Abstract
The 90-kDa heat shock protein (Hsp90) is involved in the regulation and activation of numerous client proteins essential for diverse functions such as cell growth and differentiation. Although the function of cytosolic Hsp90 is dependent on a battery of cochaperone proteins regulating both its ATPase activity and its interaction with client proteins, little is known about the real Hsp90 molecular mechanism. Besides its highly flexible dimeric state, Hsp90 is able to self-oligomerize in the presence of divalent cations or under heat shock. In addition to dimers, oligomers exhibit a chaperone activity. In this work, we focused on Mg2+-induced oligomers that we named Type I, Type II, and Type III in increasing molecular mass order. After stabilization of these quaternary structures, we optimized a purification protocol. Combining analytical ultracentrifugation, size exclusion chromatography coupled to multiangle laser light scattering, and high mass matrix-assisted laser desorption/ionization time of flight mass spectrometry, we determined biochemical and biophysical characteristics of each Hsp90 oligomer. We demonstrate that Type I oligomer is a tetramer, and Type II is an hexamer, whereas Type III is a dodecamer. These even-numbered structures demonstrate that the building brick for oligomerization is the dimer up to the Type II, whereas Type III probably results from the association of two Type II. Moreover, the Type II oligomer structure, studied by negative stain transmission electron microscopy tomography, exhibits a “nest-like” shape that forms a “cozy chaperoning chamber” where the client protein folding/protection could occur.
Keywords: Chaperone Chaperonin, Dynamic Light Scattering, Electron Microscopy (EM), HPLC, Mass Spectrometry (MS), Protein Structure, Ultracentrifugation, Characterization, Hsp90, Oligomers
Introduction
Molecular chaperones are essential for the correct folding of neo-synthesized proteins in vivo, from their emergence from the ribosomal tunnel to their acquisition of a functional state. Among chaperone proteins, the 90-kDa heat shock protein (Hsp90)2 is one of the most abundant proteins in the cytosol and is essential for cell survival (1). The Hsp90 functions require the cooperation of cochaperones and other chaperones within macro complexes (2). In contrast to most of the chaperone protein families, Hsp90 is not involved in de novo folding but operates at later stages of the folding process, assisting the maturation and activation of numerous client proteins, including steroid receptors and cell cycle kinases (3, 4). The effectiveness of Hsp90 in protecting and activating such proteins has led researchers to focus on the Hsp90 hub as a target for anti-cancer drugs (5–7). Hsp90 is ubiquitous and shows an extraordinary conservation from bacteria to higher eukaryotes. In the cell, Hsp90 amounts to 1–2% of the total proteins under nonstress conditions depending on the organism and cell type (2–25 mg/ml). Hsp90 is overexpressed under stress conditions such as heat or hypoxia (8–10). In eukaryotes, the cytosol contains two isoforms, α and β, encoded by distinct genes (11). These isoforms coexist in the cytosol; Hsp90β is constitutively expressed, whereas Hsp90α is stress-inducible. The α and β isoforms have 85% sequence identity and are associated as homodimers (12, 13).
At the structural level, Hsp90 is composed of three distinct domains: the N-terminal domain contains the common binding site for ATP and geldanamycin; the middle domain is implicated in ATP γ-phosphate binding/hydrolysis and client proteins binding (14, 15); and the C-terminal domain is mainly involved in the dimerization process and also in the binding of client proteins (16). The domains are linked via large hinge loop regions conferring both an important intrinsic flexibility and an interdomain communication within the Hsp90 dimer (17, 18). The cryo-EM structures of apo-Hsp90 revealed that the protein coexists as two conformations, a fully open “flying seagull” and a semi-open V shape (19). Furthermore, it is clear that Hsp90 flexibility is crucial for the course of its chaperoning cycle, because ATP binding seems to drive large conformational changes (18, 20–22). Besides the influence of nucleotide and/or ligand binding on its structure, the Hsp90 dimer also undergoes some large conformational changes under heat shock or in the presence of divalent cations, leading to oligomerization. Hsp90 oligomerization is a highly ordered process, and heat-induced oligomers are sensitive to modulators such as ATP and geldanamycin, suggesting a real function in the chaperoning cycle (23, 24). Hsp90 oligomers have also been observed under nonheated conditions, suggesting a basal oligomer level in the cell under physiological conditions (25, 26). We previously demonstrated that oligomers are formed in the presence of divalent cations that induce an increase of the compactness of the Hsp90 dimer, leading to its self-association (27). A recent small angle x-ray scattering study has highlighted the presence of tetramers for HtpG and eukaryote homologues, amounting to 10–20% of the quaternary structures present in solution. The V-shaped dimer would be more prone than the flying seagull to form an apo tetramer (28). The magnitude of the Hsp90 structural modifications, with respect to the flexibility of the dimer as well as the oligomerization process, enables Hsp90 to adapt to its broad spectrum of client proteins (25). Concerning their functional aspects, oligomers were able to maintain dihydrofolate reductase in a folding-competent state and to prevent firefly luciferase thermal denaturation (29). A comparative study of oligomerization of Hsp90 homologues showed that oligomerization is a prerequisite for chaperoning under stress conditions and that HtpG, Hsp90, and Grp94 oligomers were able to prevent glutathione S-transferase precipitation only in the oligomeric state (26). Moreover, it has been suggested that Hsp90 heat-induced oligomerization would be accompanied by an auto-chaperone process (30). Because Hsp90 oligomers play a chaperone function, a clear understanding of the role of oligomerization in the chaperoning cycle of the Hsp90 machinery is crucial. Furthermore, some discrepancies remain concerning the degrees of oligomerization. Little is known regarding the biochemical and biophysical properties of Hsp90 oligomers, and even less is known about their structures.
This work focuses on magnesium-induced Hsp90 oligomers. The optimization of experimental conditions reveals several oligomer types that we have termed Type I, Type II, and Type III in increasing order of molecular mass. Once the oligomers were stabilized, the prerequisite of this work was to obtain pure samples of each species in suitable quantity for their biochemical and biophysical characterization. For this purpose, we have designed and optimized several protocols based on their molecular mass difference; we used both electroelution from native-PAGE and size exclusion chromatography. For the first time, we were able to purify distinct Hsp90 oligomeric species. We have elucidated the self-association stoichiometries of Hsp90 magnesium-induced oligomers by combining complementary techniques such as analytical ultracentrifugation (AUC), size exclusion chromatography coupled to multiangle laser light scattering (SEC-MALLS), and high mass MALDI-TOF MS. We demonstrate that this self-assembly process involves the dimer as the basic unit up to the Type II oligomer, whereas Type III results from the association of two Type II oligomers. An understanding of the magnesium-induced oligomerization process allows us to explain the temperature-dependent oligomerization and reinforces the hypothesis that oligomers are involved in auto-chaperone activity. We initiated the first structural study by electron tomography on negatively stained specimens, and the reconstruction of the structure obtained for the Type II displays a typical shape reminiscent of a “cozy nest.”
EXPERIMENTAL PROCEDURES
Hsp90 Purification
Hsp90 was purified from pig brains according to the method of Yonezawa et al. (31) modified by Garnier et al. (23, 32). The samples were flash frozen in liquid nitrogen and stored at −80 °C. Protein concentration was determined by measuring UV absorption with a molar absorption coefficient (280 nm) of 124,000/M·cm in 10 mm MES-NaOH buffer, pH 6.5, considering that Hsp90 is a dimer. The absorption was corrected for light scattering using Beckman DU640B spectrophotometer software.
Purification of Magnesium-induced Hsp90 Oligomers
Sample Preparation
Hsp90 (54 μm) in 10 mm MES-NaOH buffer, pH 6.5, was supplemented with 8 mm MgCl2 (Sigma-Aldrich) and preincubated for 10 min at 25 °C. Chemical cross-linking was performed using N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (Sigma-Aldrich) at a final concentration of 2.3 mm for 30 min at 25 °C, and the Hsp90 concentration was 46 μm. The reaction was stopped by adding Tris quenching buffer (750 mm Tris-HCl, 8 mm MgCl2, 150 mm NaCl, pH 7.5). A 20-fold excess of amine containing buffer is enough to quench the reaction.
Electroelution from Native-PAGE
The prepared sample was supplemented with ⅓ Coomassie Brilliant Blue G250 2%, glycerol 30% and applied (10 × 30 μl) on a 6% native-polyacrylamide gel. Migration was performed in Tris-glycine buffer for 2.5 h at 20 mA/gel (33). Bands corresponding to each oligomer type were cut and placed in micro baskets adaptable to YM50 Centricon (Millipore). Centricons were adapted to an electroelutor system (Bio-Rad). Extensively degassed Tris-glycine buffer was used for migration (4 h at 2.5 mA/Centricon). Centricons were then centrifuged 2,000 × g, 30 min, 4 °C. The resulting concentrate was ultracentrifuged 100,000 × g, 5 min, 4 °C on a Beckman TL100. Aliquots of supernatant (50 μl) were used for electron microscopy studies.
Large Scale Purification by Size Exclusion Chromatography
A 500-μl cross-linked and quenched sample (final concentration, 29 μm) was loaded on a Superdex 200 size exclusion chromatography column, 1 cm × 210 cm (90/60/30/30 cm Tricorn columns in series, GE Healthcare) equilibrated in 10 mm MES-NaOH, 8 mm MgCl2, 150 mm NaCl, pH 6.5, with chromatography conditions controlled by an HPLC pump system (625 LC System, Waters). The column was eluted at a flow rate of 0.5 ml/min at room temperature, the eluate was monitored at 280 nm (2996 photodiode Array detector, Waters), and fractions of 500 μl were collected between 130 and 180 min of elution time. Fractions corresponding to each oligomer type were selected and stored at −80 °C. Corresponding fractions from identical runs were pooled, concentrated on an Amicon Ultra YM30 (Millipore), and stored at −80 °C. The columns were standardized with a calibration kit of proteins with known molecular masses and Stokes radii (GE Healthcare) (34, 35), using thyroglobulin (Mr = 669 kDa, Rs = 85 Å), ferritin (Mr = 440 kDa, Rs = 61 Å), and catalase (Mr = 232 kDa, Rs = 52.2 Å). Blue dextran 2000 (GE Healthcare) was used to determine the column exclusion volume.
SDS-PAGE
The samples were supplemented with Laemmli buffer (36) and loaded onto polyacrylamide 3–8% gradient gel. Migration was performed in SDS-Tris-glycine buffer at 100 V. The gels were stained with Coomassie Brilliant Blue. Molecular mass markers were from the high molecular weight-SDS marker kit (GE Healthcare).
Native-PAGE
The samples were supplemented with buffer containing Coomassie Blue G-250 in 30% glycerol and loaded onto 6/8% double layer polyacrylamide gels. Migrations were performed in Tris-glycine buffer over 2.5 or 3 h at 20 mA/gel. The gels were stained using EZ-Blue (Sigma-Aldrich). Molecular mass markers were purchased from GE Healthcare.
Analytical Ultracentrifugation
Sedimentation Velocity
Hsp90 oligomers samples were prepared as described above. Concentrated SEC-HPLC fractions 128–133, 134–139, and 145–150, corresponding to Type III, Type II, and Type I oligomers, respectively, were analyzed by sedimentation velocity. Protein concentrations ranged between 0.03 and 0.19 mg/ml for Type III oligomer, 0.22 and 0.32 mg/ml for Type II oligomer, and 0.29 and 0.46 mg/ml for Type I oligomer. Before all of the experiments, the samples were centrifuged at 100,000 × g (5 min) to remove potential aggregates formed during freezing (Air-Fuge ultracentrifuge; Beckman). The experiments were carried out at 20 °C in a Beckman Optima-XL-A analytical ultracentrifuge, using 12-mm Epon double sector centerpieces in an AN55Ti rotor. The scans were acquired in continuous mode at wavelengths selected for appropriate signals in the range of 0.1 to 1 absorption, i.e. 230 nm/20,000 rpm for Type III oligomer, 240 nm/20,000 rpm for Type II oligomer, and 280 nm/30,000 rpm for Type I oligomer. Hsp90 extinction coefficients were calculated for all of the wavelengths tested: 0.7337, 0.9538, and 4.8727 liter/g·cm at 280, 240, and 230 nm, respectively. Apparent sedimentation coefficients were determined using SEDFIT (37) and were corrected to standard conditions using SEDNTERP (38). Examining the sedimentation coefficient distributions curves generated in SEDFIT for Type I and Type II oligomers, the purest fractions (i.e. thinnest peaks) were chosen to determine the sedimentation coefficient at standard conditions and infinite dilution. Then fractions 138–139 (STII) and 147–148 (STI) were diluted in 10 mm MES-NaOH, 8 mm MgCl2, 150 mm NaCl, pH 6.5, to obtain final concentrations of 0.10, 0.20, and 0.30 mg/ml for Type I oligomer and 0.11, 0.16, and 0.22 mg/ml for Type II oligomer and run at 25,000 rpm at 20 °C. The scans were acquired in continuous mode at 230 nm.
Sedimentation Equilibrium
The experiments were performed at 4 °C with the same analytical ultracentrifuge. The measurements were done at three successive speeds (3400, 4100, and 6000 rpm) by taking scans at the appropriate wavelengths (230 and 280 nm), when sedimentation equilibrium was reached. High sedimentation (40,000 rpm) was conducted afterward for base-line correction. At 4 °C, the values of partial specific volume of Hsp90, solvent density, and viscosity, calculated with SEDNTERP, were 0.7273 ml/g, 1.00673 g/ml, and 0.01567 poise, respectively. The average molecular speed masses were determined by fitting a sedimentation equilibrium model for a single solute to individual data sets with EQASSOC programs supplied by Beckman (39). Data analysis was also performed by global analysis of several data sets obtained at different loading concentrations and speeds using the Sedphat program (40).
Multiangle Laser Light Scattering
MALLS experiments were carried out using an HPLC system (Waters) combined with a DAWN EOS System (Wyatt Technology Corp) at room temperature. Elution was monitored at 280 nm by the photodiode array detector 2996 (Waters) and a refractive index detector (2414, Waters). STI, STII, and STIII samples were injected on a double size exclusion chromatography apparatus composed of two Tricorn columns (GE Healthcare): a 1 cm × 60 cm Superdex 200 (GE Healthcare) coupled to a 1 cm × 16 cm Sepharose 6B (Sigma Aldrich). Elution was performed with 10 mm MES-NaOH, 8 mm MgCl2, and 150 mm NaCl, pH 6.5, at a flow rate of 0.5 ml/min. The data were collected and processed with Astra 4.90.08 software using the Debye fit method with a dn/dc set to 0.19 ml/g, typical of nonglycosylated proteins.
High Mass MALDI-TOF Mass Spectrometry
Sample Preparation
STI (1.21 mg/ml), STII (0.85 mg/ml), and Type III SEC-HPLC individually eluted fractions 129 (0.665 mg/ml), 130 (1.16 mg/ml), and 131 (2.42 mg/ml) were diluted in K200 cross-linking buffer (CovalX) from ½ to in a 10 μl of final volume. For control experiments, 1 μl of each mixture was mixed with 1 μl of a sinapinic acid matrix (10 mg/ml) in acetonitrile/water (1:1, v/v), trifluoroacetic acid 0.1%. After mixing, 1 μl of each sample was spotted on the MALDI plate (SCOUT 384; AnchorChip). The analysis was repeated in triplicate. After crystallization at room temperature, the plate was introduced in the MALDI mass spectrometer. The remaining 9 μl of the sample were submitted to cross-linking using a CovalX K200 MALDI MS analysis kit (CovalX AG, Zurich, Switzerland). Each sample was mixed with 1 μl of K200 stabilizer reagent (2 mg/ml) and incubated at room temperature. After 1, 3, or 6 h of incubation time, the samples were prepared for MALDI analysis as for control experiments.
Data Acquisition
The samples were analyzed by high mass MALDI-TOF immediately after crystallization. The analysis was performed using the standard nitrogen laser and focusing on different mass ranges from 0 to 1200 kDa. High mass MALDI mass spectra were obtained using a 4800 MALDI-TOF TOF instrument (Applied Biosystems, Foster City, CA) equipped with a HM2 high mass retrofit system (CovalX AG, Zürich, Switzerland). The instrument was operated in linear and positive mode by applying an accelerating voltage of 20 kV. The HM2 system was operated in the high mass mode with acceleration voltage set to 20 kV and gain voltage set to between 2.85 and 3.25 kV. The instrument was calibrated using clusters of insulin, bovine serum albumin, and IgG. For each sample, three spots were analyzed (200 laser shots/spot). The presented spectra correspond to the average of 200 laser shots. The MS data were analyzed using Complex Tracker analysis software.
Electron Tomography
Negative Staining
Electroeluted sample of Type II (∼ 0.1 mg/ml) oligomer was applied for 30 s to collodion/carbon-coated copper grids previously glow-discharged, rinsed with water or 0.05% glutaraldehyde, and stained with 1% uranyl acetate for 1 min.
Image Acquisition and Structure Reconstruction
Tilt series were acquired on a Tecnai G2 Sphera electron microscope (FEI Company, Eindhoven, The Netherlands) operating at 200 kV. Digital images were automatically recorded on a CCD camera (charge-coupled device camera; Megascan) at ×25,000 nominal magnification (pixel size, 0.46 nm) over a tilt range of −60 to +60 with 1° increments. The images were aligned, and tomograms were calculated by the ART reconstruction algorithm (41) using the plug-in TomoJ (42) integrated in the ImageJ software (National Institutes of Health, Bethesda, MD) (43). The three-dimensional volumes were imported into the three-dimensional visualization software AMIRA (Mercury Computer Systems, Mérignac, France).
RESULTS
Magnesium-induced Oligomers Have Temperature-dependent Counterparts
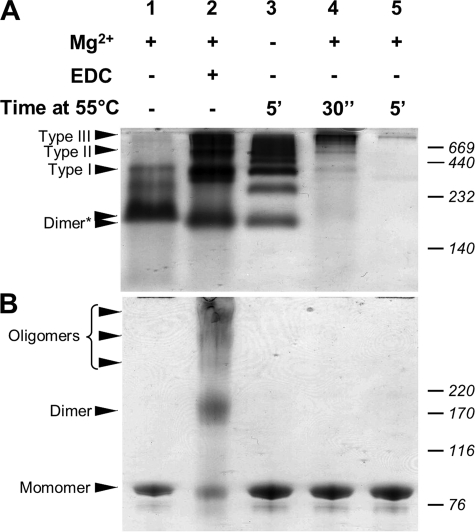
As we previously demonstrated, Hsp90 dimer undergoes a Mg2+-dependent oligomerization process involving the C-terminal domain. Indeed, magnesium (i) induces a conformational change within the Hsp90 dimer, making it more compact and (ii) leads to the oligomerization process (27). Hsp90 chaperone protein also self-oligomerizes under stress temperature (23, 24). To compare these oligomerization processes, we performed parallel studies with native and SDS-PAGE (Fig. 1). Native-PAGE analysis of Hsp90 preincubated in the presence of magnesium showed a major band for the dimer and very low intensity bands corresponding to oligomers (Fig. 1A, lane 1). Although magnesium induced an oligomerization process, these quaternary structures must be stabilized before native-PAGE. For this purpose we used EDC, a zero length cross-linker. When Hsp90 was cross-linked in the presence of magnesium, native-PAGE revealed a low intensity band for the Hsp90 dimer and at least three or more additional bands of higher molecular masses corresponding to magnesium-induced oligomers (Fig. 1A, lane 2). Heat-induced oligomers do not require cross-linking; we observed more heat shock-induced oligomeric species in this sample than magnesium-induced ones (Fig. 1A, lane 3). In fact, some bands were identical to the magnesium-induced oligomers, whereas other bands were of intermediate mass. The relative migrations of the dimer band (lanes 2 and 3) were slightly different from lane 1. For the Hsp90 sample heated for 30 s in the presence of magnesium, we observed a fast oligomerization process, with protein bands that remained at the top of the resolving gel. Increasing the incubation time did not reveal any protein bands on the native-PAGE (Fig. 1A, lanes 4 and 5). As we previously reported, magnesium and temperature had a synergic effect on the Hsp90 oligomerization process (23). SDS-PAGE analysis of Hsp90 showed only monomers (Fig. 1B, lane 1). When Hsp90 was cross-linked in the presence of magnesium, several bands were visualized, with two bands corresponding to the Hsp90 monomer and dimer, and three additional diffuse bands having higher molecular masses (Fig. 1B, lane 2). The presence of monomeric species under denaturing conditions indicated an incomplete cross-linking reaction; however, the reaction is sufficient to stabilize magnesium-induced oligomers under native conditions (Fig. 1A, lane 2). The SDS-PAGE analysis of the heated samples (with or without magnesium) showed only monomer species (Fig. 1B, lanes 3–5). PAGE analyses clearly demonstrated that (i) cross-linking is necessary to stabilize Mg2+-induced oligomers and (ii) temperature induces additional species of intermediate masses.
FIGURE 1.
Comparison of magnesium- and temperature-induced Hsp90 oligomers by PAGE. A, 6/8% double layer native-PAGE analysis. The asterisk indicates the two migration distances observed for the Hsp90 dimer. B, 3 to 8% gradient SDS-PAGE analysis.
Purified Magnesium-induced Oligomers Samples Are Suitable for Biophysical Characterization
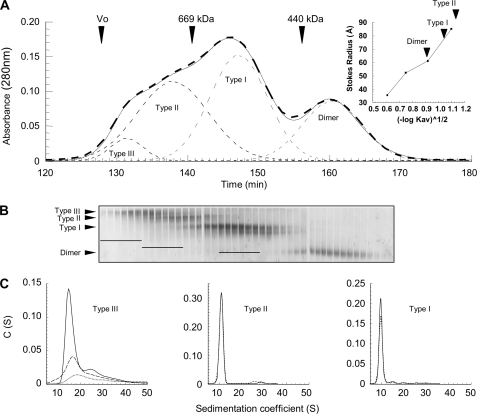
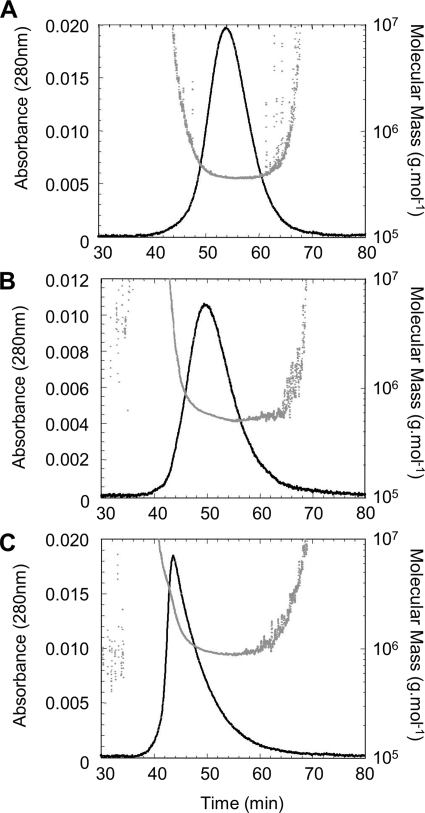
To purify each Hsp90 magnesium-induced oligomer type, we tested several hydrophobic interactions (Resource ETH, ISO, PHE, Butyl-Sepharose 4 fast flow; GE Healthcare) and ion exchange (DEAE-cellulose, Mono Q, and Mono S from GE Healthcare; hydroxyapatite from Bio-Rad) liquid chromatography, without success. Consequently, oligomers were purified according to their masses. We employed different techniques including electroelution from native-PAGE, sucrose gradient density ultracentrifugation, and size exclusion chromatography. The most resolving system was native-PAGE, which allowed the best separation between the different oligomer types. Combined with electroelution, it provided good purity for each species. However, with a very low yield, this technique was not suitable for large scale biochemical and biophysical analyses but was sufficient for EM studies. Sucrose gradient density ultracentrifugation presented an alternative means to separate oligomers. After optimization, we chose a 30–34% gradient, and eluted fractions were analyzed by native-PAGE. Despite a poor separation between the different oligomeric species, using standard proteins (thyroglobuline (19 S), catalase (11.3 S), and aldolase (7.3 S)), we roughly estimated the Hsp90 species sedimentation coefficients: 7, 11, 16, and ∼20 S for the dimer, Type I, Type II, and Type III oligomers, respectively (data not shown). These preliminary results were useful to anticipate AUC experiments. For size exclusion chromatography, we have tested several columns without success (Superose 6, Sepharose 6B and 6B CL, and Sephacryl S-300 HR (27)). These gels had adapted fractionation ranges for Hsp90 oligomers expected masses, but their resolutions were insufficient. Despite the closeness to the upper limit of the fractionation range, Superdex 200 was the most appropriate technique to improve both resolution and purification yield. The best results were obtained with a 1 × 210-cm column (Fig. 2A). The 280-nm UV absorbance elution profile displayed a multimodal curve clearly showing that the Hsp90 dimer is well separated from oligomers. Furthermore, the theoretical deconvolution of the elution profile into Gaussian curves suggested that some eluted fractions could be pure (fractions before 140 min and ∼150 min of elution time for the Type II and Type I oligomers, respectively). Calibration of the chromatography column using standards allowed us to determine Stokes radii of 59 and 77 Å for the dimer and Type I oligomer (Fig. 2A, inset). Because Type II oligomer was eluted just before thyroglobulin, we estimated its Stokes radius at ∼85–90 Å. Thus, the increase of the oligomerization was accompanied by a modest increase of the Stokes radii. All collected fractions were analyzed by native-PAGE (Fig. 2B). Combining SEC-HPLC deconvolution and native-PAGE analysis, six fractions for each oligomer type were selected (fractions 128–133, 134–139, and 145–150 for Type III, Type II, and Type I oligomers, respectively; Fig. 2B). To select the most homogeneous samples, all of the samples were analyzed by AUC sedimentation velocity. Analysis of the sedimentation coefficients distribution (C(S)) obtained by Sedfit for the Type III oligomer fractions (fractions 128–133) always showed a multimodal pattern that reflected the presence of several species with coefficients ranging from 14 to 40 S with a major species at ∼15 S. The three most enriched fractions were fractions 129–131 (Fig. 2C). Sedimentation coefficient distributions obtained for Type II and Type I fractions displayed mainly Gaussian curves reflecting mono-dispersity. The comparison of results obtained for all tested fractions allowed us to select the two purest fractions for each oligomer type (fractions 138–139 and 147–148 for Type II and Type I oligomers; Fig. 2C). We named these samples STII and STI; their purity was >90% according to the peak deconvolution. STI and STII were used for all of the following experiments.
FIGURE 2.
Hsp90 oligomers purification by size exclusion chromatography and fractions analysis. A, solid line, elution profile of 500-μl sample of cross-linked Hsp90 (27 μm) in the presence of magnesium. The thin dashed lines correspond to the elution profile deconvolution as Gaussian curves, and the bold dashed line is the sum of Gaussian peaks. Arrows indicate the elution volume of blue dextran 2000 (Vo) and of molecular mass markers with known Stokes radii. The inset shows the calibration curve of the Stokes radius as a function of the partition coefficient (Kav), and the arrows denote the partition coefficient of each Hsp90 species (Table 1). B, analysis of eluted fractions 128–170 by 6/8% double layer native-PAGE. The bars drawn on the gel indicate the six preselected fractions for each oligomer type. C, analysis of preselected fractions by sedimentation velocity (AUC). The graphs represent the apparent sedimentation coefficients distribution C(S), analyzed as described under “Experimental Procedures.” Only the purest fractions selected for the following biochemical and biophysical characterization are shown, i.e. 129, 130, and 131 for Type III (dotted, dashed, and solid lines, respectively), 138 (dashed line) and 139 (solid line) for Type II, and 147 (dashed line) and 148 (solid line) for Type I. Depending on sample concentrations, sedimentation data were acquired at 230, 240, and 280 nm for Type III, II, and I eluted fractions, respectively.
Characterization of Hsp90 Oligomers Hydrodynamic Properties and Quaternary Structures
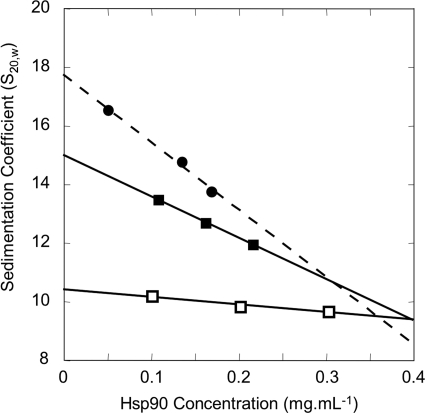
We investigated Hsp90 quaternary structure by analytical ultracentrifugation, combining velocity and sedimentation equilibrium experiments. For the S20,w0 determination of Type III oligomer, despite its heterogeneity, we nonetheless used the previous sedimentation velocity results obtained for the three selected fractions (fractions 129–131). Considering that the chemical cross-linking blocked the equilibrium, we deconvoluted the distribution curves to calculate the sedimentation coefficients and the amount of the major species. The plot of the sedimentation coefficients versus Hsp90 concentration allowed an estimation of the Type III oligomer S20,w0: 17.7 ± 2.0 S (Fig. 3). For the determination of Type I and Type II S20,w0, we used different dilutions of STI and STII, and we performed a single species analysis (Fig. 3). The STI and STII sedimentation coefficients decreased with increasing concentrations as expected for nonassociative single particle. Linear regression of apparent S20,w versus concentration permitted us to extrapolate S20,w0 at infinite dilution, yielding 10.4 ± 0.3 S for Type I and 15.0 ± 0.8 S for Type II. The calculated frictional coefficient ratios (f/f0), of 1.50 ± 0.03 and 1.58 ± 0.06 for the respective Type I and Type II oligomers show that they resembled one another in terms of steric hindrance. Sedimentation velocity can also provide information concerning the molecular mass of the protein. In fact, regarding STI data, we could estimate a 355 ± 18-kDa molecular mass, suggesting that the Type I oligomer corresponds to 2.1 ± 0.2 Hsp90 dimers. For the Type II oligomer, we found a molecular mass of 618 ± 57 kDa, indicating 3.6 ± 0.4 Hsp90 dimers, which did not enable us to draw a conclusion about the degree of oligomerization for Type II.
FIGURE 3.
Determination of standard sedimentation coefficients for each Hsp90 oligomer type. Sedimentation velocities were determined for three concentrations of each sample. □, STI (fractions 147–148); ■, STII (fractions 138–139). For the Type III oligomer (●), the estimation was done directly with previous results obtained for the raw fractions 129–131. The major peak on sedimentation coefficient distribution plot was taken into account, considering that the equilibrium is blocked because of cross-linking. Linear regressions permitted the determination of the S20,w0 for Type I and Type II (solid lines) and for Type III (dashed line). The results are presented in Table 1.
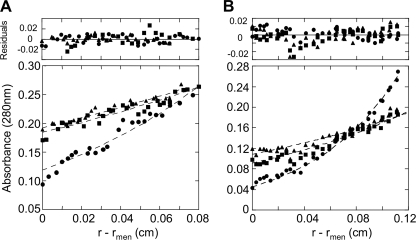
To refine these results, we carried out sedimentation equilibrium experiments using STI and STII homogeneous samples. In this context, we used three initial concentrations and three rotor speeds, resulting in nine data sets for each sample (Fig. 4). A separate analysis of each data set using the single ideal species model gave weighted average molecular masses of 316 ± 18 and 539 ± 67 kDa for Type I and Type II oligomers, respectively. These results confirm that Type I oligomer is a tetramer, as suggested by sedimentation velocity, and that Type II oligomer would most likely be an Hsp90 hexamer. Notably, in the case of STI, we observed a material loss of ∼40% over the lengthy time of analysis, and high speed data sets suggested that the Type I oligomer is altered during the AUC run.
FIGURE 4.
Hsp90 quaternary structures analysis by sedimentation equilibrium. The symbols show the experimental radial distribution of Hsp90 oligomers at sedimentation equilibrium at 3400 (▴), 4100 (■), and 6000 rpm (●) performed at 4 °C. The dashed lines represent the best fit curves of global analysis of multiple sedimentation equilibrium data with the single ideal species model of nine fitted data sets (the three speeds indicated above at three Hsp90 oligomer concentrations). A, STI sedimentation equilibrium was performed at 0.20, 0.30, and 0.40 mg/ml. Only the 0.40 mg/ml concentration is shown. B, STII sedimentation equilibrium was performed at 0.05, 0.11, and 0.16 mg/ml. Only the 0.11 mg/ml concentration is shown. r is the radial distance, and rmen is the radial distance at the meniscus. The residuals represent the variation between the experimental data and those generated by the fit with a single species. The results are presented in Table 1.
Multiangle Laser Light Scattering Experiments Suggest That the Type III Building Brick Is Not the Dimer
We performed multiangle laser light scattering experiments in line with size exclusion chromatography (SEC-MALLS). Like AUC, SEC-MALLS is a nondestructive technique allowing the analysis of samples in solution under their native state (44). Moreover, this technique accurately determines the molecular mass of macro complexes, even with low amounts of material, and is complementary to AUC. The running time of SEC-MALLS is very short compared with sedimentation equilibrium and thus avoids protein alteration, as we previously observed for the Type I oligomer. To analyze the Type III oligomer by SEC-MALLS, fractions 129–131, previously used for sedimentation velocity AUC, were pooled (STIII). A typical mass analysis provides a plot of the calculated molecular masses versus elution times for STI, STII, and STIII samples (Fig. 5). Taking into account the maximum of each UV monitored elution peak, we calculated the average molecular mass in a 4-min window (132 measurements). For STI, we obtained a very stable scattering signal throughout the peak width leading to a plateau for the calculated molecular mass. We obtained an unambiguous average molecular mass of 364.7 ± 3.7 kDa (Fig. 5A). Considering the molecular mass measured by MALLS for the dimer obtained under the same conditions, 189.6 ± 1.8 kDa (data not shown), we calculate 1.9 ± 0.1 dimers, which corroborated the AUC results and confirmed the tetrameric state of the Type I oligomer. The STII sample analysis gave a molecular mass of 587.9 ± 30.7 kDa, or 3.1 ± 0.2 dimers, sustaining previous sedimentation equilibrium results (Fig. 5B). Hence, the Type II oligomer is an Hsp90 hexamer. For STIII, the UV elution profile displays an asymmetric peak (Fig. 5C). The molecular mass curve displayed two phases, with the first one being characterized by a fast decrease of the calculated mass during peak elution to reach the second phase, a plateau at the end of elution peak. During the first phase, the molecular masses varied from 107 to 106 Da. Considering the maximum of the elution peak, the calculated average molecular mass was ∼1352 kDa, which would correspond to seven dimers. For the plateau phase, we found a minimal mass of 900 kDa, suggesting that Type III oligomerization degree is comprised of between 5 and 7 dimers, i.e. decamer to tetradecamer. Indeed, we did not observe any octameric species, indicating that beyond the Type II oligomer, the oligomerization process does not seem to follow a classical dimeric association rule.
FIGURE 5.
Hsp90 quaternary structures analysis by SEC-MALLS. STI, STII, and STIII samples properties were followed using a MALLS detector during the elution from the double size exclusion chromatography (Superdex 200 60 cm and Sepharose 6B 16 cm). The solid curve represents the UV absorbance profile at 280 nm overlaid with the scatter plot, which corresponds to the mass estimated from light scattering profile and concentration measurement versus the elution time. The results are presented in Table 1.
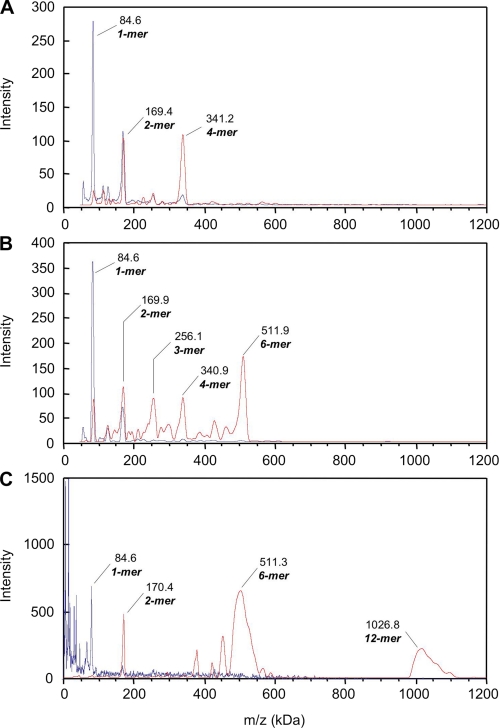
Mass Spectrometry Demonstrates That the Hsp90 Type III Oligomer Is a Dodecamer
To get accurate mass measurements concerning Type I and Type II and to elucidate the Hsp90 Type III quaternary structure, we performed high mass MALDI-TOF mass spectrometry. As controls, we analyzed our EDC cross-linked purified samples, i.e. STI, STII, and Type III SEC-HPLC eluted fractions 129, 130, and 131. For all the control experiments (Fig. 6), only peaks at m/z = 84.6 kDa and m/z ∼ 170kDa were detected for every dilution tested, corresponding to Hsp90 monomer and dimer, respectively. As we previously demonstrated by native-PAGE, AUC, and MALLS, the EDC cross-linking only stabilizes Hsp90 oligomers under native conditions. Nevertheless, our MS results confirm that under denaturing conditions our cross-linking is not total, allowing at best the stabilization of the dimeric species as we observed by SDS-PAGE. In addition to the two major monomer and dimer peaks, the STI spectrum displays two small peaks at m/z ∼ 253 kDa and m/z ∼ 340 kDa that may correspond to trimeric and tetrameric Hsp90. STI, STII, and Type III SEC-HPLC eluted fractions were over-cross-linked using the K200 kit (CovalX) over 1, 3, and 6 h. For the STI, after the 1- and 3-h incubation times, the intensity of the two major peaks increased. They corresponded to cross-linked complexes with m/z = ∼169 and 340 kDa, assignable to the Hsp90 dimer and tetramer. This increase was accompanied by a decrease of the minor peak intensity at m/z = ∼85 kDa matching the Hsp90 monomer (data not shown). The 6-h incubation time showed only a small increase of the peak intensity compared with the 3-h incubation time, suggesting that the reaction was completed. The results obtained after 6 h of STI sample cross-linking (0.15 mg/ml) are presented in Fig. 6A. The intensity pattern of the peak corresponding to the dimer (m/z = 169.4 kDa) indicated the presence of real species and not only doubly charged ions formed by the tetramer. In addition, as we previously demonstrated with AUC and MALLS experiments, STI does not contain any dimeric species. The presence of Hsp90 dimer is an artifact caused by denaturing conditions and resulted from the incomplete cross-linking of the tetramer. Thus, the major complex detected for STI at m/z = 341.2 kDa corresponded to an Hsp90 tetramer, a dimer of dimers. Concerning STII, we observed the same cross-linking reaction kinetics as for STI. The 6-h cross-linked STII (0.1 mg/ml) MS results are shown in Fig. 6B. The spectrum obtained displayed five major peaks at m/z = 84.6, 169.9, 256.1, 340.9, and 511.9 kDa. The major peak detected at m/z = 511.9 kDa corresponds to Hsp90 hexamer, a trimer of dimers, whereas the peak at m/z = 340.9 kDa matches the Hsp90 tetramer. This was due to the partial cross-linking reaction leading to a partial dissociation of the hexamer, as we observed a low intensity peak at m/z = 84.6 kDa for the monomer. From the intensity pattern of the peaks corresponding to m/z = 256.1 kDa and m/z = 169.9 kDa, the ions detected were mostly formed by doubly and triply charged ions of Hsp90 hexamer. Thus, the main complex detected by high mass MALDI-TOF MS for STII is an Hsp90 hexamer, likely corresponding to a trimer of dimers. In the spectrum of Type III after 1 h of cross-linking, two major peaks appeared at m/z = 511.3 and 1026.8 kDa (Fig. 6C). The first peak detected corresponded to an Hsp90 hexamer, or Type II, whereas the second one matched an Hsp90 dodecamer. Previous SEC-MALLS experiments gave a minimal mass of 900 kDa for STIII (pool of fractions 129, 130, and 131). Thus, the presence of hexamer for fraction 129 was due to the dissociation of the dodecamer and/or to doubly charged ions of Hsp90 dodecamer. No oligomeric species were detectable between Hsp90 hexamer and dodecamer.
FIGURE 6.
Hsp90 oligomers analyzed by high mass MALDI-TOF MS. EDC cross-linked oligomers samples STI (A), STII (B), and Type III SEC-HPLC eluted fraction 129 (C) were analyzed using the CovalX HM2 high mass detector without supplementary cross-linking (blue traces). The same samples were over cross-linked using CovalX K200 cross-linking kit and analyzed under the same conditions (red traces). The results are presented in Table 1.
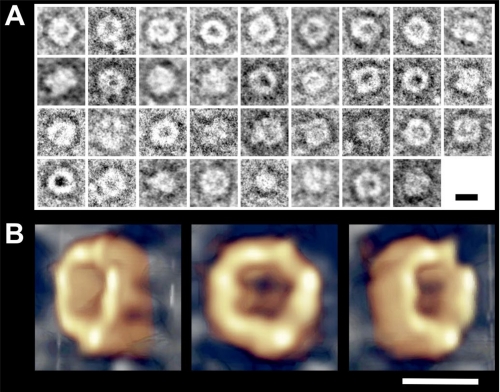
Preliminary Type II Oligomer Structure Obtained by Negative Staining and TEM Tomography
Because our purification methods led to quite homogeneous samples in terms of Hsp90 quaternary structures, we focused on the structural characterization of the oligomers. For this purpose, we attempted TEM negative staining experiments on Type I and Type II purified by electroelution from native-PAGE. Whatever the oligomer type considered, electron micrographs showed a high variability in terms of structure. As we previously observed for the Hsp90 dimer (19), oligomers seem to be highly sensitive to the ionic strength and pH of the stain. Thus, we decided to stabilize the quaternary structures by using 0.05% glutaraldehyde solution before negative staining. The glutaraldehyde concentration was optimized to reinforce the EDC cross-linked quaternary structures previously purified, thus avoiding the formation of new aberrant species. Although glutaraldehyde treatment afforded no improvement for STI, (data not shown), it significantly improved the structural homogeneity of the Type II oligomer. We observed 140–150 Å diameter O-ring shaped structures; some of them presented an accumulation of uranyl acetate in their center, whereas others seemed to exhibit a protuberance (Fig. 7A). We performed EM tomography on isolated particles to obtain accurate information about the overall Type II oligomer structure. For this, we acquired images on a CCD camera with tilt angles from −60° to +60° with an increment of 1° (121 images). The resulting Hsp90 Type II oligomer three-dimensional reconstruction is shown in Fig. 7B. The overall shape resembles a cup, a crucible, or a nest with an external diameter of ∼140 Å and an internal diameter of 50 Å. Because of the contact of the particle with the carbon film and the negative stain, we obtained little structural information at the contact area between the nest and the support. The structure of the Hsp90 Type II cozy nest oligomer is available in a supplemental movie.
FIGURE 7.
The cozy nest-shaped Hsp90 Type II oligomer structure obtained by negative stain TEM tomography. A, image gallery showing Type II oligomer particles boxed from electron micrograph (supplemental Fig. S1). B, tomographic reconstruction computed from the −60°/+60° tilt series. Three views are presented: −45°, 0°, and +45°. Scale bars, 100 Å.
DISCUSSION
Oligomers of Hsp90 have been shown to be Hsp90 quaternary structures that act as chaperones under stressed and unstressed conditions (24, 26, 29, 30). The presence of oligomeric species has been reported in the cell under physiological conditions, and these oligomers artificially dissociate into dimers when subjected to native-PAGE (25). In a previous study, we demonstrated that, in solution, divalent cations like magnesium and calcium modified Hsp90 quaternary structure. The cations increased the compactness of the Hsp90 dimer, leading to its auto-association process (27). This outcome suggested that the previously described V-shaped structure (19) would be the starting point for oligomerization. In our earlier work, we were not able to characterize the oligomers produced. Indeed, we only observed an increase of the S20,w (9–10 S) in the presence of magnesium, and AUC sedimentation velocity experiments displayed a unique wide peak, suggesting that the Hsp90 quaternary structure is highly dynamic, with subunits capable of freely and rapidly exchanging between oligomers (27). Oligomeric species are also induced by a temperature increase, so thermal stress would enhance the in vitro activity of Hsp90 and homologues (26).
In the present study, we focused on the biochemical, biophysical, and structural characterizations of magnesium-induced Hsp90 oligomers, and we developed a purification protocol. Because purification of Hsp90 oligomer species requires chemical stabilization, we initially used EDC as a zero length cross-linker. Different purification techniques were tested, and the only discriminating criteria were molecular mass and steric hindrance. For the first time, we obtained pure samples for each oligomer type, with quantities suitable for biochemical and biophysical studies. Through a multiple approach study using SEC, AUC, SEC-MALLS, and high mass MALDI-TOF MS, we accurately characterized the Hsp90 macrocomplexes, with each technique contributing a piece of the puzzle that is the Hsp90 quaternary structure. Analytical ultracentrifugation is a relevant technique to characterize auto-association processes of proteins in solution, thus reflecting their native states. Velocity experiments were initially useful to assess the purity of our fractions after SEC-HPLC purification and permitted us to select the purest samples for the entire study. With pure samples in hand, we determined the biophysical parameters of the oligomers, particularly the sedimentation coefficients (Table 1). Combined with AUC sedimentation equilibrium, we unambiguously demonstrated that Type I is a tetramer, whereas Type II could correspond to an Hsp90 hexamer. These even-numbered oligomers suggest that the building brick for Type I and Type II oligomers is the Hsp90 dimer. However, longer running times (up to 3 days) induced a protein alteration, as evidenced by the loss of 40% of material observed for the Type I oligomer. For the Type II, the molecular mass obtained gave an elevated standard deviation that did not allow us to assert its apparent hexameric state. Moreover, because of its heterogeneity, the Type III oligomer could not be analyzed by sedimentation equilibrium. Consequently, we performed MALLS analysis. Like AUC, MALLS allows protein sample analysis in solution under native conditions and gives the accurate protein molecular mass. The significant advantage of MALLS is its short running time. Moreover, before MALLS measurements, the sample separation by SEC (Superdex 200 coupled to Sepharose 6B) permitted the analysis of heterogeneous samples while excluding high molecular mass compounds and improving sample purification. This sequential SEC-MALLS configuration was very useful for the Type III analysis, and the stable laser light scattering signal enabled us for the first time to assert that Type III has a minimal molecular mass of 900 kDa, indicating at least an Hsp90 decamer. The averaged light scattering signal at the maximum of the elution peak gave a molecular mass of ∼1350 kDa, thus seven dimers on the basis of 189 kDa for the dimer analyzed by MALLS. The absence of an Hsp90 octamer (i) excludes the association of two Type I for the Type III oligomer and (ii) suggests that the Hsp90 dimer is no longer the basic building brick. Higher association order leading to the Type III oligomer is likely the result of Type I and/or Type II association. Moreover, considering an oligomerization degree ranging from five to seven dimers, i.e. decamer to tetradecamer, three hypotheses can be advanced concerning the Type III oligomeric state. The putative decamer could correspond to the association of a Type I and a Type II oligomer [T1 · T2], the dodecamer to [T2 · T2], or the tetradecamer to [T1 · T1 · T2]. The last hypothesis, requiring the association of three entities, seems to be unlikely. The MALDI-TOF mass spectrometry technique is highly sensitive in terms of mass accuracy and combined with a high mass detector is well suited to determine masses of macromolecular complexes up to 1.2 MDa. On the other hand, the ionization occurring under non-native conditions permitted us to evaluate our EDC cross-linking efficiency. High mass MALDI-TOF mass spectrometry induced the dissociation of Hsp90 oligomers into cross-linked dimers and monomers, corroborating the SDS-PAGE analysis. This result confirmed that EDC cross-linking is mild and only induced the stabilization of oligomeric states under native conditions. When we attempted to increase the stability of the quaternary structures by using the K200 cross-linking CovalX kit, the mass spectra displayed multiple peaks with masses corresponding to doubly and triply charged species as well as dissociated oligomers. Thus, the additional K200 cross-linking was incomplete. Nevertheless, a careful analysis of spectra demonstrated that STI and STII were mainly composed of species with maximum molecular masses of 341 and 512 kDa. Using high mass MALDI-TOF mass spectrometry, we confirmed the Type I tetrameric and the Type II hexameric states. Moreover, we demonstrated that the Type III is a dodecamer, which probably results from Type II dimerization.
TABLE 1.
Biochemical and biophysical parameters of Hsp90 oligomers
| Techniques | Parameters | Dimer | Type I | Type II | Type III |
|---|---|---|---|---|---|
| Ultracentrifugation (sucrose gradient) | (S) | 7 | 11 | 16 | >19 |
| Size exclusion chromatography | (kDa)a | 415 | 638 | 863c | 1060c |
| Rs (Å) | 59 | 77 | 85–90c | NDd | |
| Analytical ultracentrifugation | |||||
| Velocity | S20,w0 (S) | 6.86 ± 0.05b | 10.4 ± 0.3 | 15.0 ± 0.8 | 17.7 ± 2.0 |
| f/f0 | 1.25b | 1.50 ± 0.03 | 1.58 ± 0.06 | ND | |
| (kDa) | 355 ± 18 | 618 ± 57 | ND | ||
| Equilibrium | (kDa) | 144–175b | 316 ± 18 | 539 ± 67 | ND |
| Multiangle laser light-scattering | (kDa) | 189.6 ± 1.8 | 364.7 ± 3.7 | 587.9 ± 30.7 | >900e |
| Rg (Å) | 92 | 122 | 185 | 234 | |
| High mass MALDI-TOF MS | (kDa) | 169.9 ± 0.5 | 341.2 ± 1.1 | 511.9 ± 3.1 | 1026.8 ± 21.9 |
a Apparent molecular mass considering a globular protein.
b From Garnier et al. (27). f and f0 are the frictional coefficients.
c Extrapolation from calibration curve. Rs is the Stokes radius. S20,w0 is the corrected sedimentation coefficient at 20 °C in water at infinite dilution.
d ND, not determinable.
e Minimal molecular mass measured. Rg is the gyration radius.
To understand the Hsp90 structure-function relationships, we initiated the structural study of Hsp90 oligomers performing TEM negative stain experiments for the Type II oligomer. We observed 140–150 Å diameter O-ring shaped structures with or without stain accumulation in their centers. The same negative stain samples were then analyzed by TEM tomography. Structure reconstruction demonstrated that Type II has a crucible shape, like a nest, and suggests that Hsp90 could function with a mechanism based on an oligomerization process creating a cage-like nano-compartment where folding/protection of the target proteins can occur. A structure comparable with our Type II in terms of size (150 Å) and shape was already observed in the presence of the nonhydrolyzable ATP analogue AMP-PNP by negative stain TEM and published by Pearl and Prodromou (45). This structure was attributed to an Hsp90 dimer that adopts a closed toroidal conformation. Considering the external diameter (150 Å) and the internal cavity diameter (50 Å), the volume calculation of such toroidal structure gives ∼6 × 105 Å3. Given a theoretical volume of 1.02 × 105 Å3 for the monomer, such a particle likely corresponds to an hexamer rather than a closed dimer. We frequently observed this structure in our TEM preparations in the presence of nucleotides, i.e. ADP and AMP-PNP, and in the absence of cross-linking.
The relative migration of cross-linked and/or heated Hsp90 dimer was modified compared with untreated Hsp90. Two nonexclusive hypotheses can be proposed. First, for lane 1, at the gel loading stage all oligomers were present; then during migration the equilibrium would progressively be displaced toward dimeric species, reducing the migration distance for the dimer band. Second, as we previously demonstrated (27), Mg2+ induces an increase of Hsp90 dimer compactness that could modify its migration pattern after cross-linking. The absence of equilibrium and/or the increase of Hsp90 dimer compactness could explain the migration pattern for the heated sample. The synergic effect observed between the temperature increase and the presence of Mg2+ suggests that the dimer brick would have a similar structure, thus leading to a similar migration pattern in native-PAGE.
Native PAGE analysis showed that the temperature-dependent oligomerization process produced a large number of oligomers; indeed we observed oligomers with masses intermediate to those for Mg2+-induced oligomers. Knowing that Mg2+ induces even-numbered oligomers, the intermediate mass bands observed for heat-induced oligomerization may correspond to odd-numbered species. During heat treatment, dimers would dissociate into labile monomers, which would be captured by even-numbered Hsp90 quaternary structures, indicating an auto-chaperone activity, as has been previously suggested (24, 25, 29, 30). All of these observations strongly support previously published results demonstrating that Hsp90 oligomers are quaternary structures endowed with a chaperone activity.
The organization of Hsp90 into oligomeric structures is compatible with the lateral fixation of substrate as was observed for cdk4 (15). Moreover, some chaperone proteins adopt oligomeric structures to perform their chaperone activity, i.e. small Hsp, Hsp27, Hsp60, and Hsp100/110. The intermonomer structural cooperation between domains within the Hsp90 dimer concerning ATP hydrolysis (18) can be transposed within oligomers between adjacent monomers and/or dimers. Such quaternary structural organizations would be advantageous for the folding of client proteins. Indeed, the nest-like shape of the Type II oligomer structure we present here could be assimilated to a “folding chamber” of ∼50 Å diameter suitable for the chaperoning of client proteins, and the concentration of ATP hydrolysis within the nest would confer enough energy to Hsp90 to accomplish its chaperone function, as is well known for the Hsp60 family. Type III could result from a face-to-face or back-to-back dimerization of Type II, envisioned as a “cockleshell” or as two stacked hexameric rings, a quaternary structure comparable with the Hsp60 family.
Thus, in addition to the high intrinsic flexibility of the Hsp90 dimer, the coexistence of oligomeric species would further increase the Hsp90 structural variability, enabling the adaptation of its structure to the broad spectrum of client proteins. Unraveling the Hsp90 chaperone cycle will be achieved only when the roles of all the actors, i.e. the Hsp90 dimer and oligomers as well as cochaperones, are taken into account. To gain more resolution and to better understand the oligomerization processes and oligomer functions, structural studies using cryo-EM on oligomers structures are now in progress.
Supplementary Material
Acknowledgments
We thank Reynald Gillet for helpful discussions. We thank Julien Weber (CovalX) for high mass MALDI-TOF mass spectrometry experiments performed on Type III SEC-HPLC eluted fractions.
This work was supported by a grant from the Ligue contre le Cancer Conseil Scientifique Interrégional Grand-Ouest (to C. G.) and a Région Bretagne Ph. D. grant (to L. M.) and research grant (to D. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and a movie.
- Hsp
- heat shock protein
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate
- AUC
- analytical ultracentrifugation
- EDC
- 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride
- MALDI-TOF MS
- matrix-assisted laser desorption/ionization time of flight mass spectrometry
- MALLS
- multiangle laser light scattering
- MS
- mass spectrometry
- SEC
- size exclusion chromatography
- TEM
- transmission electron microscopy
- HPLC
- high pressure liquid chromatography
- EM
- electron microscopy
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Pearl L. H., Prodromou C. (2006) Annu. Rev. Biochem. 75, 271–294 [DOI] [PubMed] [Google Scholar]
- 2.Albanèse V., Yam A. Y., Baughman J., Parnot C., Frydman J. (2006) Cell 124, 75–88 [DOI] [PubMed] [Google Scholar]
- 3.Aherne W., Maloney A., Prodromou C., Rowlands M. G., Hardcastle A., Boxall K., Clarke P., Walton M. I., Pearl L., Workman P. (2003) Methods Mol. Med. 85, 149–161 [DOI] [PubMed] [Google Scholar]
- 4.Zhao C., Wang E. (2004) Cell Signal. 16, 313–321 [DOI] [PubMed] [Google Scholar]
- 5.Whitesell L., Lindquist S. L. (2005) Nat. Rev. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 6.Le Bras G., Radanyi C., Peyrat J. F., Brion J. D., Alami M., Marsaud V., Stella B., Renoir J. M. (2007) J. Med. Chem. 50, 6189–6200 [DOI] [PubMed] [Google Scholar]
- 7.Radanyi C., Le Bras G., Marsaud V., Peyrat J. F., Messaoudi S., Catelli M. G., Brion J. D., Alami M., Renoir J. M. (2009) Cancer Lett. 274, 88–94 [DOI] [PubMed] [Google Scholar]
- 8.Lai B. T., Chin N. W., Stanek A. E., Keh W., Lanks K. W. (1984) Mol. Cell. Biol. 4, 2802–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katschinski D. M., Le L., Schindler S. G., Thomas T., Voss A. K., Wenger R. H. (2004) Cell Physiol. Biochem. 14, 351–360 [DOI] [PubMed] [Google Scholar]
- 10.Nollen E. A., Morimoto R. I. (2002) J. Cell Sci. 115, 2809–2816 [DOI] [PubMed] [Google Scholar]
- 11.Hickey E., Brandon S. E., Sadis S., Smale G., Weber L. A. (1986) Gene 43, 147–154 [DOI] [PubMed] [Google Scholar]
- 12.Radanyi C., Renoir J. M., Sabbah M., Baulieu E. E. (1989) J. Biol. Chem. 264, 2568–2573 [PubMed] [Google Scholar]
- 13.Garnier C., Lafitte D., Jorgensen T. J., Jensen O. N., Briand C., Peyrot V. (2001) Eur. J. Biochem. 268, 2402–2407 [DOI] [PubMed] [Google Scholar]
- 14.Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. (2003) Mol. Cell. 11, 647–658 [DOI] [PubMed] [Google Scholar]
- 15.Vaughan C. K., Gohlke U., Sobott F., Good V. M., Ali M. M., Prodromou C., Robinson C. V., Saibil H. R., Pearl L. H. (2006) Mol. Cell 23, 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minami Y., Kimura Y., Kawasaki H., Suzuki K., Yahara I. (1994) Mol. Cell. Biol. 14, 1459–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips J. J., Yao Z. P., Zhang W., McLaughlin S., Laue E. D., Robinson C. V., Jackson S. E. (2007) J. Mol. Biol. 372, 1189–1203 [DOI] [PubMed] [Google Scholar]
- 18.Cunningham C. N., Krukenberg K. A., Agard D. A. (2008) J. Biol. Chem. 283, 21170–21178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bron P., Giudice E., Rolland J. P., Buey R. M., Barbier P., Díaz J. F., Peyrot V., Thomas D., Garnier C. (2008) Biol. Cell 100, 413–425 [DOI] [PubMed] [Google Scholar]
- 20.Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Nature 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiau A. K., Harris S. F., Southworth D. R., Agard D. A. (2006) Cell 127, 329–340 [DOI] [PubMed] [Google Scholar]
- 22.Vaughan C. K., Piper P. W., Pearl L. H., Prodromou C. (2009) FEBS J. 276, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnier C., Protasevich I., Gilli R., Tsvetkov P., Lobachov V., Peyrot V., Briand C., Makarov A. (1998) Biochem. Biophys. Res. Commun. 249, 197–201 [DOI] [PubMed] [Google Scholar]
- 24.Chadli A., Ladjimi M. M., Baulieu E. E., Catelli M. G. (1999) J. Biol. Chem. 274, 4133–4139 [DOI] [PubMed] [Google Scholar]
- 25.Nemoto T., Sato N. (1998) Biochem. J. 330, 989–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemoto T. K., Ono T., Tanaka K. (2001) Biochem. J. 354, 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garnier C., Barbier P., Devred F., Rivas G., Peyrot V. (2002) Biochemistry 41, 11770–11778 [DOI] [PubMed] [Google Scholar]
- 28.Krukenberg K. A., Böttcher U. M., Southworth D. R., Agard D. A. (2009) Protein Sci. 18, 1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yonehara M., Minami Y., Kawata Y., Nagai J., Yahara I. (1996) J. Biol. Chem. 271, 2641–2645 [DOI] [PubMed] [Google Scholar]
- 30.Thorne M. E., McQuade K. L. (2004) Biochem. Biophys. Res. Commun. 323, 1163–1171 [DOI] [PubMed] [Google Scholar]
- 31.Yonezawa N., Nishida E., Sakai H., Koyasu S., Matsuzaki F., Iida K., Yahara I. (1988) Eur. J. Biochem. 177, 1–7 [DOI] [PubMed] [Google Scholar]
- 32.Garnier C., Barbier P., Gilli R., Lopez C., Peyrot V., Briand C. (1998) Biochem. Biophys. Res. Commun. 250, 414–419 [DOI] [PubMed] [Google Scholar]
- 33.Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 34.Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 35.le Maire M., Rivas E., Møller J. V. (1980) Anal. Biochem. 106, 12–21 [DOI] [PubMed] [Google Scholar]
- 36.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 37.Schuck P., Rossmanith P. (2000) Biopolymers 54, 328–341 [DOI] [PubMed] [Google Scholar]
- 38.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) Computer-aided Interpretation of Analytical Sedimentation Data for Proteins in Analytical Ultracentrifugation in Biochemistry and Polymer Sciences (Harding S. E., Rowe A. J., Horton J. C. eds) Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 39.Minton A. P. (1994) Modern Analytical Ultracentrifugation, Birkhävser, Boston, MA [Google Scholar]
- 40.Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R. A., Rouault T. A., Schuck P. (2004) Anal. Biochem. 326, 234–256 [DOI] [PubMed] [Google Scholar]
- 41.Herman G. T., Lent A., Rowland S. W. (1973) J. Theor. Biol. 42, 1–32 [DOI] [PubMed] [Google Scholar]
- 42.Messaoudii C., Boudier T., Sanchez Sorzano C. O., Marco S. (2007) BMC Bioinformatics 8, 288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophotonics Int. 11, 36–42 [Google Scholar]
- 44.Bruneaux M., Rousselot M., Leize E., Lallier F. H., Zal F. (2008) Curr. Protein Pept. Sci. 9, 150–180 [DOI] [PubMed] [Google Scholar]
- 45.Pearl L. H., Prodromou C. (2001) Adv. Protein Chem. 59, 157–186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.