.bmb.a011623. [DOI] [PubMed] [Google Scholar]
- 13.Wyllie AH, Bellamy CO, Bubb VJ, Clarke AR, Corbet S, Curtis L, Harrison DJ, Hooper ML, Toft N, Webb S, Bird CC. Apoptosis and carcinogenesis. Br. J. Cancer. 1999;80(Suppl. 1):34–37. [PubMed] [Google Scholar]
- 14.Deichaite I, Berthiaume L, Peseckis SM, Patton WF, Resh MD. Novel use of an iodo-myristyl-CoA analog identifies a semialdehyde dehydrogenase in bovine liver. J. Biol. Chem. 1993;268:13738–13747. [PubMed] [Google Scholar]
- 15.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 16.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galbiati F, Guzzi F, Magee AI, Milligan G, Parenti M. Chemical inhibition of myristoylation of the G-protein Gi1 alpha by 2-hydroxymyristate does not interfere with its palmitoylation or membrane association. Evidence that palmitoylation, but not myristoylation, regulates membrane attachment. Biochem. J. 1996;313(Pt 3):717–720. doi: 10.1042/bj3130717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadler MJ, Harrison ML, Ashendel CL, Cassady JM, Geahlen RL. Treatment of T cells with 2-hydroxy-myristic acid inhibits the myristoylation and alters the stability of p56lck. Biochemistry. 1993;32:9250–9255. doi: 10.1021/bi00086a034. [DOI] [PubMed] [Google Scholar]
- 19.Paige LA, Zheng GQ, DeFrees SA, Cassady JM, Geahlen RL. Metabolic activation of 2-substituted derivatives of myristic acid to form potent inhibitors of myristoyl CoA:protein N-myristoyltransferase. Biochemistry. 1990;29:10566–10573. doi: 10.1021/bi00498a021. [DOI] [PubMed] [Google Scholar]
- 20.Luchansky SJ, Hang HC, Saxon E, Grunwell JR, Yu C, Dube DH, Bertozzi CR. Constructing azide-labeled cell surfaces using polysaccharide biosynthetic pathways. Methods Enzymol. 2003;362:249–272. doi: 10.1016/S0076-6879(03)01018-8. [DOI] [PubMed] [Google Scholar]
- 21.Laughlin ST, Agard NJ, Baskin JM, Carrico IS, Chang PV, Ganguli AS, Hangauer MJ, Lo A, Prescher JA, Bertozzi CR. Metabolic labeling of glycans with azido sugars for visualization and glycoproteomics. Methods Enzymol. 2006;415:230–250. doi: 10.1016/S0076-6879(06)15015-6. [DOI] [PubMed] [Google Scholar]
- 22.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- 24.Eisenhaber F, Eisenhaber B, Kubina W, Maurer-Stroh S, Neuberger G, Schneider G, Wildpaner M. Prediction of lipid posttranslational modifications and localization signals from protein sequences: big-Pi, NMT and PTS1. Nucleic Acids Res. 2003;31:3631–3634. doi: 10.1093/nar/gkg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armah DA, Mensa-Wilmot K. S-myristoylation of a glycosylphosphatidylinositol-specific phospholipase C in Trypanosoma brucei. J. Biol. Chem. 1999;274:5931–5938. doi: 10.1074/jbc.274.9.5931. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, McCabe JB, Vance J, Berthiaume LG. Palmitoylation of apolipoprotein B is required for proper intracellular sorting and transport of cholesteroyl esters and triglycerides. Mol. Biol. Cell. 2000;11:721–734. doi: 10.1091/mbc.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxon E, Luchansky SJ, Hang HC, Yu C, Lee SC, Bertozzi CR. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J. Am. Chem. Soc. 2002;124:14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 28.Sprung R, Nandi A, Chen Y, Kim SC, Barma D, Falck JR, Zhao Y. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res. 2005;4:950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 29.Devadas B, Lu T, Katoh A, Kishore NS, Wade AC, Mehta PP, Rudnick DA, Bryant ML, Adams SP, Li Q, Gokel GW, Gordon JI. Substrate specificity of Saccharomyces cerevisiae myristoyl-CoA: protein N-myristoyl-transferase. Analysis of fatty acid analogs containing carbonyl groups, nitrogen heteroatoms, and nitrogen heterocycles in an in vitro enzyme assay and subsequent identification of inhibitors of human immunodeficiency virus I replication. J. Biol. Chem. 1992;267:7224–7239. [PubMed] [Google Scholar]
- 30.Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical probes for the rapid detection of fatty-acylated proteins in mammalian cells. J. Am. Chem. Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 31.Utsumi T, Sato M, Nakano K, Takemura D, Iwata H, Ishisaka R. Amino acid residue penultimate to the amino-terminal gly residue strongly affects two cotranslational protein modifications, N-myristoylation and N-acetylation. J. Biol. Chem. 2001;276:10505–10513. doi: 10.1074/jbc.M006134200. [DOI] [PubMed] [Google Scholar]
- 32.Giang DK, Cravatt BF. A second mammalian N-myristoyltransferase. J. Biol. Chem. 1998;273:6595–6598. doi: 10.1074/jbc.273.12.6595. [DOI] [PubMed] [Google Scholar]
- 33.Ducker CE, Upson JJ, French KJ, Smith CD. Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol. Cancer Res. 2005;3:463–476. doi: 10.1158/1541-7786.MCR-05-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvakumar P, Smith-Windsor E, Bonham K, Sharma RK. N-myristoyltransferase 2 expression in human colon cancer: cross-talk between the calpain and caspase system. FEBS Lett. 2006;580:2021–2026. doi: 10.1016/j.febslet.2006.02.076. [DOI] [PubMed] [Google Scholar]
- 35.Basu A, Lu D, Sun B, Moor AN, Akkaraju GR, Huang J. Proteolytic activation of protein kinase C-epsilon by caspase-mediated processing and transduction of antiapoptotic signals. J. Biol. Chem. 2002;277:41850–41856. doi: 10.1074/jbc.M205997200. [DOI] [PubMed] [Google Scholar]
- 36.Huang CY, Wu YM, Hsu CY, Lee WS, Lai MD, Lu TJ, Huang CL, Leu TH, Shih HM, Fang HI, Robinson DR, Kung HJ, Yuan CJ. Caspase activation of mammalian sterile 20-like kinase 3 (Mst3). Nuclear translocation and induction of apoptosis. J. Biol. Chem. 2002;277:34367–34374. doi: 10.1074/jbc.M202468200. [DOI] [PubMed] [Google Scholar]
- 37.Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane JD, Vergnolle MA, Woodman PG, Allan VJ. Apoptotic cleavage of cytoplasmic dynein intermediate chain and p150(Glued) stops dynein-dependent membrane motility. J. Cell Biol. 2001;153:1415–1426. doi: 10.1083/jcb.153.7.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puthalakath H, Huang DC, O’Reilly LA, King SM, Strasser A. The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol. Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 40.Granville DJ, Carthy CM, Jiang H, Shore GC, McManus BM, Hunt DW. Rapid cytochrome c release, activation of caspases 3, 6, 7 and 8 followed by Bap31 cleavage in HeLa cells treated with photodynamic therapy. FEBS Lett. 1998;437:5–10. doi: 10.1016/s0014-5793(98)01193-4. [DOI] [PubMed] [Google Scholar]
- 41.Ng FW, Shore GC. Bcl-XL cooperatively associates with the Bap31 complex in the endoplasmic reticulum, dependent on procaspase-8 and Ced-4 adaptor. J. Biol. Chem. 1998;273:3140–3143. doi: 10.1074/jbc.273.6.3140. [DOI] [PubMed] [Google Scholar]
- 42.Botta D, Franklin CC, White CC, Krejsa CM, Dabrowski MJ, Pierce RH, Fausto N, Kavanagh TJ. Glutamate-cysteine ligase attenuates TNF-induced mitochondrial injury and apoptosis. Free Radic. Biol. Med. 2004;37:632–642. doi: 10.1016/j.freeradbiomed.2004.05.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.