Abstract
Mitochondrial copper metabolism and delivery to cytochrome c oxidase and mitochondrially localized CuZn-superoxide dismutase (Sod1) requires a growing number of intermembrane space proteins containing a twin Cx9C motif. Among them, Cmc1 was recently identified by our group. Here we describe another conserved mitochondrial metallochaperone-like protein, Cmc2, a close homologue of Cmc1, whose function affects both cytochrome c oxidase and Sod1. In the yeast Saccharomyces cerevisiae, Cmc2 localizes to the mitochondrial inner membrane facing the intermembrane space. In the absence of Cmc2, cytochrome c oxidase activity measured spectrophotometrically and cellular respiration measured polarographically are undetectable. Additionally, mutant cmc2 cells display 2-fold increased mitochondrial Sod1 activity, whereas CMC2 overexpression results in Sod1 activity decreased to 60% of wild-type levels. CMC1 overexpression does not rescue the respiratory defect of cmc2 mutants or vice versa. However, Cmc2 physically interacts with Cmc1 and the absence of Cmc2 induces a 5-fold increase in Cmc1 accumulation in the mitochondrial membranes. Cmc2 function is conserved from yeast to humans. Human CMC2 localizes to the mitochondria and CMC2 expression knockdown produces cytochrome c oxidase deficiency in Caenorhabditis elegans. We conclude that Cmc1 and Cmc2 have cooperative but nonoverlapping functions in cytochrome c oxidase biogenesis.
Keywords: Copper, Cytochrome Oxidase, Mitochondria, Mitochondrial Metabolism, Protein Assembly, Superoxide Dismutase (SOD), Cmc1, Cmc2, Cx9C Protein Family
Introduction
The maintenance of a regulated copper metabolism in the mitochondrial intermembrane space (IMS)2 is essential for both cellular respiration and free radical scavenging. Cytochrome c oxidase (COX), the terminal enzyme of the mitochondrial respiratory chain, and the mitochondrial localized fraction of CuZn-superoxide dismutase (Sod1) use copper as an essential cofactor. COX is a multimeric metalloenzyme that contains two copper metal centers within its catalytic core (1). The CuA site formed by two copper atoms is located in COX subunit 2 (Cox2). A third copper atom in the CuB site forms a binuclear center with heme a3 in subunit 1 (Cox1). Sod1 contains one copper atom paired with one zinc atom at its catalytic site. Although mostly cytoplasmic, ∼1–5% of cellular Sod1 localizes to the mitochondrial IMS (2).
Both COX and mitochondrial Sod1 receive copper within the mitochondrial IMS through pathways involving the assistance of evolutionary conserved enzyme-specific copper chaperones localized to this compartment (3, 4). Sod1 copper insertion requires the specific copper chaperone Ccs1 (5), a portion of which is localized to the mitochondrial IMS (2, 6). COX copper metallation requires at least three key proteins. The IMS soluble chaperone Cox17 (7) binds copper(I) ions (8) and transfers them to Sco1 (9) and Cox11 (10, 11), which facilitate copper insertion into the COX CuA and CuB active sites, respectively. Sco1 and Cox11 are anchored to the mitochondrial inner membrane through a transmembrane α-helix and expose their copper binding sides to the IMS, where copper transfer occurs (8, 12). The exact mechanism of copper transfer between Cox17 and these proteins remains unclear because physical interactions among them have not been detected (13). Importantly, at least in the case of the Cox17-Sco1-Cox2 pathway, copper transfer is coupled to thiol redox chemistry, thus adding a regulatory level into the copper transfer pathway (14).
The mitochondrial matrix contains a pool of copper bound by a non-proteinaceous ligand (15), which is the copper source for metallation of COX and mitochondrial Sod1 (16). The copper ligand from the matrix pool has also been found in the cytoplasm and it has been suggested that the copper ligand may recruit copper to mitochondria in place of a copper chaperone (16). The transporter involved in copper trafficking from the mitochondrial matrix to the IMS remains to be identified. How copper subsequently reaches Cox17 and Ccs1 in the mitochondrial IMS also remains currently unknown. Current data support the idea that several other proteins could be involved in the latter functions. Cox17 contains a twin Cx9C structural motif that is also present in a growing number of IMS proteins required for COX assembly. These proteins include the soluble Cox19 (17) and Cox23 (42) and the peripherally bound to the inner membrane Pet191 (18) and Cmc1 (19). A recent systematic analysis of the twin Cx9C protein family identified a total of 14 mitochondrial proteins, which absence produce heterogeneous effects not only in cytochrome c oxidase activity but also in cytochrome c reductase activity (20). Although no physical interactions among these proteins have been detected to date, several pieces of information obtained in Saccharomyces cerevisiae suggest that at least some of them could be part of a copper transfer pathway toward COX. The respiratory deficiency of a cox23 null mutant is rescued by copper supplementation, albeit only with concomitant overexpression of COX17 (21). Recombinant Cox19 binds copper (17), but the COX assembly defect of a cox19 null mutant strain is not rescued by copper supplementation (22). Cox19 was proposed to participate in a different part of the Cox23-Cox17 pathway (21). It is currently unknown whether Pet191 binds copper but the respiratory defect of Δpet191 cells is not rescued by exogenous copper and therefore Pet191 has not been linked to a role in mitochondrial copper metabolism (18). Concerning Cmc1, we have recently shown that the recombinant form has the ability to bind copper (19). Differently from Δcox17, Δcox23, Δcox19, and Δpet191 cells, which display a complete absence of COX, Δcmc1 cells retain ∼40% of COX wild-type levels. The Δcmc1 respiratory defect is rescued by copper supplementation (19). We have proposed that Cmc1 could be acting in the same pathway as Cox19 and Cox23 (3).
How copper distribution from the matrix pool toward COX and Sod1 is regulated also remains largely unexplored. We have reported that Cmc1 levels modulate copper delivery to mitochondrial Sod1 (19), which support a model of Cmc1 as a gatekeeper allocating copper to the two pathways leading to the metallation of COX and Sod1. Because Δcmc1 cells retain a significant amount of COX activity we had speculated that Cmc1 function could either be redundant or to modulate the function of another protein(s). Therefore, to better understand the role of Cmc1 as a COX biogenetic factor and as a copper gatekeeper within the mitochondrial IMS, we aimed to identify close homologues and functional partners of this protein.
Here, we describe the characterization of Cmc2, a conserved Cx9C protein, the BLAST reciprocal best match of Cmc1. In S. cerevisiae, Cmc2 is encoded by open reading frame YBL059C-A, whose deletion leads to a complete loss of COX and an increase in activated Sod1. Cmc2 physically interacts with Cmc1 and both proteins have non-overlapping cooperative functions. Data obtained in C. elegans and cultured human cell lines support the functional conservation of Cmc2 during evolution. We propose that Cmc2, acting together with Cmc1, regulates copper distribution toward COX, which affects the level of active mitochondrial Sod1.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
Most S. cerevisiae strains used were all in the W303 background and included the wild-type W303-1A (MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1) and the previously reported mutant Δcox14 (23). Mutant Δcmc2 cells were created by inserting a URA3 cassette in the gene YBL059C-A locus in yeast chromosome II. The URA3 gene and flanking sequences of the CMC2 gene were PCR amplified using primers: 5′-CTGTTTGGACTTCATCAATGCACTCGATAAATGCCATCAAAAGGAATATTACAAGAGAATATTTGGCCTAAATTTGTAGAGGACTCAG-3′ and 5′-GCGTACTGTCTATCTAAGATAGTCTTTAATATAGCGTCCTCGCCATATTCTTCTTCCTTAGTTTTGCTGGCCGCATC-3′. The amplicon was transformed into a W303-1A wild-type strain to create W303Δcmc2. The Δcmc2Δcox14 double mutant was obtained from crosses of the respective single mutants. A Δcmc1 mutant in the BY4741 background was previously reported (19). The growth medium compositions have been described elsewhere (24).
Characterization of Yeast Mitochondrial Respiratory Chain
Endogenous cell respiration was assayed in whole cells in the presence of galactose at 24 °C using a Clark-type polarographic oxygen electrode from Hansatech Instruments (Norfolk, United Kingdom) (25). The specific activities reported were corrected for KCN-insensitive respiration. Mitochondria were prepared from strains grown in medium containing 2% galactose, according to the method of Faye et al. (26) except that zymolyase 20T (ICN Biochemicals Inc., Aurora, OH) instead of glusulase was used for the conversion of cells to spheroplasts. Mitochondria prepared from the different strains were used for spectrophotometric assays carried out at 24 °C to measure KCN-sensitive COX activity and antimycin A-sensitive NADH cytochrome c reductase (NCCR), succinate cytochrome c reductase, and ubiquinol cytochrome c reductase activities were measured as described (25). NADH dehydrogenase was measured by monitoring the diphenyliodonium chloride (0.5 mm)-sensitive oxidation of NADH at 340 nm. Total mitochondrial cytochromes spectra were obtained as reported (27). The sedimentation properties of Cmc2 and Cmc2–3HA in sucrose gradients were analyzed as described (23).
Peptide Antibodies against Cmc1 and Cmc2
We have used the services of Open Biosystems/Thermo Scientific (Huntsville, AL) to generate affinity purified rabbit polyclonal peptide antibodies against Cmc1 (peptide, DEKYLDGERDKIVLEKINK) and Cmc2 (peptide, EEYGEDAILKTILDRQYAKK).
Expression and Purification of Recombinant Cmc2 from Escherichia coli
Intronless CMC2 sequence was obtained using the RT-PCR Introductory System kit (Promega). The sequence containing the CMC2 gene was cloned using SacII and KpnI restriction enzymes into the pET41b protein expression vector (Novagen, Madison, WI), which provided an N-terminal GST affinity tag. A PreScission Protease (GE Healthcare) cleavage site was introduced between the GST tag and CMC2 for removal of the tag. The protein expression vector was transformed into the BL21 Star protein expression strain of E. coli (Invitrogen). Cells were initially grown overnight in 5 ml of Luria broth (LB) containing kanamycin (50 μg/ml) at 37 °C. Starter cultures were then diluted into 500 ml of minimal phosphate medium with antibiotic and grown with shaking (250 rpm) at 37 °C until OD of 0.8, then induced with 0.5 mm isopropyl β-d-thiogalactopyranoside for 3 h. The cells were harvested by centrifugation at 3500 × g for 20 min and subsequently lysed by resuspension in 10 ml of lysis buffer/g of cells (20 mm KPO4, pH 7.4, 500 mm NaCl, 2 mm dithiothreitol, 1 mm benzamidine, 2 mm phenylmethanesulfonyl fluoride) containing additional DNase I (0.1 mg/ml). The cells were lysed using a EmulsiFlex®-C3 high pressure homogenizer (Avestin, Inc., Ottawa, ON, Canada), and particulates were removed from the lysate by centrifugation for 30 min at 14,000 × g in an SS-34 Sorvall rotor.
The soluble E. coli lysate containing the GST-Cmc2 recombinant fusion construct was directly loaded by fast protein liquid chromatography (1 ml/min) onto a 10-ml GSTrap FF affinity column (Amersham Biosciences) equilibrated at 4 °C in 20 mm KPO4, pH 7.4, 500 mm NaCl, 2 mm dithiothreitol. The column was subsequently washed until the absorbance at 280 nm returned to baseline. The GST-Cmc2 protein was eluted with GST elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm GSH, and 2 mm dithiothreitol). Chromatographic fractions (1 ml) containing the designated fusion protein were detected by absorbance at 280 nm and analyzed by SDS-PAGE. Fractions containing recombinant protein were pooled and subjected to a desalting column for buffer exchange into 50 mm KPO4, pH 7.4, 500 mm NaCl, 1 mm tris(carboxyethyl)phosphine. The desalted protein was incubated with 1 unit/mg of PreScission Protease overnight at 4 °C to cleave off the GST tag. After incubation with the protease, separation of cleaved and non-cleaved recombinant protein was performed by passing the sample on a 10-ml GSTrap FF affinity column (Amersham Biosciences) and collecting the flow-through, and the sample was subsequently concentrated and loaded onto a Hiload Superdex 75 (Amersham Biosciences) gel-filtration column, equilibrated in the same buffer. High purity Cmc2 was subsequently collected from the appropriate fractions and concentrated as needed.
In-gel SOD Activity
To quantify Sod1 activity in whole cells and isolated mitochondria, we used an in-gel assay as described (28). To quantify the SOD activity staining gel, the images were digitalized and densitometric analyses was performed using the histogram function of the Adobe Photoshop program.
Caenorhabditis Elegans Maintenance and CMC2 RNA Interference
Standard nematode culturing techniques were employed (29). The N2 (wild type) and NL2099 (rrf-3(pk1426) II) were used. The cmc-2 (C35D10.17) bacterial feeding RNAi construct was generated ab initio (corresponding to the spliced coding region of genomic fragment III: 4867025–4867432 bp). A full-length cDNA fragment (333 bp long), homologue to the yeast gene CMC2, was amplified by reverse transcription from wild-type N2 total RNA with specific primers carrying the PstI and ApaI restriction sites (forward, 5′-cttctctgcagatgcttccggatttgtct-3′ and reverse, 5′-cttctgggcccttctaaattttacatcctt-3′) and cloned into the bacterial expression vector pL4440 (FireLab, Addgene). The construct was verified by direct sequencing. The pL4440 vector was used as a negative control and pLT61.1 (FireLab, Addgene), a pL4440-derived plasmid carrying the unc-22 gene, was employed as control of proper double-stranded RNA expression as its silencing produces a visible phenotype.
RNAi was carried out by feeding as described previously (30, 31). Briefly, cultures of HT115(DE3) containing the empty vector (pL4440), pLT61.1, or the gene of interest were prepared in LB broth containing 100 μg/ml of ampicillin and 13 μg/ml of tetracycline and grown overnight at 37 °C. Bacteria lawns were prepared by adding 300 μl of a stationary overnight culture to a 10-cm RNAi plate (NMG agar supplemented with 1 mm isopropyl β-d-thiogalactopyranoside, 100 μg/ml of ampicillin, and 13 μg/ml of tetracycline) and growing overnight at 37 °C. One-day-old gravid adult rrf-3(pk1426) hermaphrodites were transferred from standard OP50 culture plates to each type of RNAi plate and allowed to lay eggs at 20 °C. Animals were grown on RNAi plates for three consecutive generations as described previously (32).
For COX histochemistry, synchronized silenced worms were harvested and cuticles were permeabilized for histochemical staining as described (33). COX staining was performed as reported by Grad et al. (34). Control reactions were incubated in the presence of 10 mm KCN to inhibit complex IV activity. Stained worms were centrifuged for 10 s (360 × g) and the supernatant was removed. The worm pellet was washed three times for 5 min in phosphate buffer. Stained animals were mounted and photographed with an automated Leica microscope.
Miscellaneous Procedures
Standard procedures were used for the preparation and ligation of DNA fragments, and for transformation and recovery of plasmid DNA from E. coli (35). Yeast were transformed by the method of Schiestl and Gietz (36). The one-step gene insertion method (37) was used to integrate linear plasmids at the URA3 or LEU2 locus of yeast nuclear DNA. Protein concentration was measured with Folin phenol reagent (38). Proteins were separated by SDS-PAGE using the buffer system of Laemmli (39), and Western blots were treated with antibodies against the appropriate proteins followed by a second reaction with anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (Sigma). The SuperSignal chemiluminescent substrate kit (Pierce) was used for the final detection.
Statistical Analysis
All experiments (enzymatic and respiratory assays) were done at least in triplicate. Data are presented as mean ± S.D. of absolute values or percent of control. The values obtained for wild-type and cmc2 mutant strain for the different parameters studied were compared by Student's t test. p < 0.05 was considered significant.
RESULTS
CMC2 Is Essential for Cellular Respiration and Mitochondrial Cytochrome c Oxidase Biogenesis
The open reading frame YBL059C-A in yeast chromosome II codes for the BLAST reciprocal best match of Cmc1 (Fig. 1A). For this reason the gene encoded by YBL059C-A was named CMC2. Cmc2 contains a twin Cx9C motif and shares 23% of identical residues and 50% similarity with Cmc1 (Fig. 1A).
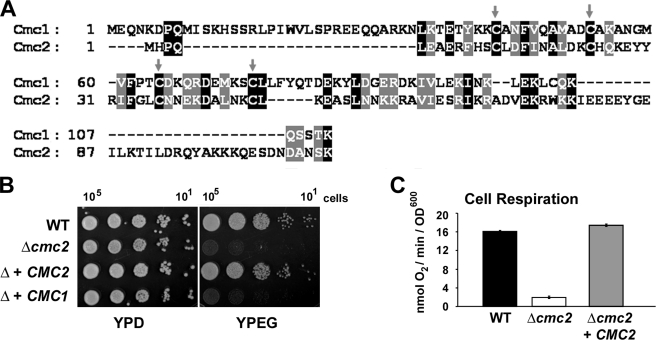
FIGURE 1.
S. cerevisiae CMC2 codes for a protein required for respiratory growth. A, sequence alignment of S. cerevisiae CMC1 and CMC2. Gray arrows indicate conserved cysteine residues. B, the respiratory competent wild-type strain W303-1A (WT), a strain carrying a null allele of cmc2 (Δcmc2 or Δ), and a Δ strain overexpressing either CMC1 or CMC2 were grown overnight in liquid YPD media. 10-Fold serial dilutions of the two strains were plated on solid YPD or YPEG medium and incubated at 30 °C. Pictures were taken after 3 days of incubation. C, KCN-sensitive endogenous cell respiration measured polarographically in the presence of galactose. The bars indicate the mean ± S.D. from at least three independent sets of measurements.
We have constructed a W303-1A strain carrying a null cmc2 allele by substituting the entire YBL059C-A reading frame with an URA3 cassette. This strain (W303Δcmc2) is unable to utilize respiratory substrates for growth (Fig. 1B) due to a virtually complete abolishment of the cells capacity to respire as shown by a polarographic measurement of KCN-sensitive endogenous cell respiration (Fig. 1C). The ability of the mutant cells to respire was restored by reinsertion of recombinant CMC2 but not by overexpression of CMC1 (Fig. 1B) suggesting that the two proteins perform different functions.
To ascertain whether the origin of the respiratory defect stemmed from a defect in the respiratory chain enzymes, we analyzed the oxidized minus reduced total mitochondrial cytochrome spectra. Comparison of the wild-type and mutant spectra revealed slight differences in the levels of cytochromes b, c, or c1. More importantly, cytochromes a and a3 were absent in the Δcmc2 mutant compared with the wild-type strain (Fig. 2A) indicating a complete COX impairment. The enzymatic defect was confirmed by measuring COX activity in isolated mitochondria, which indicated a complete absence of functional COX in the mutant as compared with the isogenic wild-type (Fig. 2B). Steady-state levels of Cox1 and Cox2 were not detected in the mutant and those of Cox3 were significantly reduced. This is most likely due to the rapid turnover of the COX catalytic core subunits when the assembly process is hindered. A less significant difference was detected in the more stable nuclear-encoded subunits, such as Cox4 (Fig. 2C).
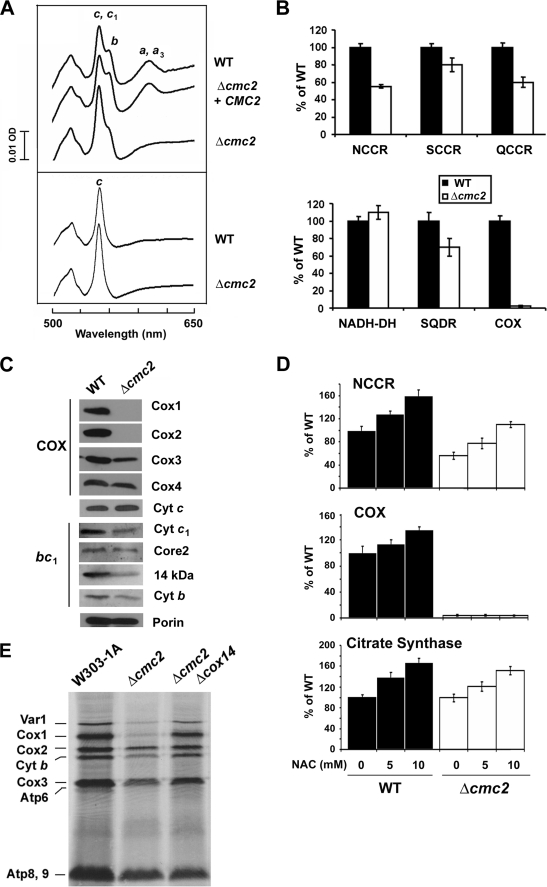
FIGURE 2.
Biochemical properties of Δcmc2 cells. A, total mitochondrial cytochrome spectra. Mitochondria from wild-type strain W303, the null mutant Δcmc2, and Δcmc2 cells overexpressing CMC2 were extracted at a protein concentration of 5 mg/ml with potassium deoxycholate under conditions that quantitatively solubilize all the cytochromes (upper panel (54)), or only with salt to exclusively extract cytochrome c (lower panel (40)). Difference spectra of the reduced (sodium dithionite) versus oxidized (potassium ferricyanide) extracts were recorded at room temperature. The absorption bands corresponding to cytochromes a and a3 have maximum at 603 nm (a and a3), the maximums for cytochrome b (b) and cytochrome c and c1 (c, and c1) are 560 and 550 nm, respectively. B, mitochondrial respiratory chain enzyme spectrophotometric measurements in isolated mitochondria. COX, NADH cytochrome c reductase (NCCR), succinate cytochrome c reductase (SCCR), ubiquinol cytochrome c reductase (QCCR), NADH dehydrogenase, succinate quinone dichlorophenol indophenol reductase (SQDR), and cytochrome c oxidase (COX) were measured as described under ”Experimental Procedures.“ C, steady-state concentrations of COX and bc1 complex subunits estimated by Western blot analyses of proteins separated in a 12% Tris glycine SDS-PAGE. D, N-acetyl-l-cysteine (NAC)-treated and untreated cells were used to spectrophotometrically measure COX, NCCR, and citrate synthase activities as described (55). The bars indicate the mean ± S.D. from three independent sets of measurements. E, in vivo mitochondrial protein synthesis in the presence of cycloheximide in the indicated strains. The mitochondrial products are identified in the margin.
In addition to abolishing COX biogenesis, the Δcmc2 mutation produces several pleiotropic effects in other segments of the mitochondrial respiratory chain (see below), whereas the oligomycin-sensitive ATPase activity in the mutant was similar to the wild-type (not shown). Cytochrome spectra showed that the amount of cytochrome b appeared decreased and the amount of cytochrome c increased. To distinguish between cytochromes c and c1, mitochondria were treated with high salt to remove cytochrome c. Spectra of the salt extracts (Fig. 2A) showed an increase in cytochrome c in Δcmc2 cells. Steady-state levels of cytochrome c were found slightly increased consistent with the spectra data (Fig. 2C). A similar increase has been observed in other COX assembly mutants including Δshy1 (40). Additionally, the Δcmc2 mutant showed decreased steady-state levels of some bc1 complex subunits (Fig. 2C) and a consistent decrease of ∼20–45% in several enzymatic assays involving the bc1 complex (Fig. 2B). A similar pleiotropic effect has been previously reported for strains carrying null alleles of cox10 (35% decreased NCCR activity (41) and cox20 (20% decreased NCCR activity (42)), whereas in some other cases including Δshy1 (40), Δpet191 (18), and Δcmc1 cells (19), NCCR activity was found 2-fold increased. The heterogeneous effects on COX and NCCR activities resulting from the absence of 14 mitochondrial Cx9C proteins were also recently reported (20). The mechanism underlying these pleiotropic effects remains uncharacterized and is currently being investigated in our laboratory.
An initial experiment to discern the primary target of Cmc2 is reported in Fig. 2D. We and others have recently reported that antioxidant treatment enhances mitochondrial biogenesis (43, 44). Chevtzoff and colleagues (44) have shown that the Ras/cAMP/protein kinase A pathway senses reactive oxygen species to signal to the Hap2,3,4,5 transcriptional system and regulate mitochondrial biogenesis. We showed that increasing mitochondrial biogenesis in this way by treatment with N-acetyl-l-cysteine in a mitochondrial ATPase mutant, we were able to partially suppress a profound pleiotropic effect on COX activity (43). Treatment of wild-type and Δcmc2 cells with 5 and 10 mm N-acetyl-l-cysteine promoted a general increase in mitochondrial biogenesis reflected in an increase in citrate synthase activity, a tricarboxylic acid cycle enzyme (Fig. 2D). COX and NCCR activities were proportionally enhanced in wild-type cells (Fig. 2D). In Δcmc2 cells, the residual NCCR activity was significantly enhanced upon N-acetyl-l-cysteine treatment, whereas COX activity remained undetectable (Fig. 2D), strongly supporting a primary role of Cmc2 in COX biogenesis.
Pulse labeling of cells in the presence of cycloheximide showed that the amount of newly synthesized Cox1 is less abundant in Δcmc2 mutant than in wild-type cells (Fig. 2E). This phenotype is likely resulting from a down-regulation of Cox1 synthesis when normal COX assembly is compromised, as reported for most COX assembly mutants (23). In all such COX assembly defective mutants, the Cox1 synthesis defect can be rescued when the original mutation is combined with a Δcox14 mutation (23). The Δcmc2Δcox14 strain displayed a wild-type pattern of mitochondrial protein synthesis (Fig. 2D) supporting the fact that Cmc2 is a COX biogenetic factor. Independently of the pleiotropic effects, the lack of spectrally and enzymatically detectable COX and the deficit of the mitochondrially translated subunits in Δcmc2 cells point to COX assembly as the primary target of Cmc2 function.
CMC2 Codes for a Mitochondrial Inner Membrane Protein Facing the Intermembrane Space
To determine subcellular localization of Cmc2, we have generated an affinity purified peptide antibody that recognizes this protein. Cell fractionation and Western blot analyses enabled us to find Cmc2 in the mitochondrial fraction but not in the postmitochondrial supernatant containing cytoplasmic proteins (Fig. 3A). The protein was absent in the Δcmc2 strain (not shown). Sonic irradiation of wild-type mitochondria solubilized cytochrome b2, a soluble protein of the intermembrane space, but not Cmc2 (Fig. 3B). Cmc2, however, was solubilized with alkaline carbonate, suggesting that it is a peripheral membrane protein (Fig. 3C). In these experiments, Shy1 was recovered in the membrane fraction, confirming earlier evidence that it is an intrinsic protein of the inner membrane (25). Cmc2 is located on the intermembrane space side of the inner membrane as evidenced by its resistance to proteinase K in mitochondria but not in mitoplasts prepared by hypotonic swelling of mitochondria (Fig. 3D). Similarly, Sco1, an inner membrane protein previously shown to face the intermembrane space (8), was digested in the mitoplasts but not in mitochondria (Fig. 3D). Mss51, an inner membrane-associated protein facing the matrix, was protected against proteinase K digestion in both mitochondria and mitoplasts (Fig. 3D). As expected, the hypotonic conditions used to disrupt the outer membrane resulted in the loss of cytochrome b2 (Fig. 3D). These results indicate that Cmc2 is membrane bound and faces the intermembrane space. To discern which membrane Cmc2 was bound, we sonicated mitochondria and separated inner and outer membranes by sucrose gradient fractionation. Porin and Cox3 were used as markers of the outer and inner mitochondrial membranes, respectively, to test the purity of the fractions. Similar to Cox3, Cmc2 was found exclusively in the inner membrane fraction (Fig. 3E). Similar results were obtained using a strain expressing either an integrative or an episomal plasmid containing a CMC2 gene fused to a C-terminal hemagglutinin (HA) tag that fully complements the respiratory deficient phenotype of Δcmc2 (not shown) and are consistent with a previously reported localization of a His-tagged version of Cmc2 to the IMS (20). We concluded that Cmc2, similar to its homologue Cmc1, is a mitochondrial protein loosely associated with the inner membrane and facing the intermembrane space.
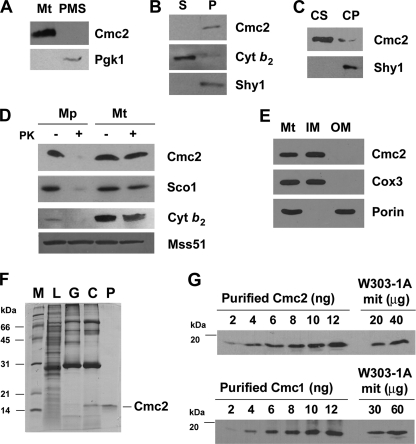
FIGURE 3.
Mitochondrial localization and abundance of Cmc2. A, Cmc2 is a mitochondrial protein. Mitochondria (Mt) and the postmitochondrial supernatant (PMS) fraction were isolated from the W303-1A strain. Samples of the two fractions corresponding to 40 μg of protein were analyzed by Western blotting using antibodies against Cmc2 and the cytosolic marker 3-phosphoglycerate kinase subunit 1 (Pgk1). B, Cmc2 is a membrane protein. Soluble (S) and membrane-bound (P) mitochondrial proteins were separated from 40 μg of total mitochondria by brief/gentle sonication followed by centrifugation at 35,000 × g for 30 min at 4 °C. Samples were analyzed by Western blotting using antibodies against Cmc2, the soluble intermembrane space protein cytochrome b2, and the inner membrane intrinsic protein Shy1. C, Cmc2 is an extrinsic protein. The pellet from B was resuspended in 0.6 m sorbitol, 20 mm HEPES buffer containing 0.1 m Na2CO3, pH 11, and 50 mm EDTA and incubated on ice for 30 min. Centrifugation at 35,000 × g for 30 min at 4 °C allows the separation of the extrinsic proteins present in the supernatant (CS) from the intrinsic proteins in the pellet (CP). The different samples were analyzed by Western blotting using antibodies against Cmc2 and Shy1. D, Cmc2 is a membrane protein facing the intermembrane space. Four aliquots of 40 μg of mitochondrial protein were pelleted and resuspended in buffer containing either 20 mm HEPES or 0.6 m sorbitol, 20 mm HEPES. One aliquot in each buffer was supplemented with a final 100 μg/ml of proteinase K (PK) and incubated on ice for 60 min. The reaction was stopped with 2 mm phenylmethylsulfonyl fluoride. Mitochondria and mitoplasts were recovered by centrifugation at 40,000 × g for 15 min at 4 °C. The pelleted fraction was resuspended in gel buffer and loaded on a 10% Tris Tricine gel. Western blots were probed with antibodies against Cmc2, Sco1, Mss51, and cytochrome b2. E, Cmc2 is an inner mitochondrial membrane protein. Isolated mitochondria were fractionated into inner and outer membranes by sonication plus sucrose gradient sedimentation as described (48). Whole mitochondria (M), inner membranes (IM), and outer membranes (OM) were analyzed by Western blotting using antibodies against porin (outer membrane marker), Cox3 (inner membrane marker), and Cmc2. F, purification of recombinant Cmc2. Recombinant Cmc2 was affinity purified using a combination of GST pulldown and size exclusion chromatography as described under ”Experimental Procedures.“ Subsequent fractions were analyzed by SDS-PAGE and Coomassie staining. L, cell lysate; G, material eluted from the GST column; C, digestion with precision protease to cleave GST off Cmc2; P, size exclusion chromatography yielding purified recombinant Cmc2, respectively. G, concentration of Cmc2 and Cmc1 in wild-type mitochondria. Western blots of the indicated amounts of W303-1A mitochondria and purified Cmc2 (panel F) and Cmc1 (19). The density of the signals was used to estimate the concentration of Cmc1 and Cmc2 in the mitochondrial samples.
Cmc2 and Cmc1 Accumulate in Equimolar Amounts in Wild-type Mitochondria
To estimate the mitochondrial concentration of Cmc1 and Cmc2, we have obtained purified recombinant proteins (Ref. 19 and Fig. 3F) and generated peptide antibodies against each protein. With these tools, we have obtained a standard curve relating known amounts of each purified protein to the signal detected by Western blot analysis (Fig. 3G). The concentration of both proteins in wild-type W303-1A mitochondria was calculated to be of ∼0.2 μg (15 pmol) of Cmc1 or Cmc2/mg of mitochondrial protein. This value is ∼2- and 5-fold lower than the reported estimates for Sco1 (27 pmol/mg of protein (45)) and Cox17 (80 pmol/mg protein (8)), respectively.
Cmc2 Physically Interacts with Cmc1
The similar peripheral association of Cmc2 and Cmc1 with the inner membrane suggested that they could be both components of a higher structure anchored in the membrane. By sucrose gradient analyses, most of the ∼13-kDa Cmc1 was previously shown to be part of a complex of ∼40 kDa only when the Western blot films were overexposed, Cmc1 was also found in higher molecular mass complexes of ∼245 and ∼475 kDa (19). We have now estimated that the amount of Cmc1 that accumulated in these higher molecular mass complexes is less than 5% of total Cmc1 (not shown) and their significance remains to be established. To determine the native molecular weight of Cmc2, we efficiently extracted the protein from the mitochondrial membranes with 0.4% lauryl maltoside and 200 mm KCl (not shown), conditions similar to those required for Cmc1 extraction (19). Subsequently, mitochondrial extracts were used to determine the sedimentation properties of Cmc2 in a 7–20% sucrose gradient. After fractionation, the samples were separated by SDS-PAGE and analyzed by Western blot. The ∼13-kDa Cmc2 protein was detected in a complex of an estimated mass of 40 kDa, co-sedimenting with Cmc1 (Fig. 4A). HA-tagged Cmc1 and Cmc2 proteins had sedimentation patterns similar to the endogenous proteins (not shown). These results suggested that Cmc1 and Cmc2 could be interacting partners.
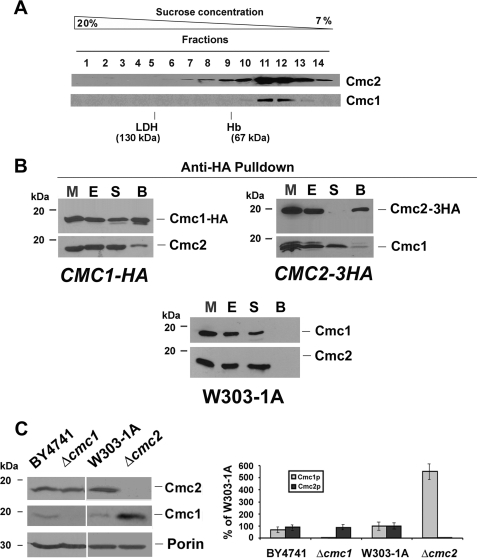
FIGURE 4.
Physical and genetic interaction of Cmc2 and Cmc1. A, sedimentation properties of Cmc1-HA in a linear 7–20% sucrose gradient were analyzed as described under ”Experimental Procedures.“ The gradient was calibrated with hemoglobin (Hb, 67 kDa) and lactate dehydrogenase (LDH, 130 kDa). Following centrifugation the gradient was collected in 14 equal fractions. Each fraction was assayed for Hb by absorption at 410 nm, and LDH activity by measuring NADH-dependent conversion of pyruvate to lactate. The distribution of Cmc1 was assayed by Western blot analysis. The mass of Cmc2 and Cmc1 were determined from the positions of the respective peaks relative to those of the markers. B, Cmc2 physically interacts with Cmc1. Mitochondria extracted from Δcmc1 and Δcmc2 cells expressed from chromosomally integrated plasmids CMC1-HA and CMC2-3HA, respectively, were extracted with 0.4% lauryl maltoside and 200 mm KCl. The extract (E) was used for immunoprecipitation assays using anti-HA-associated Sepharose-4B. The unbound material in the supernatant (S) and the material bound to the beads (B) were separated by centrifugation and analyzed by Western blotting using antibodies against Cmc1 and Cmc2. The bands shown in the Western blots were quantified by densitometry. In the lower panel, the open bars represent the percentage of the corresponding protein bound to the beads, and the filled bars represent the percentage of unbound protein recovered in the supernatant fraction. C, Cmc1 and Cmc2 interact genetically. Steady-state levels of Cmc1 and Cmc2 in the presence and absence of each protein estimated by Western blotting. The graph on the right panel shows the quantification of the Western blot presented on the left panel. The results of two independent experiments did not differ by more than 5%. The bars indicate the mean ±S.D. from three independent sets of measurements.
The possible interaction of Cmc1 with Cmc2 was examined in yeast strains, expressing either Cmc1-HA or Cmc2–3HA in Δcmc1 and Δcmc2 backgrounds, respectively. Pulldown assays of mitochondrial extracts containing Cmc1-HA using anti-HA-associated Sepharose-4B have indicated that ∼60% of Cmc1-HA and 19% of Cmc2 were adsorbed onto the beads (Fig. 4B). Likewise, when crude mitochondrial extracts containing Cmc2–3HA were adsorbed onto anti HA-Sepharose-4B beads, 95% of Cmc2–3HA and 16% of total Cmc1 were pulled down (Fig. 4B). As a negative control, Cmc1 and Cmc2 were not pulled down in a wild-type strain (Fig. 4B). These results suggest that only a fraction of Cmc2 is complexed to Cmc1 or that their interaction is transient. Analyses of the same samples failed to detect other candidates that could interact with Cmc2 such as Cox17, Cox11, and Sco1 (not shown).
Cmc2 Interacts with Cmc1 Genetically
Although Cmc1 and Cmc2 physically interact, neither CMC1 is a multicopy suppressor of Δcmc2 (Fig. 1B) nor did CMC2 overexpression suppress the respiratory defect of Δcmc1 cells (not shown). We have observed, however, that, whereas the steady-state levels of Cmc2 remain unaltered in Δcmc1 cells, the absence of Cmc2 induces a 5–6-fold increase in the steady-state levels of Cmc1 (Fig. 4C). These data suggest that Cmc1 and Cmc2 do not have completely overlapping functions although they act cooperatively.
Exogenous Copper Does Not Suppress the cmc2 Mutant Phenotype
Although the respiratory defect of Δcmc1 cells was suppressed by exogenous copper (19), this was not true for Δcmc2 cells (not shown). Similarly, overexpression of COX copper assembly factors including Cmc1 (Fig. 1B), Cox11, and Sco1 (not shown) was not effective in restoring respiratory growth of Δcmc2 even when copper was supplemented to the medium. Reciprocally, overexpression of CMC2 in strains carrying null alleles of cmc1, sco1, cox11, cox17, cox19, cox23, and pet191 did not rescue the respiratory defect of these strains even in the presence of exogenous copper (not shown).
Mitochondrial CuZn-SOD Activity Is Increased in Δcmc2
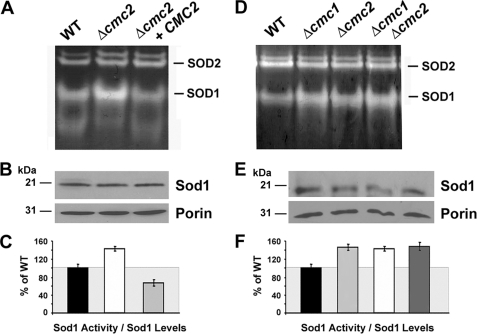
COX and Sod1 have been proposed to use the same mitochondrial matrix copper pool for their metallation (15). We have previously shown that Cmc1 levels regulate the activation of Sod1 (19). In Δcmc1 cells, Sod1 activity is increased, whereas in cells overexpressing Cmc1, Sod1 activity is decreased (19). These observations allowed us to propose that from its location on the IMS face of the inner mitochondrial membrane, Cmc1 could be involved in controlling copper traffic from the matrix to the respective COX and Sod1 metallochaperones located in the IMS (3, 19). Now we have explored whether Cmc2 is involved in a similar role. No significant difference in total (cytoplasmic plus mitochondrial) SOD1 activity was observed between wild-type and Δcmc2 cell extracts (not shown). However, as shown in Fig. 5A, Δcmc2 cells have a significant 1.5-fold increase in the mitochondrial fraction of Sod1 activity as compared with the wild-type despite no increase in the steady-state levels of the protein (Fig. 5B). The activities were normalized to the steady-state levels of Sod1 (Fig. 5C). Interestingly, overexpression of CMC2 in a Δcmc2 strain resulted in a significant decrease in mitochondrial Sod1 activity down to ∼60% of wild-type values, although the steady-state level of the protein remained unchanged (Fig. 5, A–C). To explore whether the effect on mitochondrial SOD1 by Cmc1 and Cmc2 was additive, we estimated SOD1 activity in a double mutant Δcmc1Δcmc2 strain, which showed a similar increase in SOD1 activity as in the single mutants (Fig. 5, D–F). This observation additionally indicates that the absence of Cmc2 governs the levels of Sod1 activity despite the increased Cmc1 protein levels detected in Δcmc2 cells. We can envision that Cmc1 and Cmc2 could cooperate in a pathway controlling the trafficking of copper toward COX and thus indirectly affecting Sod1 metallation.
FIGURE 5.
Mitochondrial superoxide dismutase 1 activity depends upon Cmc2 levels. A, SOD activity in mitochondria isolated from wild-type, Δcmc2 mutant, and CMC2 overexpressing cells. B, the same samples were solubilized in SDS-sample buffer and separated on a 12% PAGE gel. Mitochondrial Sod1 and porin, a loading control, were visualized by immunoblotting. C, quantification of experiment in panels A and B. D and E, similar to A–C but including mitochondria isolated from wild-type, Δcmc1, and Δcmc2 single mutants and a Δcmc1Δcmc2 double mutant strain. The images were digitalized and densitometry was performed using the histogram function of the Adobe Photoshop program. Data from three independent experiments were included. *, indicates p < 0.01.
Cmc2 Is Conserved from Yeast to Human
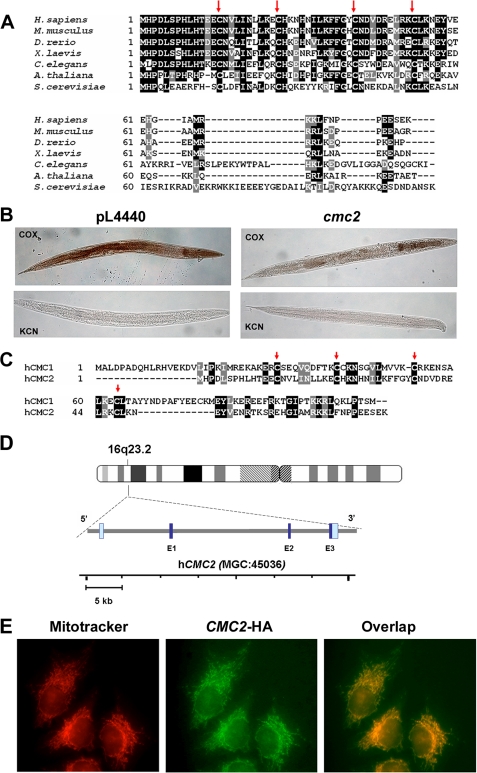
COX biogenesis factors containing the twin Cx9C motif, including Cox17, Cox19, Cox23, and Pet191, are conserved from yeast to humans (46). Similarly, we previously showed that Cmc1 is a conserved protein that localizes to mitochondria in human cells (19). The S. cerevisiae Cmc2 also has homologues in mammals, fruit flies, worms, plants, and other fungi. All homologues contain the signature conserved twin Cx9C motif as indicated by the red arrows in Fig. 6A.
FIGURE 6.
Properties and expression of metazoans homologues of yeast CMC2. A, CMC2 is conserved across kingdoms, from yeast to human. Sequence alignment of S. cerevisiae CMC2 homologues from human (Homo sapiens), fish (Danio rerio), frog (Xenopus laevis), fruit fly (Drosophila melanogaster), worm (C. elegans), and plant (Arabidopsis thaliana). Red arrows indicate conserved cysteine residues. B, histochemical staining for cytochrome c oxidase activity in cmc2-silenced C. elegans. Worms fed HT115DE(3) bacteria transformed with empty vector pL4440 were used as negative controls. Silenced (cmc2) and control (pL4440) rrf-3(pk1426) nematodes were fixed, permeabilized, and stained for COX. Control reactions contained the COX-specific inhibitor KCN. Photographs of representative examples of the whole controls and silenced nematodes are shown. C, sequence alignment of human CMC1 and CMC2 proteins. D, localization and genomic structure of the human hCMC2 gene based on the result of a BLAST search of the human genome for homology to the hCMC2 cDNA sequence. The dark blue and gray bars depict the exon and intron regions, respectively. E, subcellular localization of CMC2 in human HeLa cells. Cells were transiently transfected with a cDNA coding for a hemagglutinin-tagged CMC2. The protein was visualized by indirect immunofluorescence using Mitotracker Red and a fluorescently conjugated antibody to human to the hemagglutinin epitope of CMC2-HA (green). A merged image is shown on the right.
We have used the free-living soil nematode, C. elegans, to investigate the role of CMC2 in a multicellular organism. The worm CMC2 protein shares 28% identity and 49% similarity with its yeast counterpart (Fig. 6A). We developed a transient knockdown model of CMC2 deficiency by RNAi technology and used it to investigate the levels of COX activity in silenced animals by histochemical staining. Control and cmc2 knockdown worms were fixed and stained for COX activity. The COX-dependent oxidation of diaminobenzidine produces a brown stain that is more evident in the pharyngeal muscles and intestine, with a fainter staining in the body wall muscles; lack of color indicates the absence of activity. The specificity of the reaction was ascertained by performing parallel experiments with animals treated with 10 mm KCN to inhibit COX activity. COX activity was strongly reduced in both the intestine and body wall of L3 larvae from cmc2-silenced nematodes, and decreased to a lesser extent in the pharyngeal muscles (Fig. 6B).
A human cDNA clone (IMAGE 5399193) codes for a putative homologue of yeast Cmc2 and was obtained for further studies. As in yeast, the BLAST best reciprocal match of human CMC2 is CMC1, proteins that share significant similarity (Fig. 6C). The cDNA sequence was mapped to locus MGC45036 on chromosome 16q23.2. The human gene, here referred to as hCMC2, consists of 3 exons separated by 2 introns (Fig. 6D). The subcellular location of hCMC2 was studied by transient expression of the protein tagged with a C-terminal HA epitope in human HeLa cells. Immunohistochemical assays for the HA epitope in the transformed cells show a typical mitochondrial network pattern that co-localized with Mitotracker Red staining (Fig. 6E).
DISCUSSION
Biogenesis of COX and Sod1 in the mitochondrial intermembrane space requires copper trafficking from the mitochondrial matrix pool to the intermembrane space chaperones involved in metal delivery to these enzymes. Although an inner membrane transporter has not yet been identified, several proteins have been proposed to play a role in a putative pathway leading to copper delivery to Cox17. These include soluble proteins Cox23 and Cox19 and the inner membrane-bound protein Cmc1. A common characteristic of all these proteins is that they contain a twin Cx9C structural motif, which is necessary for their import through the Mia40 pathway (20, 47). Cmc1 levels were shown to regulate activation of Sod1, which suggested that Cmc1 could participate directly or indirectly in regulating copper distribution toward COX and Sod1. In this study, we show that Cmc2, a twin Cx9C conserved protein and best BLAST reciprocal match of Cmc1, physically and genetically interacts with Cmc1, is essential for COX biogenesis, and modulates the levels of functional mitochondrial Sod1.
CMC2 codes for a COX biogenesis factor. Although cmc1 null mutants produced 20–40% of the fully assembled and functional COX depending on the genetic background (19), Δcmc2 cells display a complete absence of COX, similar to most COX assembly mutants.
Although the precise function of Cmc2 is unknown, it is tempting to propose that it could be an upstream element of a pathway regulating copper delivery to COX. Several pieces of information could support this hypothesis. 1) Cmc2 shares homology with Cox17 and other IMS proteins containing twin Cx9C structural motifs required for COX assembly, including Cox19 and Cox23, and particularly with Cmc1. 2) The identical submitochondrial localization of Cmc1 and Cmc2, the physical interaction detected between the two proteins, and the fact that Cmc1 is overexpressed in the absence of Cmc2 would suggest the possibility that both proteins play cooperative functions. The partial COX assembly defect of cmc1 mutants could be explained by a redundant function with a yet uncharacterized protein or if Cmc1 played a role in catalyzing or regulating the function of another protein, such as Cmc2.
We have reported that the respiratory defect of Δcmc1 cells is rescued by copper supplementation to the medium as previously shown for cox17 mutants (7) and cox23 mutants overexpressing COX17 (21), which suggested a role of Cmc1 in mitochondrial copper homeostasis (19). In Δcmc2 cells, exogenous copper does not suppress their respiratory growth defect. In addition, CMC2 overexpression does not suppress the COX assembly defect of Δcmc1 cells. Reciprocally, CMC1 overexpression does not restore respiratory growth and COX assembly to Δcmc2 cells, even when grown in the presence of exogenous copper. These results are taken to conclude that Cmc1 and Cmc2 play cooperative but nonredundant functions in COX biogenesis.
A redox switch-dependent copper transfer mechanism has been proposed for the relevant pair of proteins leading to metallation of the two copper enzymes in the IMS, COX and Sod1. Copper transfer from human COX17 to SCO1 is coupled to electron transfer in vitro (14). The transfer of copper from SCO1 to COX2 has been proposed to occur in a reaction involving redox changes in cysteine thiols present in the copper binding domain (49). In humans, SCO1 and SCO2 are both required for copper transfer to COX2, and SCO2 additionally acts as a thiol-disulfide oxidoreductase to oxidize the copper-binding cysteines of SCO1 (50). Concerning Sod1 activation, the chaperone Ccs1 forms a Ccs1-Sod1 heterodimer that facilitates copper transfer (51, 52). Ccs1-mediated copper insertion into Sod1 also catalyzes the formation of a critical disulfide bond in Sod1, which seems to be important for regulation of enzyme activity and prevention of misfolding or aggregation (53). The pathways leading to transfer of copper from the matrix pool to the metallochaperones Cox17 and Ccs1 are unknown but could involve similar mechanisms. Given the growing number of mitochondrial IMS Cx9C proteins relevant for COX assembly, we and others have speculated that they could be part of the same pathway directly or indirectly facilitating copper transfer to COX. Interestingly, the steady-state levels of at least two of these proteins, Cmc1 (19) and Cmc2 reported here, modulate mitochondrial Sod1 activation. These observations could fit in a model in which the Cmc1/2 proteins are involved in facilitating copper trafficking from the matrix copper pool to the IMS and subsequently to COX. In the absence of Cmc1 and Cmc2, copper flow toward COX would be limited and the metal more readily available for Sod1 activation. However, at this stage, it remains unclear whether Cmc1 and Cmc2 participate in copper homeostasis, redox homeostasis, both, or play some other role in COX biogenesis.
In conclusion, in this work we have described Cmc2, a twin Cx9C protein partner of Cmc1, as a COX biogenesis factor that acts in concert with Cmc1 to additionally affect mitochondrial Sod1 activity. Cmc2 homologues are absent in bacteria but exist in other fungi, plants, and animals, including humans. Two pieces of evidence supported the functional equivalence of the S. cerevisiae Cmc2 and homologues from multicellular organisms, from invertebrates to mammals. 1) Silencing of cmc2 in the nematode C. elegans resulted in a COX deficiency phenotype and 2) HA-tagged human CMC2 co-localizes with a mitochondrial marker in human HeLa cells. The human gene must be considered as a new candidate when screening for mutations responsible for mitochondrial disorders associated with COX deficiency. Future work will focus on determining whether human CMC2 and CMC1 physically interact and functionally cooperate in COX and mitochondrial Sod1 biogenesis in a way similar to their yeast counterparts.
Acknowledgment
We thank Dr. K. Hell for providing antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant GM071775A (to A. B.), a Research Grant from the Muscular Dystrophy Association (to A. B.), Italian Telethon Grant GGP06256 (to L. S.), and an American Heart Association Predoctoral Fellowship (to D. H.).
- IMS
- intermembrane space
- COX
- cytochrome c oxidase
- Sod1
- copper-zinc superoxide dismutase
- Sod2
- manganese superoxide dismutase
- HA
- hemagglutinin
- NCCR
- NADH cytochrome c reductase
- GST
- glutathione S-transferase
- SOD
- superoxide dismutase
- RNAi
- RNA interference
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1995) Science 269, 1069–1074 [DOI] [PubMed] [Google Scholar]
- 2.Sturtz L. A., Diekert K., Jensen L. T., Lill R., Culotta V. C. (2001) J. Biol. Chem. 276, 38084–38089 [DOI] [PubMed] [Google Scholar]
- 3.Horn D., Barrientos A. (2008) IUBMB Life 60, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobine P. A., Pierrel F., Winge D. R. (2006) Biochim. Biophys. Acta 1763, 759–772 [DOI] [PubMed] [Google Scholar]
- 5.Culotta V. C., Klomp L. W., Strain J., Casareno R. L., Krems B., Gitlin J. D. (1997) J. Biol. Chem. 272, 23469–23472 [DOI] [PubMed] [Google Scholar]
- 6.Field L. S., Furukawa Y., O'Halloran T. V., Culotta V. C. (2003) J. Biol. Chem. 278, 28052–28059 [DOI] [PubMed] [Google Scholar]
- 7.Glerum D. M., Shtanko A., Tzagoloff A. (1996) J. Biol. Chem. 271, 14504–14509 [DOI] [PubMed] [Google Scholar]
- 8.Beers J., Glerum D. M., Tzagoloff A. (1997) J. Biol. Chem. 272, 33191–33196 [DOI] [PubMed] [Google Scholar]
- 9.Glerum D. M., Shtanko A., Tzagoloff A. (1996) J. Biol. Chem. 271, 20531–20535 [DOI] [PubMed] [Google Scholar]
- 10.Hiser L., Di Valentin M., Hamer A. G., Hosler J. P. (2000) J. Biol. Chem. 275, 619–623 [DOI] [PubMed] [Google Scholar]
- 11.Carr H. S., George G. N., Winge D. R. (2002) J. Biol. Chem. 277, 31237–31242 [DOI] [PubMed] [Google Scholar]
- 12.Carr H. S., Maxfield A. B., Horng Y. C., Winge D. R. (2005) J. Biol. Chem. 280, 22664–22669 [DOI] [PubMed] [Google Scholar]
- 13.Horng Y. C., Cobine P. A., Maxfield A. B., Carr H. S., Winge D. R. (2004) J. Biol. Chem. 279, 35334–35340 [DOI] [PubMed] [Google Scholar]
- 14.Banci L., Bertini I., Ciofi-Baffoni S., Hadjiloi T., Martinelli M., Palumaa P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobine P. A., Ojeda L. D., Rigby K. M., Winge D. R. (2004) J. Biol. Chem. 279, 14447–14455 [DOI] [PubMed] [Google Scholar]
- 16.Cobine P. A., Pierrel F., Bestwick M. L., Winge D. R. (2006) J. Biol. Chem. 281, 36552–36559 [DOI] [PubMed] [Google Scholar]
- 17.Rigby K., Zhang L., Cobine P. A., George G. N., Winge D. R. (2007) J. Biol. Chem. 282, 10233–10242 [DOI] [PubMed] [Google Scholar]
- 18.Khalimonchuk O., Rigby K., Bestwick M., Pierrel F., Cobine P. A., Winge D. R. (2008) Eukaryot. Cell 7, 1427–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn D., Al-Ali H., Barrientos A. (2008) Mol. Cell. Biol. 28, 4354–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., Westermann B., Herrmann J. M., Riemer J. (2009) J. Mol. Biol. 393, 356–368 [DOI] [PubMed] [Google Scholar]
- 21.Barros M. H., Johnson A., Tzagoloff A. (2004) J. Biol. Chem. 279, 31943–31947 [DOI] [PubMed] [Google Scholar]
- 22.Nobrega M. P., Bandeira S. C., Beers J., Tzagoloff A. (2002) J. Biol. Chem. 277, 40206–40211 [DOI] [PubMed] [Google Scholar]
- 23.Barrientos A., Zambrano A., Tzagoloff A. (2004) EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers A. M., Pape L. K., Tzagoloff A. (1985) EMBO J. 4, 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrientos A., Korr D., Tzagoloff A. (2002) EMBO J. 21, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faye G., Kujawa C., Fukuhara H. (1974) J. Mol. Biol. 88, 185–203 [DOI] [PubMed] [Google Scholar]
- 27.Tzagoloff A., Akai A., Needleman R. B. (1975) J. Biol. Chem. 250, 8228–8235 [PubMed] [Google Scholar]
- 28.Flohé L., Otting F. (1984) Methods Enzymol. 105, 93–104 [DOI] [PubMed] [Google Scholar]
- 29.Wood W. B.the Community of C. elegans Researchers (eds) (1988) The Nematode Caenorhabditis elegans, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30.Timmons L., Court D. L., Fire A. (2001) Gene 263, 103–112 [DOI] [PubMed] [Google Scholar]
- 31.Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. (2001) Genome Biol. 2, RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rea S. L., Ventura N., Johnson T. E. (2007) PLoS Biol. 5, e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie G., Jia Y., Aamodt E. (1995) Genet. Anal. 12, 95–100 [DOI] [PubMed] [Google Scholar]
- 34.Grad L. I., Sayles L. C., Lemire B. D. (2007) Methods Mol. Biol. 372, 51–66 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 36.Schiestl R. H., Gietz R. D. (1989) Curr. Genet. 16, 339–346 [DOI] [PubMed] [Google Scholar]
- 37.Rothstein R. J. (1983) Methods Enzymol. 101, 202–211 [DOI] [PubMed] [Google Scholar]
- 38.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 39.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 40.Mashkevich G., Repetto B., Glerum D. M., Jin C., Tzagoloff A. (1997) J. Biol. Chem. 272, 14356–14364 [DOI] [PubMed] [Google Scholar]
- 41.Nobrega M. P., Nobrega F. G., Tzagoloff A. (1990) J. Biol. Chem. 265, 14220–14226 [PubMed] [Google Scholar]
- 42.Hell K., Tzagoloff A., Neupert W., Stuart R. A. (2000) J. Biol. Chem. 275, 4571–4578 [DOI] [PubMed] [Google Scholar]
- 43.Soto I. C., Fontanesi F., Valledor M., Horn D., Singh R., Barrientos A. (2009) Biochim. Biophys. Acta 1793, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevtzoff C., Yoboue E. D., Galinier A., Casteilla L., Daignan-Fornier B., Rigoulet M., Devin A. (2010) J. Biol. Chem. 285, 1733–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beers J., Glerum D. M., Tzagoloff A. (2002) J. Biol. Chem. 277, 22185–22190 [DOI] [PubMed] [Google Scholar]
- 46.Solans A., Zambrano A., Barrientos A. (2004) Preclinica 2, 336–348 [Google Scholar]
- 47.Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. (2005) Cell 121, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 48.Manthey G. M., Przybyla-Zawislak B. D., McEwen J. E. (1998) Eur. J. Biochem. 255, 156–161 [DOI] [PubMed] [Google Scholar]
- 49.Banci L., Bertini I., Calderone V., Ciofi-Baffoni S., Mangani S., Martinelli M., Palumaa P., Wang S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leary S. C., Sasarman F., Nishimura T., Shoubridge E. A. (2009) Hum. Mol. Genet. 18, 2230–2240 [DOI] [PubMed] [Google Scholar]
- 51.Lamb A. L., Torres A. S., O'Halloran T. V., Rosenzweig A. C. (2000) Biochemistry 39, 14720–14727 [DOI] [PubMed] [Google Scholar]
- 52.Lamb A. L., Torres A. S., O'Halloran T. V., Rosenzweig A. C. (2001) Nat. Struct. Biol. 8, 751–755 [DOI] [PubMed] [Google Scholar]
- 53.Furukawa Y., Torres A. S., O'Halloran T. V. (2004) EMBO J. 23, 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzagoloff A., Akai A., Needleman R. B., Zulch G. (1975) J. Biol. Chem. 250, 8236–8242 [PubMed] [Google Scholar]
- 55.Barrientos A. (2002) Methods 26, 307–316 [DOI] [PubMed] [Google Scholar]