Abstract
The mammalian target of rapamycin (mTOR) regulates growth via promoting translation and transcription. Here, employing an mTOR active-site inhibitor WYE-125132 (WYE-132), we have performed quantitative phospho-proteomics and identified a Ser-75-containing phosphopeptide from Maf1, a known repressor of RNA polymerase III (Pol III) transcription. Treatment of cancer cells with WYE-132 or the rapamycin analog CCI-779 led to a rapid loss of the phosphorylation at Ser-75, whereas this effect was not seen in cells treated with cytotoxic agents or unrelated inhibitors. WYE-132-induced Maf1 dephosphorylation correlated with its accumulation in the nucleus and a marked decline in the cellular levels of pre-tRNAs. Depletion of cellular Maf1 via small interfering RNA increased basal pre-tRNA and rendered tRNA synthesis refractory to mTOR inhibitors. Maf1 mutant proteins carrying S75A alone or with S60A, T64A, and S68A (Maf1-S75A, Maf1–4A) progressively enhanced basal repression of tRNA in actively proliferating cells and attenuated amino acid-induced tRNA transcription. Gene alignment revealed conservation of all four Ser/Thr sites in high eukaryotes, further supporting a critical role of these residues in Maf1 function. Interestingly, mTOR inhibition led to an increase in the occupancy of Maf1 on a set of Pol III-dependent genes, with concomitant reduction in the binding of Pol III and Brf1. Unexpectedly, mTORC1 itself was also enriched at the same set of Pol III templates, but this association was not influenced by mTOR inhibitor treatment. Our results highlight a new and unique mode of regulation of Pol III transcription by mTOR and suggest that normalization of Pol III activity may contribute to the therapeutic efficacy of mTOR inhibitors.
Keywords: Phosphorylation/Kinases/Serine-Threonine, Phosphorylation/Transcription Factors, Signal Transduction/Protein Kinases, Transcription/Regulation, mTOR, mTOR Complex (mTORC), RNA Polymerase III, Maf1, RNA Polymerase III Transcription, mTOR Kinase Inhibitor
Introduction
The mammalian target of rapamycin (mTOR)3 is a central metabolic sensor that coordinates cell growth and proliferation with the availability of growth factors, nutrients, and energy sufficiency. mTOR exists in two multiprotein complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (1–3). Under conditions of rapid growth and proliferation, mTOR stimulates several anabolic processes, including mRNA translation, transcription, and lipid biosynthesis. The mTORC1 is well known to enhance cap-dependent translation initiation through the direct phosphorylation of S6K1 and 4E-BP1 in response to mitogen and nutrient stimulation (4). The more recently discovered mTORC2 can directly phosphorylate AKT and conventional protein kinase C and is involved in maintenance of actin cytoskeleton in a yet to be defined mechanism. mTOR is a major signaling component of the phosphatidylinositol 3-kinase/AKT pathway that is most frequently dysregulated in cancer (3, 5–7).
In addition to the direct control of translational apparatus by mTORC1, several reports have implicated the mTOR signaling in the molecular events occurring the nucleus, such as RNA polymerase (Pol) I transcription, essential for the ribosomal biogenesis and accumulation of cell mass (8–10). The genetic and biochemical studies in budding yeast also revealed the regulation of nutrient-dependent Pol III transcription by TOR pathway (11–13). Very few studies, however, addressed and identified the mechanisms by which mTOR regulates activity of Pol III in cancer cells. Pol III is the largest RNA polymerase that synthesizes diverse classes of non-coding RNAs, including tRNAs, 5S rRNA, U6 snRNA, and other small untranslated RNAs required for multiple cellular processes (14). Pol III products are overexpressed in transformed cells and tumors (15–18), but the direct causal link between Pol III-dependent transcription and cancer progression was only recently reported. Brf1, a TFIIIB subunit, was identified as a driver of transformed phenotype in certain cell types (19). Brf1 positively regulates recruitment of Pol III to its target genes, and it is a critical point of convergence of multiple signaling pathways, including Ras, c-Myc, p53, Rb, and more recently described PTEN (14, 20, 21).
The Pol III-associated transcription repressor Maf1 was originally discovered in budding yeast. In this organism, under optimal growth conditions, Maf1 is phosphorylated in a protein kinase A and TOR/SCH9-dependent manner, whereas under stress or nutrient limiting conditions it becomes dephosphorylated and translocates to the nucleus to repress Pol III transcription (13, 22, 23). Several studies have indicated that human Maf1 has a similar function as a negative regulator of Pol III transcription (24–27). Although little is known whether mTOR directly impinges on Maf1 function in human cancer cells, recent data show that Maf1 is a phosphoprotein and is dephosphorylated in response to serum starvation, methyl methanesulfonate, or rapamycin treatment (24). Nevertheless, the nature of the phospho-sites and the regulatory mechanisms has not yet been defined.
To date, a plethora of studies has widely utilized rapamycin and its analogs to elucidate mTORC1-dependent biological responses in cancer cells. Given only the partial inactivation of mTORC1 by rapamycin, we sought to identify the previously uncharacterized mTOR signaling events. In the present study we have performed a phospho-proteomic study of cancer cells in response to mTOR inactivation by WYE-125132 (WYE-132), a highly potent, specific inhibitor of mTORC1 and mTORC2 (28). We report the identification of Ser-75 of Maf1 as a site, phosphorylation of which is strongly suppressed by WYE-132 and the rapamycin analog CCI-779. In diverse cancer cell lines studied, pharmacological inactivation of mTORC1 leads to a rapid and marked decrease in the cellular Pol III-dependent tRNA transcription. Furthermore, we provide evidence that mTORC1-dependent phosphorylation of Maf1 at Ser-75 and additional sites functionally contribute to the dynamic regulation of the repressive activity of Maf1 toward Pol III transcription. Our results support a model that mTOR signaling fine-tunes the rates of Pol III transcription in mammalian cells to provide the growth stimulatory signals.
EXPERIMENTAL PROCEDURES
Plasmids, Reagents, and Chemicals
The mammalian expression cytomegalovirus promoter vectors containing FLAG-tagged Maf1 (#EX-V1690-M11) and eGFP-Maf1 (#EX-V1690-M29) were obtained from GeneCopoeia. An existing 1-nucleotide discrepancy with the data base version at position 706 of Maf1 was corrected by site-directed mutagenesis, so that Arg-236 was converted to glycine. The mutations S75A and S60A/T64A/S68A/S75A were introduced into Maf1 sequence in a FLAG-Maf1vector with QuikChange II or QuikChange Lightning multisite-directed mutagenesis kits (Stratagene) according to the manufacturer's instructions. Primer sequences used for the mutagenesis are available upon request. The sequences of all vectors were verified by DNA sequencing. mTOR inhibitors CCI-779 and WYE-132 were provided by Wyeth Chemical and Pharmaceutical Development. All other chemicals/inhibitors were from Sigma unless otherwise specified. All inhibitors were dissolved in dimethyl sulfoxide (DMSO) as concentrated stocks and diluted before use in culture media for cell-based assays.
Cell Culture, Proliferation Assays, and Transfections
Human tumor lines MDA-MB-361, MG63, HCT116, U87MG, HT29, and human embryonic kidney cell line HEK293 were obtained from American Type Culture Collection (ATCC). Cells were cultured in a 37 °C incubator with 5% CO2 using standard cell culture methods. All culture media, supplements, and transfection reagents were purchased from Invitrogen. For transient transfections, expression vectors were transfected into cells using the Lipofectamine 2000 reagent. For siRNA knockdown experiments with Maf1 siRNA Pool, cells were seeded at 30% confluence on 6- or 12- well culture plates and transfected with 50 nm siRNA using Lipofectamine RNAiMax (Invitrogen). siRNA transfections were repeated 48 h later, and the cells were then incubated for additional 24 h followed by the drug treatment. For the knockdown of the endogenous Maf1 using 3′-UTR-targeting siRNA and reintroduction of the Maf1 expression constructs, cells were transfected with Maf1 3′-UTR siRNA for 48 h followed by transient transfection of Maf1 expression plasmids for another 24 h. The Maf1 ON-TARGETplus siRNA pool and non-targeting control ON-TARGETplus siRNA pools were obtained from Dharmacon. An ON-TARGETplus siRNA against 3′-UTR of Maf1 (sense, CAGCUGGACCGCAGAGUUUUU) was synthesized by Dharmacon. For amino acid stimulation experiments, HEK293 cells were transfected with Maf1 constructs for 24 h and starved of amino acids by a 2-h incubation in amino acid-free medium containing Earle's balanced salt solution, minimum Eagle's medium (MEM) vitamins, MEM nonessential amino acids, 0.2% NaHCO3, 1% glucose, and 10% dialyzed fetal bovine serum. Next, cells were pretreated with WYE-132 or vehicle control for 15 min and refed with amino acids (Sigma) for 1 h.
Immunoblot Analysis
Total cellular lysates were prepared using NuPAGE-LDS sample buffer (Invitrogen). Equal amounts of proteins were subjected to immunoblotting analysis using NuPAGE electrophoresis system (Invitrogen). Anti-FLAG antibody (M2) was from Sigma. Antibodies for phospho-S6K1-Thr-389, total S6K1, phospho-AKT-Ser-473, and total AKT were obtained from Cell Signaling Technology. Antibodies for total Maf1 (sc-98715/FL-256) were obtained from Santa Cruz Biotechnology, and the β-actin antibody was from Chemicon-Millipore. Immunoblots were probed with appropriate primary and secondary antibodies following the manufacturer's instructions and detected using enhanced chemiluminescence (GE Healthcare). For calf intestinal alkaline phosphatase treatment, cells were lysed in buffer without phosphatase inhibitors containing 25 mm Hepes, pH 7.5, 100 mm NaCl, 1.5 mm MgCl2, 0.5 mm EGTA, 0.25 mm EDTA, 1% Nonidet P-40, 10 mg/ml aprotinin, 10 mg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride, 1 mm Microcystin LR. To dephosphorylate proteins, lysates were treated with calf intestinal alkaline phosphatase (New England Biolabs) at 30 °C for 30 min or at 37 °C for 1 h according to the manufacturer's instructions.
RNA Isolation and Quantitative Reverse Transcription-PCR (qRT-PCR) Analysis
Total RNA, including small RNAs species, was extracted from cells using the miRNeasy kit (Qiagen). Samples were treated with DNase twice; on-column DNase digestion was performed during RNA purification using the RNase-free DNase Set (Qiagen), and a second, in-solution DNase digestion was carried out using TURBO DNA-free (Ambion). The RNA was reverse-transcribed into cDNA by using iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using the iQ SYBR Green Supermix kit (Bio-Rad) on a Bio-Rad CFX96 system according to the manufacturer's instructions. Glyceraldehyde-3-phosphate dehydrogenase was used as a reference gene. To quantify the level of short-lived tRNA precursors, primers against intron-containing tRNA genes were designed. All primers were validated on genomic DNA to ensure that amplification efficiencies were comparable. Primer sequences used for qRT-PCR are listed in supplemental Table S1. Maf1 mRNA levels were quantified with a Maf1-inventoried assay (Hs00361082_g1), a glyceraldehyde-3-phosphate dehydrogenase endogenous control assay (4333764F), and a TaqMan Gene Expression Master Mix (Applied Biosystems) on a Bio-Rad CFX96 system. For SYBR Green or TaqMan detection, the relative expression levels were calculated by using the comparative ΔΔCt method (29) and according to the Applied Biosystems 2004 “Guide to Performing Relative Quantitation of Gene Expression Using Real-time Quantitative PCR.” Data from real-time PCR are expressed as “-fold changes” with the error bars, where the error bars represent data ranges obtained from the calculation of standard deviations of the ΔΔCt values. Each cDNA sample was run in triplicate, and the corresponding no-RT RNA sample was included as a negative control.
ChIP Assays
MG63 or HEK 293 cells treated with or without WYE-132 for the indicated times and cross-linked with formaldehyde according to the Genpathway protocol (31). Additional chromatin immunoprecipitations and qPCR analysis (FactorPath Query assays) were performed by Genpathway. Immunoprecipitations were performed using the following antibodies: mTOR (sc-1549, Santa-Cruz or ab32028, Abcam), RPC39 (RNA Pol III, sc-23913, Santa-Cruz), Brf1 (B2743–50A, US Biological), Maf1 (sc-98715/FL256, Santa Cruz), Raptor (#2280, Cell Signaling), or IgG control (I5006, Sigma). Sequences of primers used for qPCR are shown in supplemental Table S1. Negative controls represent primer pairs for the well characterized untranscribed genomic regions Untr12 (Untr12) and Untr4 (Untr4).
Identification and Quantification of Phosphorylation Sites by Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and LC-MS/MS
Untreated MDA361 cells (3 × 108) were grown in custom Dulbecco's modified Eagle's medium lacking lysine and arginine (Invitrogen) supplemented with 10% dialyzed fetal bovine serum, penicillin/streptomycin, and an l-lysine (L8662, Sigma) and l-arginine (A8094, Sigma) mixture for “light” cultures. In parallel, MDA361 cells (3 × 108) were grown in the same Dulbecco's modified Eagle's medium minus lysine and arginine medium that contained “heavy” l-Lys-2HCl (U-13C6,15N2)/l-Arg-HCl (U-13C6,15N4) (CNLN-291, CNLM-539, Cambridge Isotope Laboratories) amino acids and treated with WYE-132 for 6 h. Each cell population was lysed in 80 ml of urea buffer (25 mm HEPES, pH 7.5, 100 mm NaCl, 20 mm β-glycerophosphate, 1.5 mm MgCl2, 0.5 mm EGTA, 0.25 mm EDTA, 10 mm sodium orthovanadate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride, 1 μm Microcystin LR, 9 m urea, and Halt Phosphatase Inhibitor Mixture (Pierce)), sonicated, and centrifuged at 12,000 × g for 15 min at 10 °C. Next, equal amounts of light and heavy samples were combined and subjected to proteolytic digestion with trypsin. An additional two-step phosphopeptide enrichment procedure was performed that included a strong cation exchange fractionation followed by the titanium dioxide (TiO2) chromatography, as described previously (32). Liquid chromatography-MS/MS experiments were performed on a reversed-phase Magic C18 nanocolumn coupled with an LTQ-MS mass spectrometer (ThermoFisher Scientific). Two biological replicates were performed for mass spectrometric analysis. MS/MS raw spectra were searched against the human component of the NCBI database using Biowork 3.3 (ThermoFisher). The relative ratio of the intensity of the heavy versus the light peptides was used to express the degree of phosphorylation of a given protein.
Immunofluorescence Analysis
For immunofluorescence experiments, cells were grown in 6-well plates in complete growth media on glass coverslips coated with collagen I (BD Biosciences) for 24 h before staining. The cells were fixed with 3.7% formaldehyde in phosphate-buffered saline for 15 min and then were permeabilized with methanol. Maf1 was visualized with primary rabbit antibody (diluted 1:100, sc-98715/FL-256, Santa Cruz). Anti-rabbit IgG conjugated to Alexa Fluor 594 (diluted 1:1000) was used to visualize the proteins. Coverslips were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen). Localization was evaluated by confocal laser microscopy at 63× magnification (Carl Zeiss).
RESULTS
Identification of Maf1 as an mTOR-regulated Phosphoprotein
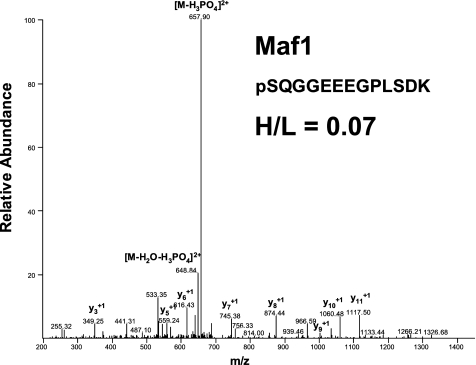
To identify novel mTOR substrates, we performed a global cellular phospho-proteome analysis by the SILAC-based mass spectrometry of MDA361 breast cancer cells after treatment with vehicle or the mTOR inhibitor WYE-132. We detected a series of phosphopeptides modulated by 2-fold or more in response to WYE-132 treatment. The identified phosphoproteins included many known mTOR downstream substrates, such as PRAS40, eIF4B, and 4E-BP1, as well as a group of previously uncharacterized proteins. One of the novel phosphopeptides that was strongly reduced by WYE-132 treatment corresponded to a transcriptional regulator Maf1 (Fig. 1). Relative quantification of the intensity ratio of the heavy-labeled over the light-labeled phosphopeptides (H/L) for the Ser-75-containing peptide yielded a value of 0.07, indicating that phosphorylation of Maf1 at Ser-75 was inhibited by >99% under drug-treated conditions. The identification of Maf1 as an mTOR-regulated phosphoprotein implicates mTOR in the broader regulatory mechanisms governing Pol III activity in cancer cells.
FIGURE 1.
MS/MS spectra of Maf1 phosphopeptide identified by SILAC. The sequence of a tryptic peptide matched to Maf1 and the SILAC ratio (heavy-labeled/light-labeled (H/L)) for Maf1 peptide is shown for the corresponding spectra.
mTORC1 Is Required for Optimal tRNA Transcription in Various Cancer Cells
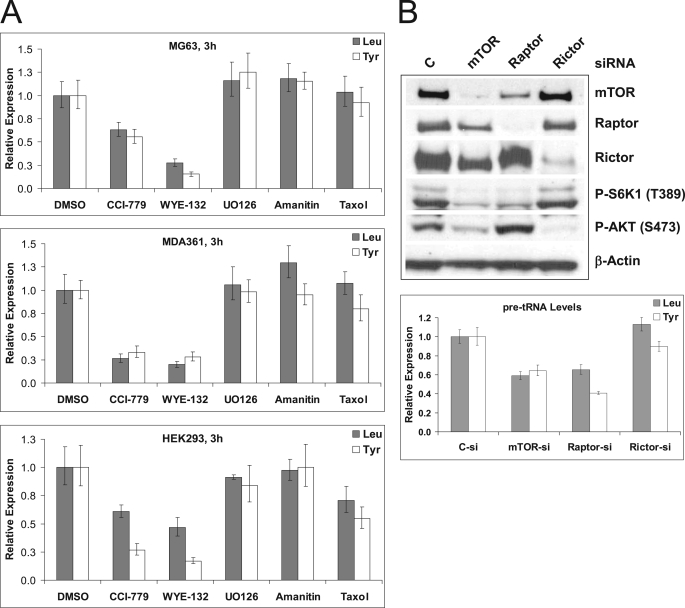
Because Maf1 is a known negative regulator of Pol III transcription and the TOR pathway is involved in control of RNA polymerase III transcription in yeast, we first investigated whether mTOR is required for Pol III transcription in human cancer cells by employing both the mTORC1-selective rapamycin analog CCI-779 and WYE-132. Actively proliferating cells of MG63 (osteosarcoma), MDA361 (breast adenocarcinoma), and HEK293 (embryonic kidney) were treated with 0.5 μm CCI-779 or WYE-132 for 3 h and were analyzed by qRT-PCR to measure the level of tRNA precursors (pre-tRNAs). WYE-132 induced a marked reduction in the synthesis of pre-tRNALeu by 72, 80, and 53% in MG63, MDA361, and HEK293 cells, respectively (Fig. 2A). Treatment with CCI-779 resulted in a similar but less pronounced decrease in pre-tRNALeu levels in these cells, indicating an involvement of mTORC1. The expression of a second tRNA precursor, pre-tRNATyr, encoded by the gene located in a separate chromosomal region was also reduced by both drugs in all cell lines tested (Fig. 2A). Conversely, unrelated control drugs UO126, α-amanitin, and taxol did not significantly reduce tRNA transcription (Fig. 2A), indicating that the inhibitory effect of mTOR inhibitors on tRNA gene transcription is unlikely due to the secondary response of cells to the growth delay or to the general inhibition of transcription. In additional assays, mTOR inhibitors similarly down-regulated pre-tRNA levels in U87MG (brain), HCT116 (colon), and HT29 (colon) cells (supplemental Fig. S1), indicating the generality of this phenomenon in multiple cancer types.
FIGURE 2.
mTOR activity is required for Pol III transcription. A, MG63 (top), MDA361 (middle), or HEK293 (bottom) cells were treated with vehicle-DMSO, 0.5 μm CCI-779, 0.5 μm WYE-132, 5 μg/ml U0126, 10 μg/ml α-amanitin, or a 100 ng/ml taxol for 3 h. qRT-PCR analysis was used to measure the expression of tRNALeu (gray-shaded bars) or tRNATyr (open bars) precursors as described under “Experimental Procedures.” The expression levels of each gene were first normalized with control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data expressed as -fold differences over control untreated samples. The experiment was performed in triplicate. B, MG63 cells were transfected with control (C), mTOR, raptor, or rictor siRNA pools for 72 h as described under “Experimental Procedures.” Left, total lysates were immunoblotted with mTOR, raptor, rictor, P-S6K, P-AKT, and β-actin. Right, expression of precursor tRNALeu (shaded bars) or tRNATyr (striped bars) was measured by qRT-PCR. Glyceraldehyde-3-phosphate dehydrogenase was used for normalization. The values represent -fold differences in expression upon siRNA depletion relative to those of the control siRNA. The data shown are representative of three independent experiments. Error bars represent the range around the mean -fold changes as described under “Experimental Procedures.”
To confirm the requirement of mTORC1 activity for Pol III transcription, MG63 cells were depleted for mTOR, Raptor, or Rictor (Fig. 2B). Similar to the chemical inhibitors, both mTOR- and Raptor-depleted MG63 cells exhibited reduced pre-tRNAs, whereas depletion of Rictor had negligible effects (Fig. 2B). Collectively, the results in Fig. 2 identify mTORC1 as a positive regulator of Pol III transcription in a panel of diverse cancer cells.
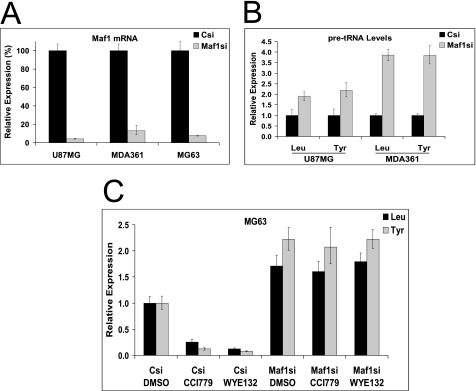
Maf1 Depletion Increases Basal tRNA Transcription and Renders tRNA Synthesis Resistant to mTOR Inhibition
Because of our findings on the involvement of mTOR in control of Pol III activity and the decrease in Maf1 phosphorylation by WYE-132, we next examined whether Maf1 is required for the mTOR inhibitor effects on Pol III suppression. Transfection of Maf1 siRNA in U87MG, MDA361, and MG63 cells resulted in an efficient reduction of Maf1 mRNA by >95, >85, and >90%, respectively (Fig. 3A). Depletion of Maf1 significantly increased the basal tRNALeu and tRNATyr precursors in all three cell lines (Fig. 3B), emphasizing the Maf1 role as a repressor of Pol III function also in cancer cells. We then tested whether Maf1 is required for the suppressive effects of mTOR inhibitors on tRNA synthesis. siRNA-transfected MG63 cells were treated with 0.5 μm CCI-779 or WYE-132 for 3 h. As expected, in the control siRNA-transfected cells, both CCI-779 and WYE-132 inhibited pre-tRNA levels. Remarkably, in the Maf1-depleted cells, levels of pre-tRNALeu and pre-tRNATyr were no longer inhibited by mTOR inhibitors (Fig. 3C). These results suggest that Maf1 functions downstream of mTORC1 to control Pol III activity.
FIGURE 3.
Effects of Maf1 knockdown on Pol III-dependent transcription and response to mTOR inhibitors. A, indicated tumor lines were transfected with siRNA pools specific for Maf1 or with control (Csi) for 72 h. Efficiency of the knockdown was measured by the qRT-PCR using the Taqman assay. Relative expression (%) is a percent mRNA remaining, which is calculated as the amount of Maf1 mRNA in Maf1-depleted cells compared with the non-targeting control siRNA samples. B, tRNALeu and tRNATyr precursor levels were quantified using qRT-PCR. The qRT-PCR results are presented as expression values relative to the control siRNA pool. Graphs are representative of three independent experiments. C, MG63 cells were transfected with Maf1 siRNA or control pools for 72 h, after which these cell populations were exposed to the vehicle-DMSO, 0.5 μm CCI-779, and WYE-132 for an additional 3 h. tRNALeu and tRNATyr levels were quantified using qRT-PCR.
mTORC1 Is Required for Maf1 Phosphorylation and Its Nuclear Exclusion
Given the identification of Maf1 as a potential mTOR substrate in our SILAC experiment as well as the suppressive effects of mTOR inhibitors on Pol III transcription in cancer cells, we investigated mTOR-dependent phosphorylation of Maf1. Maf1 was reported as a doublet on SDS-PAGE, with dephosphorylated or hypophosphorylated protein being the faster migrating form (downward band shift) (25, 26). Because of the lack of a robust Maf1 antibody suitable for immunoblotting, FLAG-Maf1 was transfected into HEK293 for examination of Maf1 mobility shift. We performed SDS-PAGE analysis of the lysates from DMSO-, CCI-779-, and WYE-132-treated cells along with a control sample pretreated with calf alkaline phosphatase. Compared with the control lysate, 1 h of calf alkaline phosphatase treatment resulted in a downward band shift of FLAG-Maf1 that is indistinguishable from that of CCI-779- or WYE-132-treated cells (Fig. 4A). This result indicates that the faster migrating form observed in the mTOR inhibitor-treated cells represents a more completely dephosphorylated Maf1.
FIGURE 4.
Maf1 phosphorylation status correlates with cellular mTOR signaling. A, MG63 cells were transfected with wild-type FLAG-Maf1 for 24 h and treated with vehicle-DMSO (Cont), 0.5 μm CCI-779, or 0.5 μm WYE-132 for an additional 3 h followed by lysis in NuPAGE-LDS buffer. For calf intestinal alkaline phosphatase (CIAP) treatment, total lysates from vehicle-treated cells were prepared in a buffer lacking phosphatase inhibitors (Phos Inh.) and treated for 1 h at 37 °C. Lysates were probed with FLAG or β-actin antibodies. B, indicated cell lines were transiently transfected FLAG-Maf1 expression vector. 24 h post-transfection cells were treated with 0.5 μm CCI-779, 0.5 μm WYE-132, 5 μg/ml U0126, 10 μg/ml α-amanitin, 100 ng/ml taxol, or DMSO-control for 3 h, as described in Fig. 2. Total cellular lysates were subjected to immunoblotting with antibodies for FLAG, phospho-S6K1, phospho-AKT, total AKT, phospho-ERK (extracellular signal-regulated kinase), and β-actin. Dephosphorylation of the Maf1 was monitored by its shift in migration on SDS-PAGE. The arrowheads indicate migration of the phosphorylated versus hypophosphorylated form of Maf1. C, HEK293 cells were subjected to amino acid withdrawal (−AA) for 2 h and then incubated with amino acids (+AA) for 1 h with or without WYE-132. Lysates were immunoblotted with FLAG, P-S6K1, total 4E-BP1, or β-actin antibodies.
Employing this technique, we next examined Maf1 mobility shift in the FLAG-Maf1-transfected MG63 and MDA361 cells. Treatment of these cells with CCI-779 or WYE-132 for 3 h caused an identical downward band shift in FLAG-Maf1, and this was accompanied by the suppression of other known mTOR biomarkers, including P-S6K1-Thr-389 and P-AKT-Ser-473 (Fig. 4B). As expected, several unrelated inhibitors that target MEK1 (UO126), RNA polymerase II (α-amanitin) or microtubule (taxol), did not induce FLAG-Maf1 band shift (Fig. 4B). To confirm a specific requirement for mTOR in Maf1 phosphorylation, MG63 cells transfected with FLAG-Maf1 were similarly treated with additional commercially available drugs. Aside from WYE-132 and CCI-779, only LY294002, wortmannin, and a wortmannin analog WAY-266175 (33, 34) achieved the most complete shift of Maf1 to the faster migrating form (supplemental Fig. S2), which is likely due to the dual suppression of phosphatidylinositol 3-kinase and mTOR by these agents.
To further confirm Maf1 phosphorylation via mTORC1, we examined Maf1 phosphorylation in response to amino acids. Amino acids rapidly induced P-S6K and P-4E-BP1 and caused an upward band shift of Maf1 consistent with a decreased mobility due to phosphorylation (Fig. 4C). As expected, amino acid-induced phosphorylation of Maf1, S6K1, and 4E-BP1 were completely blocked by WYE-132 (Fig. 4C). Collectively, these results demonstrate that Maf1 phosphorylation is specifically and positively controlled by mTORC1.
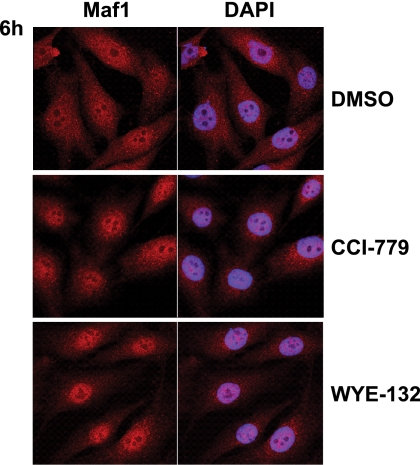
Human Maf1 was shown to localize to the nucleus when introduced into HeLa cells (27). Yeast Maf1, however, exhibits largely cytoplasmic localization under normal growth conditions (22). We, therefore, examined cellular distribution of the endogenous Maf1 employing a newly developed Maf1 antibody. The staining specificity of the antibody was first verified by the marked reduction of Maf1 signal in Maf1-depleted cells as analyzed by immunofluorescence and Western blotting (supplemental Fig. S3, A and B). Immunofluorescence staining of MG63 cells revealed that Maf1 was evenly distributed throughout the cell. Treatment of MG63 with 0.5 μm CCI-779 or WYE-132 significantly increased the amounts of Maf1 in the nucleus, with the nuclear-Maf1 being more pronounced in the WYE-132-treated cells (Fig. 5). These results suggest that mTOR-mediated phosphorylation/dephosphorylation events can modulate Maf1 nuclear-cytoplasmic dynamics and are consistent with a role of Maf1 in repressing Pol III transcription in the nucleus.
FIGURE 5.
mTOR inhibitors cause nuclear accumulation of Maf1. MG63 cells were treated with 0.5 μm CCI-779 or 0.5 μm WYE-132 for 6 h. Merge with DAPI shows the localization of Maf1 in the nucleus. Left, cells were analyzed by immunofluorescence confocal microscopy using Maf1 antibody. Right, nuclei were counterstained with DAPI. Red, Alexa Fluor 594; Blue, DAPI.
Phosphorylation of Maf1 Ser-75 and an Additional Serine/Threonine Residue(s) Is Required for mTORC1 Control of Pol III Transcription
To investigate the functional relevance of mTOR-dependent Maf1 phosphorylation, Maf1 mutant harboring the nonphosphorylatable substitution of Ser-75 to Ala was generated. When transiently expressed in HEK293 or MG63 cells, FLAG-Maf1-S75A (Maf1-S75A) displayed a downshift in gel mobility as compared with the wild-type protein (Fig. 6, A and C). Notably, this downward shift of Maf1-S75A was not as complete as the one observed under mTOR inhibitor-treated conditions, suggesting a possibility that phosphorylation of Maf1 at additional sites other than Ser-75 may also involve mTOR. Next, we examined the effects of Maf1-S75A on Pol III transcription relative to the wild-type Maf1 or a vector control. To overcome potential interference from the endogenous Maf1, its expression was first reduced using siRNA directed against Maf1 3′-UTR before introducing exogenous Maf1. As expected, Maf1 3′-UTR siRNA efficiently depleted the endogenous Maf1 in MG63 (Fig. 6B) cells but did not affect the ectopically introduced Maf1 alleles (Fig. 6C). In agreement with the proposed role of Maf1 as a repressor of Pol III transcription, introduction of the wild-type Maf1 suppressed transcription of tRNALeu and tRNATyr precursors by 40 and 55%, respectively (Fig. 6D). Introduction of Maf1-S75A caused a more pronounced repression of both pre-tRNALeu and pre-tRNATyr (Fig. 6D), indicating that by mimicking a hypophosphorylated state, S75A may function as a more active repressor of Pol III transcription.
FIGURE 6.
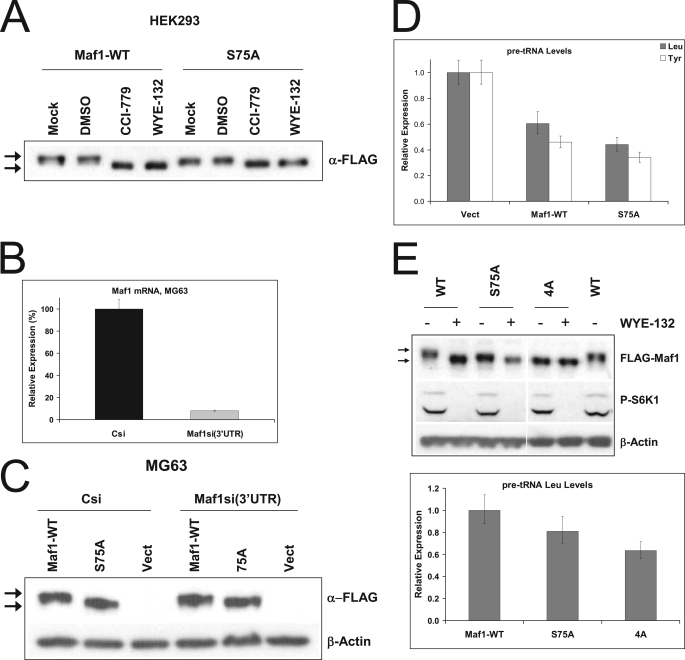
Maf1 phospho-mutants decrease basal Pol III-dependent transcription. A, HEK293 cells were transiently transfected with either vector control, wild-type FLAG-Maf1 (Maf1-WT) or a mutant FLAG-Maf1-S75A (S75A). 24 h after the transfection cells were incubated in the absence (Mock) or presence of DMSO-control, 0.5 μm CCI-779, or 0.5 μm WYE-132 for 3 h. Samples were immunoblotted with FLAG antibodies. B–D, Maf1 was depleted from MG63 cells using 3′-UTR-targeting siRNA and either vector control (Csi), wild-type FLAG-Maf1 (Maf1-WT), or a mutant FLAG-Maf1-S75A (S75A) were re-expressed in these cells for 24 h as described under “Experimental Procedures.” B, control siRNA (Csi) or Maf1-depleted cells that were transfected with empty vector were tested for the level of Maf1 mRNA by qRT-PCR using a Maf1 Taqman assay. C, immunoblotting using FLAG antibody was used to confirm re-expression of Maf1 alleles and to monitor electrophoretic mobility of Maf1. Arrows indicate bands containing phosphorylated or dephosphorylated Maf1. D, precursor tRNALeu and tRNATyr levels in vehicle-treated cells from C were quantified using qRT-PCR. E, MG63 cells were transfected with the vectors expressing either FLAG-tagged wild-type Maf1 (WT), FLAG-Maf1-S75A (S75A), and a quadruple mutant FLAG-Maf1–4A (4A) after depletion of endogenous Maf1 with 3′-UTR-targeting siRNA. For the analysis of a relative shift in Maf1 mobility, cells were treated with 0.5 μm WYE-132 for 3 h. Top, total lysates were probed with FLAG and P-S6K1 antibodies. Arrows indicate phosphorylated or hypophosphorylated forms of Maf1. Bottom, qRT-PCR was then used to measure the amounts of precursor tRNALeu in cells expressing mutant alleles relative to cells containing wild-type Maf1.
The conversion of Maf1-S75A to a partially dephosphorylated form as well as a moderate potentiation by Maf1-S75A in its Pol III repression suggested the presence of additional phospho-sites that might be relevant for mTORC1 control of Pol III. Inspection of PosphoSitePlus data base (30) revealed three potentially important phosphorylation sites, Ser-60, Thr-64, and Ser-68 in human Maf1. To assess the contribution of these phospho-sites to Pol III transcription, a combined alanine substitutions at all four residues, S60A/T64A/S68A/S75A (Maf1–4A), was constructed. After depletion of endogenous Maf1, MG63 cells were transfected with wild-type Maf1, Maf1-S75A, and Maf1–4A and were either treated with WYE-132 to visualize the gel mobility or assayed for Pol III transcription activity. The Maf1–4A protein migrated noticeably faster than the S75A and was no longer shifted further down by WYE-132 (Fig. 6E). Significantly, Maf1–4A caused a further decrease in the pre-tRNALeu levels as compared with that by Maf1-S75A (Fig. 6E). We have observed a similar suppressive effect of Maf1 mutant alleles on the expression of 5 S rRNA gene that is also transcribed by Pol III (supplemental Fig. S4A).
The involvement of Maf1 in the negative regulation of certain Pol I- and Pol II-dependent transcripts raised the possibility that Maf1 phosphorylation could be functionally important for the additional subset of genes. However, single or quadruple phospho-site mutants did not reduce the expression of 45 S precursor rRNA and had only a minor effect on TBP mRNA levels (supplemental Fig. S4, B and C). Together, these results suggest that cooperative phosphorylation of Ser-75 and an additional residue(s) is required for the optimal Pol III activity under conditions of unstressed growth but is not sufficient to repress Pol I/Pol II transcription.
Maf1 Phosphorylation Is Required for mTORC1-dependent, Amino Acid-stimulated Pol III Transcription
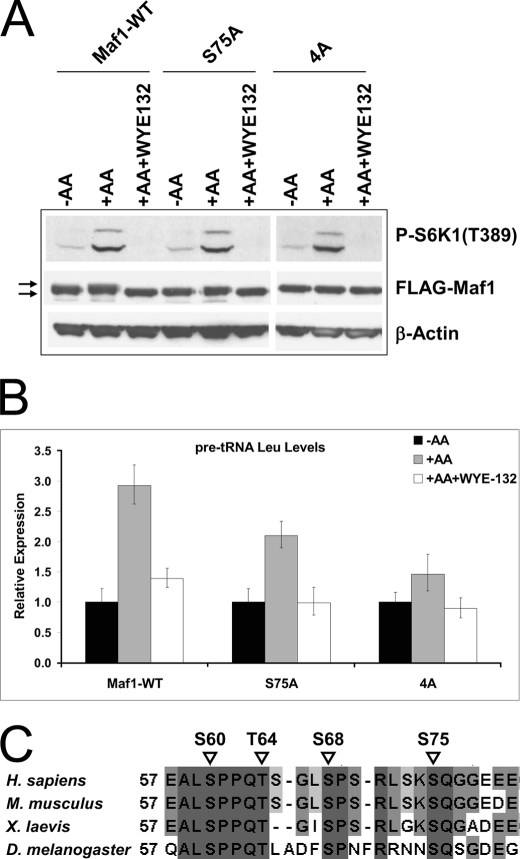
The canonical mTORC1 is known to be acutely activated by amino acids. To further assess the contribution of the Maf1 multisite phosphorylation events in amino acid-induced mTORC1 signaling and Pol III transcription, we introduced the exogenous wild-type Maf1, Maf1-S75A, and Maf1–4A into HEK293 cells. The transfected cells were amino acid-starved and then restimulated with amino acids without or with WYE-132. As with the earlier experiment in Fig. 4C, amino acids induced an upward shift of the wild-type Maf1. This shift was less obvious in cells with Maf1-S75A and virtually eliminated in cells expressing Maf1–4A mutant (Fig. 7A). This result demonstrates that phosphorylation events at Ser-75 and additional serine/threonine residues are targeted by the amino acid-sensitive signaling function of mTORC1. We observed that amino acids stimulated a 3-fold increase in pre-tRNA in the wild-type Maf1 cells. Importantly, the amino acid-induced pre-tRNA synthesis was significantly attenuated in the Maf1-S75A cells and was most dramatically reduced in the Maf1–4A cells (Fig. 7B). These findings together with the results in Fig. 6E highlight a functional requirement for mTOR-dependent Maf1 phosphorylation on Ser-75 and additional residues in mTORC1 control of Pol III transcription.
FIGURE 7.
Maf1 phospho-mutants attenuate amino acid-stimulated Pol III transcription. A, HEK293 cells were transfected with wild-type FLAG-Maf1 (Maf1-WT), FLAG-Maf1-S75A (S75A), and a quadruple mutant FLAG-Maf1–4A (4A). Cells expressing Maf1 constructs were shifted to amino acid-free medium (−AA) for 2 h followed by 1 h of amino acid stimulation with (+AA+WYE-132) or without WYE-132 as described in Fig. 4C. Lysates were immunoblotted with FLAG, P-S6K1, total 4E-BP1, or β-actin antibodies. Arrows indicate phosphorylated or hypophosphorylated forms of Maf1. B, experiments are as in A; cells expressing Maf1 alleles were stimulated with amino acids in the presence or absence of WYE-132. Pre-tRNALeu levels were quantified using qRT-PCR. C, protein sequence alignment of the Maf1 region from different species is shown. Alignment and percentage identity shading was done with ClustalW2/Jalview (EMBL-EBI). Numbers designate amino acid positions. Dark gray residues show amino acids identical between all investigated species. Positions of phosphorylation sites are marked with triangles.
The biological significance of Maf1 phosphorylation sites prompted us to examine whether similar residues are present in other organisms. Interestingly, alignment of Maf1 sequences from higher eukaryotes revealed that Ser-60, Thr-64, Ser-68, and Ser-75 are well conserved among Homo sapiens, Mus musculus, Xenopus laevis, and Drosophila melanogaster (Fig. 7C), suggesting the evolutionary importance of these sites in regulation of Maf1 function.
Association of Maf1, Pol III, Brf1, and mTORC1 with Pol III Promoters
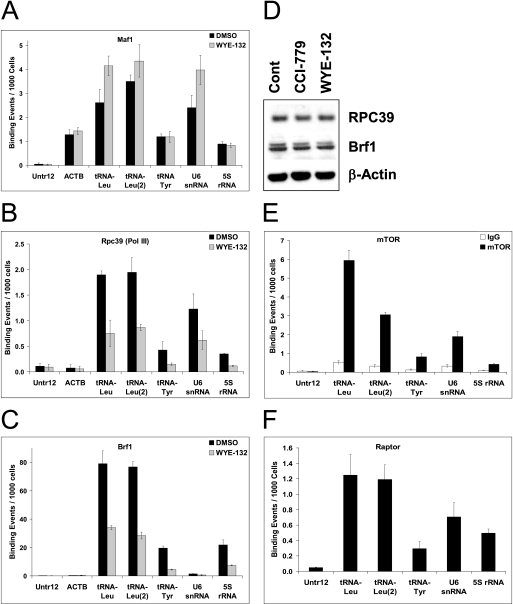
Given that Maf1 is a repressor of Pol III activity and a proximal mTOR target, we investigated whether mTORC1-dependent phosphorylation modulates its binding to Pol III-transcribed promoters. ChIP analysis of the actively proliferating MG63 cells detected specific Maf1 binding to two independent tRNALeu genes residing in different chromosomal locations, tRNATyr, U6 snRNA, and 5 S rRNA as well as the Pol II-transcribed actin region but not at a silent region of the genome Untr12 (Fig. 8A). Low, but significantly above the background level of Maf1, occupancy of actin is consistent with Maf1 binding to a set of Pol II promoters (24). Most importantly, acute WYE-132 treatment resulted in a small but reproducible increase in Maf1 recruitment to tRNALeu and U6 snRNA genes (Fig. 8A), suggesting that dephosphorylated Maf1 becomes a more efficient repressor that binds Pol III targets. Considering the possibility that the enhanced Maf1 binding may directly interfere with the recruitment of Pol III and TFIIIB to these templates, we next examined the occupancy of Pol III and Brf1 of the same subset of Pol III-transcribed genes. Employing the antibody against the Pol III subunit RPC39, Pol III was specifically detected at the two tRNALeu genes, tRNATyr, U6 snRNA, and 5 S rRNA but not at the Untr12 or the actin gene. Strikingly, WYE-132 reduced the Pol III occupancy of the corresponding genes by ∼2–2.5-fold (Fig. 8B). In control cells, we detected a robust recruitment of Brf1 to tRNALeu, tRNATyr, and 5 S rRNA genes but not to U6 snRNA, actin, or Untr12 (Fig. 8C). This is consistent with the report that Brf2 instead of Brf1 is required for U6 snRNA expression (35, 36). WYE-132 strongly diminished binding of Brf1 to the tRNALeu, tRNATyr, and 5 S rRNA regions, showing a more than 2-fold decrease for these genes (Fig. 8C). Immunoblot analysis indicated that WYE-132 did not reduce the expression of Pol III or Brf1 in these cells (Fig. 8D). Taken together, the decreased recruitments of Pol III and Brf1 to the target genes could be the primary mechanism for the suppressive effects of WYE-132 on Pol III transcription.
FIGURE 8.
Effect of pharmacological inactivation of mTOR on Maf1, Pol III, Brf1, and mTORC1 occupancy at Pol III promoters. MG63 cells were treated with vehicle-DMSO or 0.5 μm WYE-132 for 3 h and processed for ChIP assays as described under “Experimental Procedures.” Chromatin from DMSO (black bars) or WYE-132 (gray shaded bars)-treated samples were immunoprecipitated with Maf1 (FL-256) (A), RPC39 (Pol III subunit) (B) or Brf1 (C) antibodies, and occupancy of the indicated regions was determined by qPCR. Untr12 is a negative-control genomic region. Values represent the averages of transcription binding events detected per 1000 Cells. Error bars represent S.D. of triplicate assays of an individual experiment. Graphs are representative of one of three independent experiments. D, MG63 cells were treated with vehicle (DMSO), 0.5 μm CCI-779, or 0.5 μm WYE-132 for 3 h. Lysates were subjected to the immunoblotting using RPC39, Brf1, or β-actin antibodies. E–F, quantitative ChIP analysis with mTOR (E), raptor (F), or control IgG antibodies is shown. Immunoprecipitated DNA was analyzed by qPCR using primers specific for the indicated regions as described in A–C.
Considering the role of mTOR in transcriptional activation of Pol III targets and mTOR-mediated Maf1 phosphorylation, we tested whether mTOR itself may directly associate with Pol III templates. When ChIP assay was performed on MG63 cells, both mTOR and Raptor were specifically associated with two independent tRNALeu genes, U6 snRNA, tRNATyr and to a lesser extend with tRNATyr or 5 S rRNA (Fig. 8, E and F). No binding was detected by a control IgG or at the Untr12 region. WYE-132, however, did not alter the mTORC1 occupancy of these Pol III targets (data not shown), indicating that mTORC1 binding to these regions is constitutive. An additional mTOR ChIP study with HEK293 cells produced a similar finding (supplemental Fig. S5). Overall, these data indicate that in actively proliferating cells, mTORC1 interacts with endogenous Pol III-transcribed genes in a constitutive fashion.
DISCUSSION
Recent discovery of mTORC1/mTORC2 small molecule inhibitors (37–39) has yielded considerable efforts in deciphering the complexity of cellular processes that are governed by mTOR. SILAC-based identification of a novel phosphorylation site in Maf1 whose phosphorylation is profoundly decreased in breast cancer cells treated with WYE-132 led us to propose a more immediate role of mTOR in regulating RNA polymerase III transcription than previously anticipated. Specifically, suppressive effects of pharmacological mTOR inhibitors in a broad panel of cancer lines coupled with siRNA depletion studies implicate mTORC1 as a critical positive regulator of Pol III outputs. Generally, more pronounced inhibition of Pol III transcription by WYE-132 as compared with CCI-79 could be explained by 1) existence of “rapamycin-insensitive” mTORC1 substrates involved in Pol III transcription and 2) more complete suppression of mTOR catalytic activity by WYE-132. Our results are also in agreement with the rapamycin effect on Pol III-dependent transcription recently described by Woiwode et al. (21). Expanding upon the genetic link between mTOR and Maf1, we demonstrated that Maf1-depleted cells become refractory to the Pol III inhibition in response to acute exposure to CCI-779 or WYE-132. Thus, it appears that Maf1 acts downstream of mTORC1 to transmit regulatory signals to the Pol III machinery.
The identification of Maf1 in a phospho-proteomics screen as well as its profound conversion to a faster migrating form upon exposure to specific pharmacological mTOR inhibitors in different cell lines raises the possibility that Maf1 is an immediate target of mTOR. Further salient evidence in support of this idea comes from the additional manipulations of the mTORC1 signaling. Specific activation of mTORC1 during conditions of amino acid stimulation causes a decrease in Maf1 gel mobility, an indication of increased phosphorylation. Conversely, mTOR or raptor siRNA-mediated knockdown increases Maf1 gel mobility (data not shown), consistent with the effect of mTOR inhibitors. We have attempted but failed to detect a direct phosphorylation of Maf1 by immunoprecipitated mTOR enzyme in vitro (data not shown). Nevertheless, this negative observation should be interpreted with caution due to the technical complexity in the assays with mTOR complexes in vitro. Although we were not able to demonstrate a direct phosphorylation of Maf1 by mTOR, our data do not exclude another kinase(s) of the mTORC1 axis for phosphorylation of these sites. Recent reports suggest that Sch9, a yeast kinase homologous to mammalian S6K1, regulates Pol III transcription in a Maf1-dependent manner (12, 40). In support of a role for S6K1 in Maf1 phosphorylation in cancer cells, we have observed that a rapamycin-resistant S6K1 can partially protect Maf1 from the WYE-132-induced dephosphorylation (supplemental Fig. S6). It, therefore, remains formally possible that mTORC1 and/or S6K1 alone or in concert with an additional kinase(s) directly contributes to Maf1 phosphorylation and functional regulation.
Whereas the sequence surrounding Ser-75 (SPSLSKSQGGE) in Maf1 does not resemble any known mTOR phosphorylation sites or the consensus residues that could be phosphorylated by S6K1 (or other AGC kinases), we cannot exclude the possibility that mTOR (or S6K1) relies more on protein conformation than on exact amino acid context to identify cognate phosphorylation sites. Intriguingly, we noticed that human Maf1 contains residues (AVREDFKDLK, 138–147) which are somewhat similar to the mTOR signaling (TOS) motif that is important for binding of mTOR substrates to the raptor-mTOR complex. A further investigation is clearly needed to establish whether Ser-75 of human Maf1 is a direct mTOR substrate and if not so, to identity another mTOR-regulated kinase(s) responsible for Ser-75 phosphorylation in cancer cells.
In addition to our report, Ser-75-containing Maf1 peptide was also discovered in three large proteomic studies that were directed to search for potential ATM/ATR kinase substrates (41, 42) or for the mitosis-specific phosphorylation events (43). Whether Maf1 represents a bona fide substrate of the checkpoint kinases and plays a role in DNA damage is not known. It appears that mTOR kinase inhibitors CCI-779 and WYE-132 do not block ATM/ATR-dependent responses (data not shown), making it unlikely that Ser-75 dephosphorylation involves inhibition of ATM/ATR activity in the current assay conditions. Our data also emphasize the requirement of the mTOR-mediated phosphorylation for the proper intracellular localization of Maf1. Specifically, nuclear accumulation of Maf1 indeed correlates with a concomitant dephosphorylation of Maf1 in the presence of mTOR inhibitors, highlighting a potential similarity between yeast and mammalian cells in the way mTOR prevents Maf1 nuclear localization under conditions of active growth. Nevertheless, sequence alignment between human and yeast Maf1 shows that phosphorylation sites analyzed here do not overlap with amino acid residues in yeast proteins, which are known to be phosphorylated by protein kinase A and/or Sch9, the yeast ortholog of human S6K1 (supplemental Fig. S7). In fact, these phosphorylation sites in yeast Maf1 (Ser-90, -101, -177, -178, -179, -209, and -210) reside in a region that is not conserved in the human protein. At the same time, our finding that the rapamycin-resistant S6K1 partially reverses WYE-132-induced Maf1 dephosphorylation (supplemental Fig. S6) supports a role of S6K1 in mTORC1-dependent phosphorylation of human Maf1. Future studies will clarify the identity of the kinase(s) that directly phosphorylates Maf1.
Does the mTOR-dependent Ser-75 phosphorylation play a role in Maf1 function? We showed that an S75A, a nonphosphorylatable Maf1 allele, partially mimics the effects of mTOR inhibitors in both the Maf1 gel mobility and Pol III transcription assays, strongly suggesting that phosphorylation of Ser-75 attenuates its Pol III-repressive function. Notably, the magnitude of the downward shift in Maf1 gel mobility in this mutant correlates with the severity of Pol III transcriptional repression in full growth medium or under amino acid stimulation conditions. Although Maf1-S75A demonstrated a stronger Pol III repression than the wild-type Maf1, it appears that dephosphorylation of additional residues also contributes to the full repressive activity of Maf1. Indeed, our findings that 1) a quadruple substitution mutant, Maf1–4A, exhibits the most pronounced suppression of basal or amino acid-induced Pol III synthesis compared with that of Maf1-S75A and 2) Maf1–4A downward shift in gel migration is virtually complete and is no longer farther down-shifted by WYE-132 suggest that mTOR and/or unknown mTOR-regulated kinase might be required for the phosphorylation of multiple sites on Maf1. Furthermore, our studies involving amino acid stimulation not only demonstrate a direct link between specific activation of mTORC1 and phosphorylation of Maf1 but also provide novel evidence that amino acid availability modulates Pol III transcription in general. Whereas several studies have described the effects of amino acids on Pol III transcription in yeast (44, 45), to our knowledge this is the first report showing the activating action of amino acids on tRNA synthesis in human cells. The fact that WYE-132 effectively blocks amino acid-stimulated tRNA synthesis further underscores the pivotal role for mTOR kinase in the regulation of nutrient-responsive Pol III transcription. Despite the inhibitory effects of Maf1–75A or Maf1–4A on Pol III transcription, these substitutions did not result in a further decrease in overall cell growth in two-dimensional cultures as compared with that of Maf1-WT (supplemental Fig. S8). Hence, it remains to be determined whether phosphorylation of Maf1 is critical for tumor growth in vivo or in anchorage-independent settings.
Little is understood of the precise mechanism by which human Maf1 represses Pol III transcription. First, endogenous Maf1 associates with Pol III, TFIIIB, and likely with TFIIIC complexes in human cells (25–27). Second, a recent in vitro study has demonstrated that human Maf1 represses Pol III activity by blocking the recruitment of Pol III to the TFIIIB-TFIIIC-DNA complexes and to a lesser degree by interfering with the binding of TFIIIB to DNA-bound TFIIIC-DNA (46). Our results indicate that mTOR activity is required for a normal recruitment of both RNA Pol III and Brf1 to their target genes, which provides a mechanistic explanation for the inhibitory effects of mTOR inhibitors on cellular Pol III transcription. Paradoxically, we found only a small increase in Maf1 occupancy of a subset of Pol III genes during acute treatment with WYE-132. One interpretation may reflect a significantly heterogeneous basal occupancy by Maf1 in current cell culture condition, and it is possible that mTOR inhibition promotes Maf1 recruitment in only a subset of de-repressed promoters and/or cells. An additional explanation could be that Maf1-mediated repression may rely on a mechanism that does not require tight binding of Maf1 to the DNA. In this respect, recent work has demonstrated that human Maf1 is able to inhibit Pol III activity in vitro only when polymerase is free in solution, whereas Pol III, which is bound to preinitiation or elongation complexes, is largely resistant to Maf1 (46). This and the fact that Maf1 is present on a subset of genes under normal growth conditions even in the absence of the drug suggests a transient nature of Maf1-mediated repression, which may be difficult to capture by a conventional ChIP assay. At the same time, unequal modulation of Maf1 association with tRNA genes in repressive conditions could be interpreted by the differential transcriptional regulation of these tRNAs. For example in yeast, Maf1-mediated repression is not uniform for different genes; binding of Maf1 to tRNAMet gene promoter seems to be only slightly enhanced by rapamycin (22).
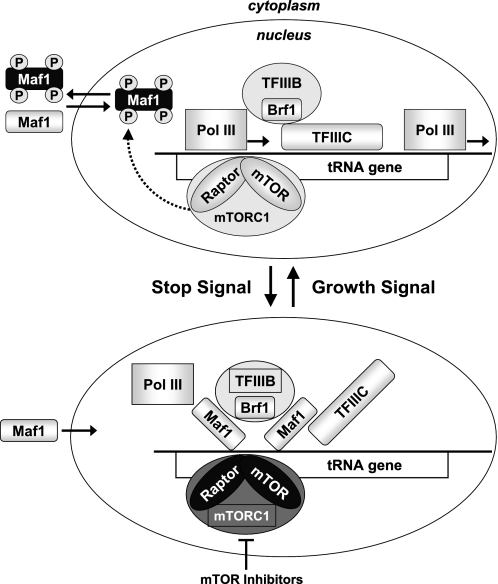
Additional support for the more immediate role of mTOR in control of Pol III output in cancer cells comes from our analysis of mTORC1 occupancy at Pol III-transcribed promoters. Although unexpected, this finding is in accord with the previously described mTOR localization in the nuclei of cancer lines (47–49) and is also consistent with the recently described association of TOR with 35 S and 5 S rDNA promoters in yeast (13, 50). Further experiments are required to determine whether mTOR interacts directly with Pol III itself, TFIIB, or Maf1. Based on our findings, we propose a model (Fig. 9) in which a nuclear pool of mTOR within mTORC1 associates with Pol III promoters to continuously monitor and adjust Pol III transcription rates according to the growth and metabolic demands of the cell. In conditions when the most active burst of Pol III-dependent synthesis is required, such as amino acid or growth factor stimulation, mTOR-mediated phosphorylation of Maf1 at sites including Ser-75 serves as a signal to assemble active TFIIIB and Pol III complexes on DNA. In this scenario Maf1 may be more completely inactivated by phosphorylation at multiple residues by mTOR itself and/or by another mTOR-regulated kinase(s) leading to its exclusion from the nucleus. Unfavorable growth conditions or mTOR inhibition by drugs such as WYE-132 or CCI-779 can induce dephosphorylation of Maf1, resulting in its nuclear accumulation. Thus, the mTOR inhibitor-invoked dephosphorylation of Maf1 represents a key mechanistic step in Pol III repression by Maf1, leading to a genome-wide repression of selected Pol III templates.
FIGURE 9.
Schematic model depicting the role for mTOR-mediated phosphorylation of Maf1 in the regulation of Pol III transcription. Shown is a proposed mechanism whereby mTOR regulates Maf1 activity in cancer cells. Top, actively proliferating cells utilize mTOR-mediated phosphorylation of Maf1 to maintain optimal activity of Pol III apparatus. Bottom, inactivation of mTOR by pharmacological inhibitors or due to unfavorable growth conditions leads to enhanced nuclear retention of dephosphorylated Maf1 and repression of new rounds of Pol III synthesis.
Acknowledgments
We thank Wei-Guo Zhang for technical assistance and Drs., James Gibbons, Arie Zask, and Robert Abraham for helpful discussions and general project support.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Table S1, and Figs. S1–S8.
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- mTORC2
- mTOR complex 2
- CCI-779
- cell cycle inhibitor 779
- ChIP
- chromatin immunoprecipitation
- Pol III
- RNA polymerase III
- SILAC
- stable isotope labeling in culture
- siRNA
- small interfering RNA
- DAPI
- 4,6-diamidino-2-phenylindole
- qRT
- quantitative reverse transcription
- MS
- mass spectometry
- snRNA
- small nuclear RNA
- UTR
- untranslated region.
REFERENCES
- 1.Alessi D. R., Pearce L. R., García-Martínez J. M. (2009) Sci. Signal. 2, pe27. [DOI] [PubMed] [Google Scholar]
- 2.Dunlop E. A., Tee A. R. (2009) Cell. Signal. 21, 827–835 [DOI] [PubMed] [Google Scholar]
- 3.Guertin D. A., Sabatini D. M. (2007) Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 4.Ma X. M., Blenis J. (2009) Nat. Rev. Mol. Cell Biol. 10, 307–318 [DOI] [PubMed] [Google Scholar]
- 5.Engelman J. A. (2009) Nat. Rev. Cancer 9, 550–562 [DOI] [PubMed] [Google Scholar]
- 6.Carracedo A., Pandolfi P. P. (2008) Oncogene 27, 5527–5541 [DOI] [PubMed] [Google Scholar]
- 7.Yuan T. L., Cantley L. C. (2008) Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannan K. M., Brandenburger Y., Jenkins A., Sharkey K., Cavanaugh A., Rothblum L., Moss T., Poortinga G., McArthur G. A., Pearson R. B., Hannan R. D. (2003) Mol. Cell. Biol. 23, 8862–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James M. J., Zomerdijk J. C. (2004) J. Biol. Chem. 279, 8911–8918 [DOI] [PubMed] [Google Scholar]
- 10.Mayer C., Zhao J., Yuan X., Grummt I. (2004) Genes Dev. 18, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaragoza D., Ghavidel A., Heitman J., Schultz M. C. (1998) Mol. Cell. Biol. 18, 4463–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J., Moir R. D., Willis I. M. (2009) J. Biol. Chem. 284, 12604–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y., Tsang C. K., Zheng X. F. (2009) EMBO J. 28, 2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White R. J. (2008) Trends Genet. 24, 622–629 [DOI] [PubMed] [Google Scholar]
- 15.Scott M. R., Westphal K. H., Rigby P. W. (1983) Cell 34, 557–567 [DOI] [PubMed] [Google Scholar]
- 16.White R. J., Stott D., Rigby P. W. (1989) Cell 59, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 17.White R. J., Stott D., Rigby P. W. (1990) EMBO J. 9, 3713–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter A. G., Sourvinos G., Allison S. J., Tosh K., Scott P. H., Spandidos D. A., White R. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12619–12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall L., Kenneth N. S., White R. J. (2008) Cell 133, 78–89 [DOI] [PubMed] [Google Scholar]
- 20.Marshall L., White R. J. (2008) Nat. Rev. Cancer 8, 911–914 [DOI] [PubMed] [Google Scholar]
- 21.Woiwode A., Johnson S. A., Zhong S., Zhang C., Roeder R. G., Teichmann M., Johnson D. L. (2008) Mol. Cell. Biol. 28, 4204–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oficjalska-Pham D., Harismendy O., Smagowicz W. J., Gonzalez de Peredo A., Boguta M., Sentenac A., Lefebvre O. (2006) Mol. Cell 22, 623–632 [DOI] [PubMed] [Google Scholar]
- 23.Roberts D. N., Wilson B., Huff J. T., Stewart A. J., Cairns B. R. (2006) Mol. Cell 22, 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson S. S., Zhang C., Fromm J., Willis I. M., Johnson D. L. (2007) Mol. Cell 26, 367–379 [DOI] [PubMed] [Google Scholar]
- 25.Reina J. H., Azzouz T. N., Hernandez N. (2006) PLoS ONE 1, e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodfellow S. J., Graham E. L., Kantidakis T., Marshall L., Coppins B. A., Oficjalska-Pham D., Gérard M., Lefebvre O., White R. J. (2008) J. Mol. Biol. 378, 481–491 [DOI] [PubMed] [Google Scholar]
- 27.Rollins J., Veras I., Cabarcas S., Willis I., Schramm L. (2007) Int. J. Biol. Sci. 3, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu K., Shi C., Toral-Barza L., Lucas J., Shor B., Kim J. E., Zhang W. G., Mahoney R., Gaydos C., Tardio L., Kim S. K., Conant R., Curran K., Kaplan J., Verheijen J., Ayral-Kaloustian S., Mansour T. S., Abraham R. T., Zask A., Gibbons J. (2010) J. Cancer Res. 70, 621–631 [DOI] [PubMed] [Google Scholar]
- 29.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 30.Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., Zhang B. (2004) Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 31.Alexiadis V., Ballestas M. E., Sanchez C., Winokur S., Vedanarayanan V., Warren M., Ehrlich M. (2007) Biochim. Biophys. Acta 1769, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J., Shakey Q., Liu W., Schuller A., Follettie M. T. (2007) J. Proteome Res. 6, 4684–4689 [DOI] [PubMed] [Google Scholar]
- 33.Yu K., Toral-Barza L., Shi C., Zhang W. G., Zask A. (2008) Cancer Biol. Ther. 7, 307–315 [DOI] [PubMed] [Google Scholar]
- 34.Zask A., Kaplan J., Toral-Barza L., Hollander I., Young M., Tischler M., Gaydos C., Cinque M., Lucas J., Yu K. (2008) J. Med. Chem. 51, 1319–1323 [DOI] [PubMed] [Google Scholar]
- 35.Saxena A., Ma B., Schramm L., Hernandez N. (2005) Mol. Cell. Biol. 25, 9406–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schramm L., Pendergrast P. S., Sun Y., Hernandez N. (2000) Genes Dev. 14, 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu K., Toral-Barza L., Shi C., Zhang W. G., Lucas J., Shor B., Kim J., Verheijen J., Curran K., Malwitz D. J., Cole D. C., Ellingboe J., Ayral-Kaloustian S., Mansour T. S., Gibbons J. J., Abraham R. T., Nowak P., Zask A. (2009) Cancer Res. 69, 6232–6240 [DOI] [PubMed] [Google Scholar]
- 38.Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feldman M. E., Apsel B., Uotila A., Loewith R., Knight Z. A., Ruggero D., Shokat K. M. (2009) PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R. (2009) Genes Dev. 23, 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 42.Stokes M. P., Rush J., Macneill J., Ren J. M., Sprott K., Nardone J., Yang V., Beausoleil S. A., Gygi S. P., Livingstone M., Zhang H., Polakiewicz R. D., Comb M. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig J. R., 2nd, Oliver S. G., McLaughlin C. S. (1977) Mol. Gen. Genet. 158, 117–122 [DOI] [PubMed] [Google Scholar]
- 45.Clarke E. M., Peterson C. L., Brainard A. V., Riggs D. L. (1996) J. Biol. Chem. 271, 22189–22195 [DOI] [PubMed] [Google Scholar]
- 46.Cabart P., Lee J., Willis I. M. (2008) J. Biol. Chem. 283, 36108–36117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Shu L., Hosoi H., Murti K. G., Houghton P. J. (2002) J. Biol. Chem. 277, 28127–28134 [DOI] [PubMed] [Google Scholar]
- 48.Bachmann R. A., Kim J. H., Wu A. L., Park I. H., Chen J. (2006) J. Biol. Chem. 281, 7357–7363 [DOI] [PubMed] [Google Scholar]
- 49.Kim J. E., Chen J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14340–14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Tsang C. K., Watkins M., Bertram P. G., Zheng X. F. (2006) Nature 442, 1058–1061 [DOI] [PubMed] [Google Scholar]