Abstract
Induction of type I interferons by the transcription factor IRF3 is essential in the initiation of antiviral innate immunity. Activation of IRF3 requires C-terminal phosphorylation by the upstream kinases TBK1-IKKi, where IRF3 phosphorylation promotes dimerization, and subsequent nuclear translocation to the IFNβ promoter. Recent studies have described the ubiquitin-editing enzyme A20 as a negative regulator of IRF3 signaling by associating with TBK1-IKKi; however, the regulatory mechanism of A20 inhibition remains unclear. Here we describe the adaptor protein, TAX1BP1, as a key regulator of A20 function in terminating signaling to IRF3. Murine embryonic fibroblasts (MEFs) deficient in TAX1BP1 displayed increased amounts of IFNβ production upon viral challenge compared with WT MEFs. TAX1BP1 inhibited virus-mediated activation of IRF3 at the level of TBK1-IKKi. TAX1BP1 and A20 blocked antiviral signaling by disrupting Lys63-linked polyubiquitination of TBK1-IKKi independently of the A20 deubiquitination domain. Furthermore, TAX1BP1 was required for A20 effector function because A20 was defective for the targeting and inactivation of TBK1 and IKKi in Tax1bp1−/− MEFs. Additionally, we found the E3 ubiquitin ligase TRAF3 to play a critical role in promoting TBK1-IKKi ubiquitination. Collectively, our results demonstrate TBK1-IKKi to be novel substrates for A20 and further identify a novel mechanism whereby A20 and TAX1BP1 restrict antiviral signaling by disrupting a TRAF3-TBK1-IKKi signaling complex.
Keywords: Interferon, Pathogen-associated Molecular Pattern (PAMP), Signal Transduction, Ubiquitin Ligase, Ubiquitination, A20, IKKi, Rig-I, TBK1, Antiviral Signaling
Introduction
The hallmark of an innate immune response against virus infection is the induction of a rapid, nonspecific antiviral state that coordinates the rapid elimination of the virus and the activation of a secondary T-cell-mediated adaptive immune response (1–4). This fundamental aspect of innate antiviral immunity is mediated by type I interferons (IFNα and IFNβ), key cytokines produced in the course of a viral infection, which, upon secretion, engage the IFNαβ receptor on neighboring cells to activate a signaling cascade that culminates in the production of interferon-stimulated genes that collectively promote an antiviral state by blocking virus replication and spread (3, 5). IFN production is tightly regulated to prevent persistent IFN production, which has been associated with a variety of immunological disorders, including systemic lupus erythematosus and Sjögren syndrome (6–8).
During acute viral infection, double-stranded RNA is often generated during replicative cycles and serves as a pathogen-associated molecular pattern. These pathogen-associated molecular patterns are detected by pattern recognition receptors, which activate antiviral signaling cascades leading to IFN production (1, 2, 5). Two major classes of pattern recognition receptors have been described for detecting pathogen-associated molecular patterns derived from viral nucleic acids: Toll-like receptors (TLRs)2 and the Rig-I-like helicases. TLRs are membrane-bound and are found on the cell surface or in endosomal compartments. Recognition of double-stranded RNA by TLR3 results in the recruitment of the adaptor molecule TRIF, leading to NF-κB activation via RIP1/TRAF6-mediated activation of the canonical IκB kinases (IKK) and IRF3 activation via TRAF3-mediated activation of the TBK1-IKKi kinases. The Rig-I-like helicases are cytoplasmic sensors of viral nucleic acids. Upon double-stranded RNA detection, these receptors reconform to associate with IPS-1 (also known as MAVS, VISA, or Cardif) (9–12) via CARD-CARD domains found in both molecules. IPS-1 further complexes with other adaptor proteins, including WDR5, TRADD, FADD, RIP1, and STING (also known as MITA or ERIS) to activate the NF-κB and IRF3 transcription factors (13–18). Signal bifurcation toward NF-κB is thought to be mediated in part by a TRAF6-IKK complex, whereas IRF3 activation involves the TRAF3-TBK1-IKKi module (1, 2, 12). Ultimately, NF-κB and IRF3 activate IFN production by binding to the enchancer/promoter region of the IFNβ gene (1–3, 5, 19).
The ubiquitin-editing enzyme A20 (also known as TNFAIP3) has been shown to negatively regulate antiviral signaling pathways leading to IRF3 activation; however, its mechanism of inhibition is poorly defined (20–22). In this report, we identify the A20-interacting protein, TAX1BP1 (Tax1-binding protein 1) (also known as T6BP or TXBP151) as a novel negative regulator of antiviral signaling pathways. A20 and TAX1BP1 cooperate to terminate antiviral signaling by antagonizing Lys63-linked polyubiquitination of TBK1 and IKKi. TAX1BP1 probably functions as an adaptor molecule for A20 because A20 is unable to interact with TBK1 or IKKi in the absence of TAX1BP1. Furthermore, our results suggest that A20 inhibits IRF3 signaling via a novel mechanism independent of A20 DUB function.
EXPERIMENTAL PROCEDURES
Cell Culture, Reagents, and Antibodies
293T cells (ATCC), Tax1bp1−/− MEFs (23), and Traf3−/− MEFs (provided by Drs. J. Bethea and G. Cheng) were grown in DMEM supplemented with fetal bovine serum (10%) and penicillin/streptomycin (1%). 293-TLR4/MD2-CD14 cells (provided by Dr. K. Tolba) were cultured in Dulbecco's modified Eagle's medium plus fetal bovine serum (10%), hygromycin B (50 μg/ml), and blasticidin (10 μg/ml). Poly(I-C) was purchased from Invivogen. PMA, ionomycin, cycloheximide, lipopolysaccharide, and anti-FLAG M2 antibodies were purchased from Sigma. Other antibodies used in this study were IRF3, GFP, TRAF6, and TRAF3 (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)); phospho-IRF3 and Myc (Upstate); TBK1, TAX1BP1, and β-actin (Abcam); A20 (BD Biosciences); IKKi (Imgenex); ubiquitin (Stressgen); Lys63-linked ubiquitin (Biomol); and HA (Roche Applied Science). Control scrambled, TAX1BP1, A20, TRAF6, and TRAF3 siGENOME SMARTpool® small interfering RNAs (siRNAs) were purchased from Dharmacon/Thermo Scientific.
Plasmid Constructs
Plasmids encoding TAX1BP1, A20, NF-κB-TATA luc, and ISRE luc reporter DNAs have been described elsewhere (23). Constructs for ΔRig-I, ΔMDA5, IPS-1, TBK1, IKKi, IRF3, IRF3SA, IRF7, TRIF, and IFNβ luc reporter DNAs have been described previously (16, 24). NFAT-TA-luc reporter DNA, HA-ubiquitin WT, and Lys63-only plasmids have been described previously (25, 26). HA-TRAF3 and FLAG-A20 1–367 DNA constructs were provided by Dr. S.-C. Sun and Dr. R. Beyaert, respectively.
Transfections and Reporter Assays
293T cells, 293-TLR4/MD2-CD14 cells, and MEFs were transfected with FuGENE 6 or FuGENE HD (Roche Applied Science). siRNA (60 pmol) transfections were performed using Lipofectamine 2000 (Invitrogen). Reporter assays were performed 24 h after DNA transfection unless otherwise indicated using a dual luciferase assay kit (Promega). Results for firefly luciferase activity were normalized to Renilla luciferase activity. Data are expressed as mean -fold increase ± S.E. relative to control levels from a representative experiment performed 3–4 times in duplicate or triplicate. An asterisk indicates a p value of <0.05 as determined by Student's t test.
ELISA
ELISAs for mouse IFNβ were performed using supernatants from cells where values are expressed as pg/ml ± S.E. as calculated from a standard curve derived from recombinant IFNβ provided in the ELISA kit (PBL Interferon Source).
Immunoblotting, Co-immunoprecipitations, and Ubiquitination Assays
Whole cell lysates were generated by lysing cells in radioimmune precipitation buffer on ice, followed by centrifugation. Cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membranes, and subjected to immunoblotting. For co-IPs, lysates were diluted 1:1 in radioimmune precipitation buffer and precleared with protein A-agarose beads for 30 min at 4 °C. Precleared lysates were further incubated at 4 °C with the indicated antibodies (1–3 μl): 2 h for antibodies against transfected proteins, overnight for antibodies against endogenous proteins. The protein-antibody complex was captured by the addition of protein A-agarose beads (40 μl) for 2h at 4 °C. Three washes with radioimmune precipitation buffer followed by the addition of 2× LSB allowed for the bound proteins to be eluted. Detection of TBK1-IKKi conjugated to ubiquitin was achieved by a similar co-IP method, where potential ubiquitinated TBK1-IKKi-interacting proteins were removed by stringent washes using radioimmune precipitation buffer containing 1 m urea as described (26).
Native Gel Dimerization Assay
293T cells recovered from 6-well dishes were harvested in 100 μl of Triton X lysis buffer (20 mm Tris-Cl, pH 7.6, 1% Triton X-100, 150 mm NaCl, 5% glycerol, 40 mm β-glycerophosphate, 1 mm EDTA) containing protease inhibitors. 10 μg of protein was mixed with 2× native PAGE sample buffer (125 mm Tris-Cl, pH 6.8, 30% glycerol, bromphenol blue) and subjected to electrophoresis on non-denaturing 7.5% polyacrylamide gels as described (22).
Virus Infections
293T cells were infected with VSV-ΔM at a multiplicity of infection of 0.1. MEFs were infected with either VSV-ΔM or Sendai virus at a multiplicity of infection of 1.0. VSV-ΔM is a vesicular stomatitis virus containing a mutation in its matrix protein compromising its function in controlling cellular mRNA nuclear export (27).
RESULTS
TAX1BP1 Is a Negative Regulator of Antiviral Signaling
Our work and that of others has demonstrated that the NF-κB inhibitory function of A20 is dependent on TAX1BP1 (23, 28). Furthermore, TAX1BP1-deficient MEFs display a similar phenotype to A20-deficient MEFs with respect to NF-κB activation, suggesting a cooperative role between A20 and TAX1BP1 (23, 28, 29). Because A20 has been shown to block signaling to IRF3, we sought to determine if TAX1BP1 could similarly inhibit IRF3. Indeed, reporter assays evaluating IFNβ promoter activation in 293T cells revealed that expression of TAX1BP1 potently blocked IFNβ activation mediated by virus infection (supplemental Fig. 1A). In addition to the production of type I interferons, virus infection also triggers the synthesis of proinflammatory cytokines, many of which are regulated by NF-κB transcription factors. To determine if TAX1BP1 regulated virus-mediated NF-κB activation, reporter assays measuring NF-κB activity were also performed. As shown in supplemental Fig. 1B, overexpression of TAX1BP1 blocked virus-mediated activation of NF-κB.
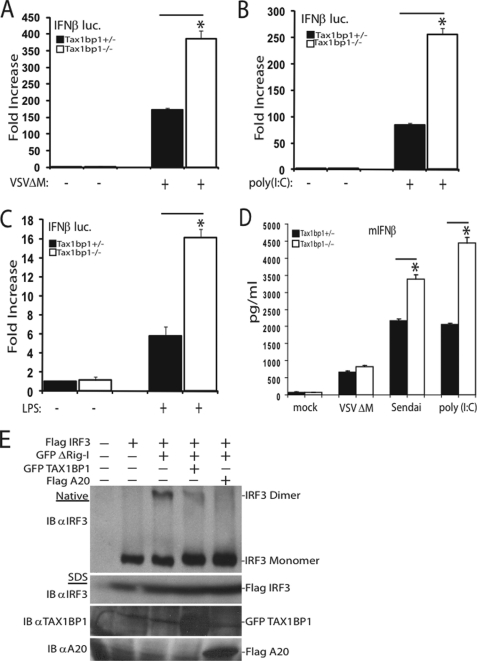
To determine whether TAX1BP1 inhibition of IFNβ was specific to virus infection, we examined other agonists capable of eliciting IRF3 activation. Our results demonstrate that TAX1BP1 also blocked poly(I-C) and lipopolysaccharide-mediated activation of IFNβ (supplemental Fig. 1, C and D). To confirm the effects of TAX1BP1 on the negative regulation of virus-mediated activation of IRF3, reporter assays measuring the activity of the IFNβ promoter were performed using siRNAs targeting TAX1BP1. Efficient knockdown of endogenous TAX1BP1 was confirmed by Western blotting (supplemental Fig. 1E). Knockdown of TAX1BP1 by siRNA enhanced virus-mediated activation of IFNβ (supplemental Fig. 1F). To further confirm the role of TAX1BP1 in blocking IRF3, IFN activation was measured in cells lacking TAX1BP1. Tax1bp1−/− MEFs exhibited enhanced levels of IFNβ promoter activation after virus infection, poly(I-C) treatment, or lipopolysaccharide treatment (Fig. 1, A–C). Furthermore, the inhibitory effects of TAX1BP1 were specific toward NF-κB and IRF3 transcription factors because NFAT activation was not inhibited by TAX1BP1 (supplemental Fig. 1G). To corroborate our findings that TAX1BP1 is a negative regulator of IRF3, IFNβ protein production was evaluated by ELISA in MEFs lacking TAX1BP1. Upon virus infection or poly(I-C) transfection, Tax1bp1−/− MEFs exhibited enhanced production of IFNβ compared with WT MEFs (Fig. 1D). To confirm that the increased production of IFNβ in Tax1bp1−/− MEFs was due specifically to a loss of TAX1BP1, Tax1bp1−/− MEFs were reconstituted with TAX1BP1 and evaluated for IFNβ production following virus infection. Knock-out MEFs reconstituted with TAX1BP1 were able to suppress virus-mediated production of IFNβ, indicating that the inhibition of IRF3 was indeed due to TAX1BP1 (supplemental Fig. 1H).
FIGURE 1.
TAX1BP1 is a negative regulator of antiviral signaling. A–C, Tax1bp1+/− or Tax1bp1−/− MEFs were transfected with an IFNβ luc reporter (200 ng) and pRL-tk (20 ng). Cells were virally infected (A), treated with poly(I-C) (25 μg/ml) (B), or treated with lipopolysaccharide (1 μg/ml) (C). IFNβ luciferase assays were performed 16 h after the respective stimulations. D, Tax1bp1+/− or Tax1bp1−/− MEFs were mock-infected, virally infected, or transfected with poly(I-C) (6 μg) for 16 h. Supernatants were subjected to IFNβ ELISA. E, 293T cells were transfected with the indicated plasmids (1 μg of FLAG-IRF3, GFP-TAX1BP1, or FLAG-A20 and 100 ng of GFP-ΔRig-I), and lysates were subjected to native PAGE (top) or SDS-PAGE (bottom) and immunoblotted (IB) with the indicated antibodies. Error bars, S.E.
Activation of IRF3 is mediated by its phosphorylation and subsequent dimerization. To determine if TAX1BP1 blocked IFNβ production by inhibiting IRF3 dimerization, we performed a native PAGE dimerization assay. TAX1BP1 expression inhibited IRF3 dimerization (Fig. 1E) and phosphorylation (supplemental Fig. 1I). Furthermore, knockdown of TAX1BP1 via siRNA enhanced IRF3 dimerization (supplemental Fig. 1J). Clearly, TAX1BP1 functions as a negative regulator of IFNβ.
TAX1BP1 Blocks IFNβ Activation by Targeting TBK1-IKKi
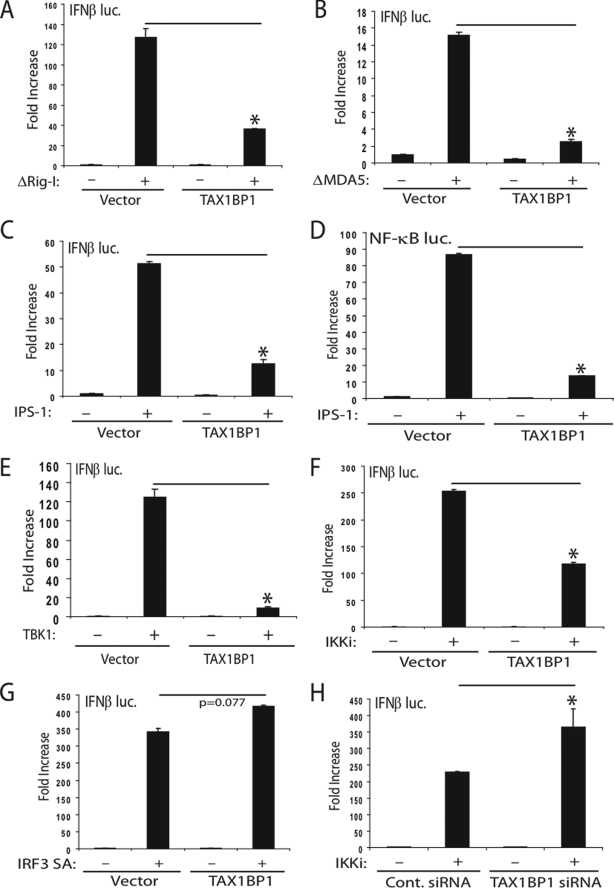
Next, we overexpressed key antiviral signaling molecules to determine where TAX1BP1 functions to block IFNβ. Rig-I and MDA5 are cytoplasmic sensors of viral nucleic acid that both contain RNA binding domains and CARD domains. Overexpression of the Rig-I CARD domain (ΔRig-I) potently activated the IFNβ reporter and was significantly blocked by TAX1BP1 (Fig. 2A). Consistently, ΔRig-I enhanced the activation of the IFNβ promoter in cells expressing TAX1BP1 siRNA (supplemental Fig. 2A). Similar results were obtained in TAX1BP1-deficient MEFs (supplemental Fig. 2B). Furthermore, reconstitution of TAX1BP1 in Tax1bp1−/− MEFs restored the suppression of ΔRig-I mediated activation of IFN (supplemental Fig. 2C). TAX1BP1 also blocked MDA5-mediated IFNβ activation (Fig. 2B). Both Rig-I and MDA5, via their N-terminal CARD domains, interact with IPS-1 to transduce signals to IRF3 and NF-κB. Overexpression of TAX1BP1 blocked IPS-1-mediated IRF3 (Fig. 2C) and NF-κB activation (Fig. 2D). Consistently, siRNA-mediated knockdown of TAX1BP1 further enhanced IPS-1-mediated activation of IRF3 (supplemental Fig. 2A). Next, we examined the effect of TAX1BP1 on TBK1-IKKi-mediated activation of IFNβ. TAX1BP1 overexpression blocked activation of the IFNβ reporter by TBK1-IKKi (Fig. 2, E and F) but not by a constitutively activated form of IRF3 (IRF3 SA) (Fig. 2G). Knockdown of TAX1BP1 by siRNA yielded a more pronounced activation of the IFNβ reporter in response to IKKi (Fig. 2H) or TBK1 (supplemental Fig. 2A) overexpression. TAX1BP1 also inhibited TBK1 activation of an interferon-stimulated response element promoter, recapitulating that TAX1BP1 is indeed a negative regulator of IFN (supplemental Fig. 2D). TLR3 and TLR4 activate IRF3 and NF-κB pathways via the adaptor TRIF. Overexpression of TAX1BP1 blocked TRIF-mediated IRF3 (supplemental Fig. 2E) and NF-κB activation (supplemental Fig. 2F). Conversely, knockdown of TAX1BP1 by siRNA resulted in enhanced TRIF-mediated IFNβ activation (supplemental Fig. 2A).
FIGURE 2.
TAX1BP1 inhibits IFNβ activation at the level of TBK1-IKKi. A–C, 293T cells were transfected with IFNβ luc reporter (100 ng) and pRL-tk (10 ng) plasmids together with ΔRig-I (A), ΔMDA5 (B), or IPS-1 (C) (1 μg) and a plasmid expressing TAX1BP1 (1 μg). D, 293T cells were transfected as described in C except an NF-κB luc reporter (100 ng) was used. E–G, 293T cells were transfected as in A–C using plasmids (1 μg each) encoding either TBK1 (E), IKKi (F), or a constitutively active form of IRF3 (IRF3 SA) (G) with or without TAX1BP1 DNA (1 μg). H, 293T cells were transfected with either control (Cont.) scrambled or TAX1BP1 siRNA. After 24 h, cells were transfected with IFNβ luc, pRL-tk, and IKKi plasmids as in F. Error bars, S.E.
As expected, A20 overexpression blocked IKKi-mediated activation of IRF3 (supplemental Fig. 2G), whereas A20 depletion resulted in enhanced IKKi-mediated IFNβ activation (supplemental Fig. 2H). Together, these results suggest that TAX1BP1 inhibits antiviral signaling upstream of IRF3, at the level of TBK1-IKKi. Previous studies have indicated that A20 also inhibits IRF3 activation by targeting TBK1-IKKi (21). Therefore, TAX1BP1 and A20 may cooperate to block virus-mediated activation of IRF3.
TAX1BP1 Interacts with TBK1-IKKi
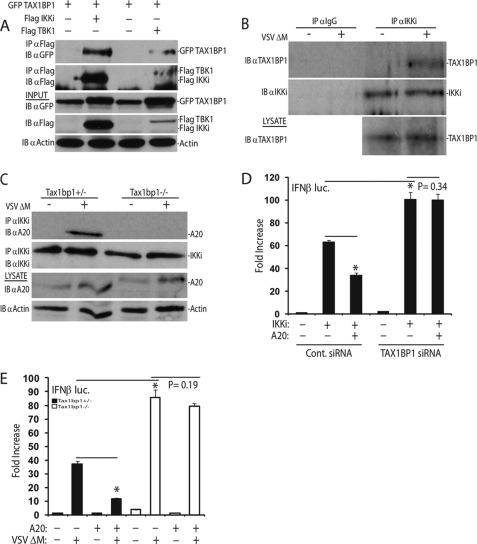
Because TAX1BP1 blocked signaling to the IFNβ promoter at the level of TBK1-IKKi, we next examined if TAX1BP1 interacted with these kinases. Co-IP experiments were performed in 293T cells transfected with epitope-tagged constructs of TAX1BP1, IKKi, and TBK1. As shown in Fig. 3A, TAX1BP1 interacted with both IKKi and TBK1. The interaction between TAX1BP1 and IKKi or TBK1 was confirmed by co-IPs using endogenous proteins. TAX1BP1 interaction with IKKi was entirely dependent upon virus infection (Fig. 3B). Low levels of TAX1BP1 were found to be persistently associated with TBK1, although the interactions were substantially increased upon virus infection (supplemental Fig. 3A). These results are similar to published data that demonstrated an A20 interaction with TBK1-IKKi upon overexpression (21) and are consistent with a recent screen identifying TBK1 as an A20-interacting protein (30).
FIGURE 3.
A20 requires TAX1BP1 to terminate antiviral signaling. A, 293T cells were transfected with the indicated plasmids (1 μg each). Co-IP and immunoblots were performed with the indicated antibodies. B, IKKi was immunoprecipitated from whole cell lysates derived from mock-infected and virally infected wild-type MEFs. Bound proteins were detected via immunoblotting, and lysates were probed as indicated. C, Tax1bp1+/− or Tax1bp1−/− MEFs were mock-infected or virally infected for 16 h, and lysates were subjected to immunoprecipitation and/or immunoblotting using the indicated antibodies. D, 293T cells were transfected with either control scrambled siRNA or TAX1BP1 siRNA. After 24 h, cells were transfected with IFNβ luc (100 ng) and pRL-tk (10 ng) DNA and plasmids encoding IKKi (1 μg) and A20 (200 ng) as indicated. E, Tax1bp1+/− or Tax1bp1−/− MEFs were transfected with an IFNβ luc reporter (200 ng) and pRL-tk (20 ng) and a plasmid encoding A20 (1 μg) as indicated. After 36 h, cells were either mock-infected or infected with virus for 16 h, followed by luciferase assays. IB, immunoblot; IP, immunoprecipitation. Error bars, S.E.
A20 Requires TAX1BP1 to Terminate Antiviral Signaling
Previous work from our laboratory and others has demonstrated that A20 requires TAX1BP1 to target and inactivate specific NF-κB regulatory proteins, including RIP1 and TRAF6 (23, 28). To assess whether A20 required TAX1BP1 to target IKKi or TBK1 for inactivation, we analyzed IKKi/TBK1-A20 interactions in Tax1bp1−/− MEFs. A20 interacted with IKKi in response to virus infection in control MEFs; however, in the absence of TAX1BP1, the IKKi-A20 interaction was lost (Fig. 3C). In addition, A20 interacted with TBK1 following poly(I-C) treatment in control MEFs but not in Tax1bp1−/− MEFs (supplemental Fig. 3B). Because TAX1BP1 was clearly required for the recruitment of A20 to IKKi and TBK1, we hypothesized that TAX1BP1 was essential for A20 to terminate antiviral signaling. Thus, we performed IFNβ reporter assays to determine if A20 required TAX1BP1 to exert its inhibitory function. As shown in Fig. 3D, A20 effectively blocked IKKi-mediated activation of IRF3 in the presence of control siRNA but not TAX1BP1 siRNA. Furthermore, A20-mediated inhibition of the IFNβ reporter in response to virus activation was impaired in TAX1BP1-deficient MEFs (Fig. 3E).
Previous studies from our laboratory have shown that TAX1BP1 recruits the E3 ligases Itch and RNF11 via two C-terminal zinc finger domains and associated PPXY motifs to terminate NF-κB signaling (25, 26). To determine if TAX1BP1 required its two zinc fingers or PPXY motifs to inhibit IRF3 activation, Tax1bp1−/− MEFs were reconstituted with either wild-type TAX1BP1 or TAX1BP1 containing point mutations in the zinc fingers or PPXY motifs. TAX1BP1 containing point mutations in either of its PPXY motifs or both zinc fingers could not suppress virus-mediated activation of an IFNβ reporter (supplemental Fig. 3C).
TAX1BP1 and A20 Inhibit TBK1-IKKi Lys63-linked Polyubiquitination
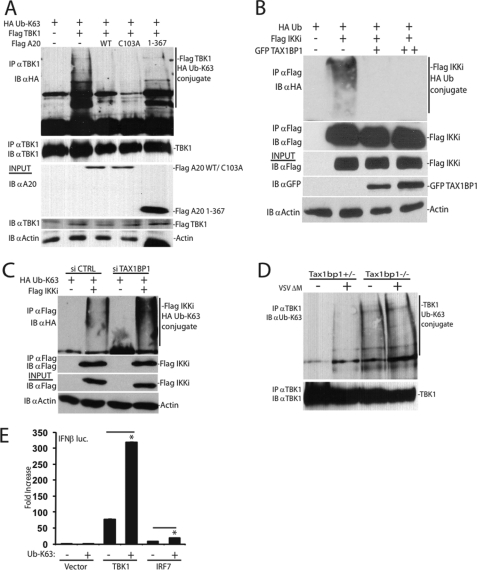
Saitoh et al. (21) have published that A20 inhibits IFNβ by targeting TBK1-IKKi; however, the mechanism of inhibition remains to be determined. A20 has been proposed to function as a ubiquitin-editing enzyme for RIP1 by cleaving Lys63-linked polyubiquitin chains and adding Lys48-linked polyubiquitin chains, ultimately resulting in the termination of NF-κB signaling (31, 32). Recent studies have also indicated that TBK1 and IKKi undergo polyubiquitination, which has been proposed to promote IRF3 activation (33, 34). Because A20 interacted with TBK1-IKKi, we next examined if TBK1-IKKi polyubiquitination was modulated by A20. Both TBK1 (Fig. 4A) and IKKi (supplemental Fig. 4A) when overexpressed were clearly polyubiquitinated via Lys63-linked ubiquitin chains. Interestingly, both wild-type A20 and an A20 DUB mutant (C103A) inhibited TBK1 and IKKi Lys63-linked polyubiquitination. However, an A20 deletion mutant (residues 1–367) lacking all of its C-terminal zinc fingers was unable to inhibit Lys63-linked polyubiquitination of TBK1 (Fig. 4A) and IKKi (supplemental Fig. 4A). Similar to A20, TAX1BP1 also inhibited IKKi polyubiquitination (Fig. 4B). In agreement with the above data, knockdown of TAX1BP1 using siRNA yielded enhanced IKKi polyubiquitination (Fig. 4C). To further corroborate the siRNA results, we evaluated endogenous levels of Lys63-specific linked ubiquitin chains conjugated onto TBK1 or IKKi in Tax1bp1−/− MEFs. Consistent with the above results, Lys63-linked polyubiquitination of endogenous TBK1 (Fig. 4D, compare lane 1 versus lane 3) or IKKi (supplemental Fig. 4B, compare lane 1 versus lane 3) was enhanced in MEFs lacking TAX1BP1. To gain better insight into the functional relevance of Lys63-linked polyubiquitination of TBK1-IKKi, we evaluated the effect of Lys63-linked ubiquitin overexpression in TBK1-mediated activation of IFNβ. As shown in Fig. 4E, Lys63-linked ubiquitin significantly enhanced the activation of IFN by TBK1. In the TNFR pathway, TAX1BP1 regulates A20 ubiquitin-editing function, resulting in the addition of Lys48-linked ubiquitin chains on RIP1 to trigger its proteasomal degradation (31). TAX1BP1, however, did not promote the degradation of TBK1, as determined by cycloheximide chase assays in Tax1bp1−/− MEFs, suggesting that it did not promote Lys48-linked polyubiquitination of TBK1 (supplemental Fig. 4C). Taken together, our results suggest that A20 and TAX1BP1 restrict antiviral signaling by inhibiting TBK1-IKKi Lys63-linked polyubiquitination.
FIGURE 4.
TAX1BP1 and A20 target TBK1-IKKi for deubiquitination. A, 293T cells were cotransfected with epitope-tagged constructs as indicated (1 μg of FLAG-TBK1, FLAG-A20 WT, FLAG-A20 C103A, and FLAG A20 1–367 and 500 ng of HA-Ub Lys63-only). Co-IPs and immunoblots evaluating TBK1 ubiquitination and immunoprecipitation were performed using the indicated antibodies. B, epitope-tagged constructs (1 μg of FLAG-IKKi, 500 ng of HA-Ub, and 1 or 2 μg of GFP-TAX1BP1) were transfected into 293T cells. Co-IPs and immunoblots evaluating IKKi ubiquitination were performed as in A. C, 293T cells were transfected with control scrambled siRNA or siRNA targeting TAX1BP1. After 24 h, cells were cotransfected with the indicated plasmids (1 μg of FLAG-IKKi and 500 ng of HA-Ub Lys63-only), and IKKi polyubiquitination was determined as in B. D, Tax1bp1+/− or Tax1bp1−/− MEFs were mock-infected or virally infected for 16 h, and lysates were subjected to immunoprecipitation and/or immunoblotting as indicated. E, 293T cells were cotransfected with IFNβ luc (100 ng) and pRL-tk (10 ng) DNA and plasmids encoding TBK1 or IRF7 (1 μg) and Lys63-only ubiquitin (500 ng) as indicated. WT, wild type; IB, immunoblot; IP, immunoprecipitation.
TRAF3 Is Required for TBK1-IKKi Ubiquitination
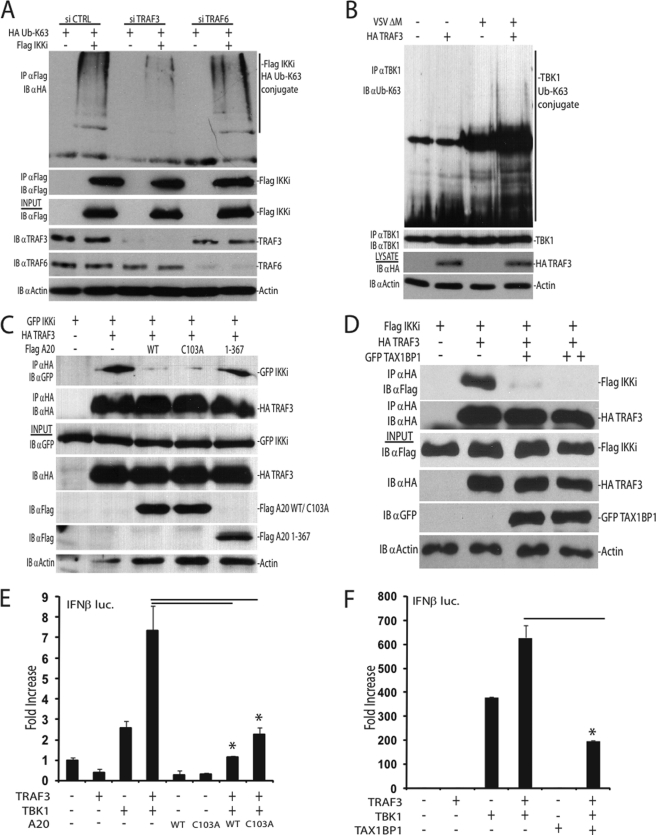
Because our results indicate that TAX1BP1 and A20 inhibit TBK1-IKKi ubiquitination, we next performed studies to identify molecules that regulate TBK1-IKKi ubiquitination. TRAF3 is a critical antiviral signaling molecule that has been proposed to function upstream of TBK1-IKKi (35–37). Because TRAF3 may also function as a ubiquitin ligase (38–40), we hypothesized that it may play a role in TBK1-IKKi ubiquitination. Using an siRNA approach to knock down TRAF3 expression, the ubiquitination status of IKKi was examined. Interestingly, upon TRAF3 knockdown, Lys63-linked polyubiquitination of IKKi was impaired (Fig. 5A). TRAF3-dependent ubiquitination of IKKi was specific because TRAF6 knockdown had little effect on IKKi ubiquitination (Fig. 5A). Furthermore, the endogenous Lys63-linked polyubiquitination of TBK1 was impaired in Traf3−/− MEFs (Fig. 5B). These results suggest that TRAF3 is critical for the polyubiquitination of TBK1-IKKi.
FIGURE 5.
A20 and TAX1BP1 disrupt a TRAF3-TBK1-IKKi signaling module. A, 293T cells were transfected with either control scrambled siRNA, TRAF3 siRNA, or TRAF6 siRNA. After 24 h, cells were transfected with plasmids as indicated (1 μg of FLAG-IKKi, 500 ng of HA-Ub Lys63-only). Cell lysates were subjected to immunoprecipitation and immunoblotting with the indicated antibodies. B, TRAF3-deficient MEFs were transfected with either empty vector DNA or a plasmid expressing TRAF3 (2.5 μg). After 36 h, MEFs were mock-infected or virally infected for 16 h. Lysates were subjected to immunoprecipitation and/or immunoblotting with the indicated antibodies. C and D, A20 and TAX1BP1 disrupt TRAF3-IKKi interactions. C, 293T cells were transfected with the indicated plasmids (1 μg of GFP-IKKi, HA-TRAF3, FLAG-A20 WT, FLAG-A20 C103A, or FLAG-A20 1–367). Whole cell lysates were subjected to co-IPs and/or immunoblotting with the indicated antibodies. D, 293T cells were cotransfected with HA-TRAF3 and FLAG-IKKi (1 μg each) and either 1 or 2 μg of GFP-TAX1BP1 plasmid DNAs as indicated. Lysates were subjected to co-IPs and immunoblotting as in C. E and F, 293T cells were transfected with IFNβ luc (100 ng) and pRL-tk (10 ng) DNA together with the indicated plasmids (1 μg each). WT, wild type; IB, immunoblot; IP, immunoprecipitation. Error bars, S.E.
TAX1BP1 and A20 Disrupt a TRAF3-TBK1-IKKi Complex
A20 has been shown previously to inhibit antiviral signaling independent of its DUB domain (20–22). In agreement with these findings, our results demonstrated that an A20 mutant impaired in DUB activity (C103A) was able to inhibit IKKi-mediated activation of IRF3 (supplemental Fig. 2G). Surprisingly, WT A20 and the A20 DUB mutant were equally capable of inhibiting TBK1 and IKKi polyubiquitination (Fig. 4A and supplemental Fig. 4A), suggesting that A20 inhibits TBK1-IKKi ubiquitination in a DUB-independent manner. Therefore, as an alternative mechanism of inhibition, we hypothesized that A20 may disrupt the interactions between TRAF3 and TBK1-IKKi, thus inhibiting TRAF3-dependent ubiquitination of TBK1-IKKi. Indeed, both WT A20 and the C103A mutant disrupted the binding between TRAF3 and IKKi, whereas an A20 mutant lacking C-terminal zinc fingers did not (Fig. 5C). Similarly, overexpression of TAX1BP1 also disrupted the interactions between TRAF3 and IKKi (Fig. 5D), whereas the absence of TAX1BP1 resulted in persistent TRAF3-TBK1 interactions (supplemental Fig. 5A). The disruption of TRAF3 and IKKi binding by TAX1BP1 was specific because TAX1BP1 had no effect on the binding between IKKi and IRF3 (supplemental Fig. 5B). Because A20 and TAX1BP1 disrupted a TRAF3-IKKi complex, we next determined the functional effects. Overexpression of TRAF3 alone is unable to induce IFNβ promoter activity; however, it synergizes with TBK1 or IKKi to enhance IFNβ activation (36). Therefore, we investigated if A20 or TAX1BP1 disrupts the synergy between TRAF3 and TBK1 in IFNβ reporter assays. Indeed, TRAF3 enhanced the activation of the IFNβ reporter by TBK1, and both A20 and TAX1BP1 inhibited the synergistic effect (Fig. 5, E and F). Collectively, these results reveal that TRAF3 is important for TBK1-IKKi ubiquitination and activation. Furthermore, A20 and TAX1BP1 function together to inhibit TBK1-IKKi polyubiquitination by antagonizing the interactions between TRAF3 and TBK1-IKKi.
DISCUSSION
Excessive production of type I interferons can be detrimental to host cells and must be tightly regulated by the innate arm of the immune system (3, 6). Indeed, host cells use a wide array of mechanisms to ensure that antiviral signaling pathways are activated transiently and are terminated when viral infections are cleared (41). Negative regulators, including NLRX1, SIKE, Atg5-Atg12, DAK, and CYLD, are constitutive or steady-state inhibitors of antiviral signaling, keeping aberrant IFN activation in check via mechanisms involving sequestration or competition (42–46). Other physiological regulators, such as RNF125, RNF5, Pin1, ISG15, ISG56, RBCK1, Ro52/TRIM21, gC1qR, DUBA, OTUB1/2, and optineurin, inhibit type I interferon production via negative feedback mechanisms (39, 40, 47–55). A20 is one such physiological inhibitor of IFN; however, its mechanism of action has remained elusive. A20 is a gene rapidly induced by tumor necrosis factor α, a key cytokine produced during virus infection (32, 41). Recent studies by our laboratory and others have indicated that A20 requires at least three additional subunits, TAX1BP1, Itch, and RNF11, to form a ubiquitin-editing complex that inhibits NF-κB signaling (23, 25, 26, 28). Although TAX1BP1 functions as an adaptor molecule for A20, the roles of Itch and RNF11 in the function of A20 are less clear. Nevertheless, we examined the role of TAX1BP1 and A20 in regulating antiviral signaling in this report.
Our results have clearly demonstrated that, similar to A20, TAX1BP1 is a negative regulator of antiviral signaling. Both TAX1BP1 and A20 interacted with the TBK1-IKKi kinases and inhibited antiviral signaling by targeting these two proteins. By Western blotting, we have observed a band shift of TAX1BP1 in the presence of either TBK1 or IKKi (Fig. 3A), suggesting that TAX1BP1 may be phosphorylated by these kinases. Similar results were observed for A20 co-expressed with TBK1-IKKi (21). Because A20 phosphorylation by IKKβ resulted in enhanced A20 effector function in the context of NF-κB signal regulation (56), it is tempting to speculate that A20 and/or TAX1BP1 phosphorylation by TBK1-IKKi may also modulate their function in antiviral signaling. Future studies will address this possibility. Although A20 has been previously demonstrated to interact with TBK1-IKKi, a satisfactory mechanism of inhibition of antiviral signaling by A20 has been lacking. TAX1BP1 probably functions as an adaptor molecule for A20 to terminate antiviral signaling because A20 recruitment to IKKi/TBK1 and A20-mediated inhibition of antiviral signaling was impaired in the absence of TAX1BP1 (Fig. 3, C–E). Because TAX1BP1 harbors a novel ubiquitin-binding motif (28), TAX1BP1 probably recruits A20 to ubiquitinated TBK1-IKKi that are targeted for inactivation. However, what determines the specificity toward ubiquitinated TBK1-IKKi by TAX1BP1 remains unclear because other components involved in IRF3 signal transduction are modified by Lys63-linked polyubiquitin chains (41). TAX1BP1 harboring point mutations in its PPXY motifs or zinc fingers was defective in blocking virus-mediated IFN activation (supplemental Fig. 3C). Because these motifs are essential in protein-protein interactions, it is plausible that other, yet to be identified proteins may participate with TAX1BP1 and A20 to terminate IRF3 signaling.
Lys63-linked polyubiquitination of key substrates in IRF3/7 signaling has critical implications in activating type I interferons (32, 38, 41, 46, 57, 58). Our results indicated that A20 and TAX1BP1 counteract TBK1-IKKi Lys63-linked polyubiquitination (Fig. 4, A–C). Interestingly, both TBK1 and IKKi were polyubiquitinated under basal conditions in Tax1bp1−/− MEFs, whereas in control MEFs, their ubiquitination occurred only upon virus infection (Fig. 4D and supplemental Fig. 4B). These results may be explained by the persistent TRAF3-TBK1 (E3-substrate) association in Tax1bp1−/− MEFs, as opposed to interactions dependent on virus infections in wild-type MEFs (supplemental Fig. 5A). Because Tax1bp1−/− MEFs displayed high levels of basal Lys63-linked polyubiquitination of TBK1 that may promote IRF3 activation (Fig. 4E), it is surprising that Tax1bp1−/− MEFs did not spontaneously activate IFN (Fig. 1, A–D). We hypothesize that Lys63-linked polyubiquitination of TBK1-IKKi is necessary but not sufficient to activate IFN. Experiments utilizing knock-out cells reconstituted with TBK1-IKKi containing lysine substitutions in their ubiquitin acceptor sites will be important in understanding the physiological roles of TBK1-IKKi ubiquitination. Although TBK1-IKKi Lys63-linked polyubiquitination was inhibited by TAX1BP1 and A20, TAX1BP1 did not trigger the degradation of TBK1 (supplemental Fig. 4C), consistent with a report indicating that A20 does not degrade either TBK1 or IKKi (22). Surprisingly, A20-mediated inhibition of IKKi/TBK1 polyubiquitination was independent of its DUB function (Fig. 4A and supplemental Fig. 4A). Previous studies have demonstrated that A20 blocked antiviral signaling independently of its DUB domain, where either an A20 deletion mutant lacking the OTU domain or a point mutant (C103A) deficient in DUB function still blocked IRF3 activation (20–22). Our results are in agreement with these data because the A20 C103A mutant blocked IKKi-mediated activation of IRF3 as effectively as the wild-type A20 (supplemental Fig. 2G) and inhibited TBK1-IKKi polyubiquitination as robustly as wild-type A20. Consistent with a new report detailing a mechanism by which A20 terminates NF-κB signaling via a similar fashion (59), our results clearly suggest that A20 inhibits antiviral signaling via a DUB-independent mechanism. Instead, our results indicate that inhibition of TBK1-IKKi ubiquitination is dependent on the C-terminal zinc finger domains (Fig. 4A and supplemental Fig. 4A), known to be important for TAX1BP1 interaction and E3 ligase activity (31, 60). This novel DUB-independent mechanism of inhibition employed by A20 contrasts with mechanisms utilized by other deubiquitinating enzymes involved in terminating antiviral signaling. Indeed, inhibition of IFN via the deubiquitinases CYLD and DUBA require their DUB domains/catalytic cysteine residues to deubiquitinate their cognate substrates (39, 46).
To further dissect the mechanism of inhibition of antiviral signaling by A20 and TAX1BP1, we focused on TBK1-IKKi ubiquitination. Our results indicated that TRAF3 was required for the Lys63-linked polyubiquitination of TBK1-IKKi (Fig. 5, A and B). Previous studies have demonstrated the importance of TRAF3 in restricting antiviral signaling (35, 36). However, it is not clear from our studies if TRAF3 directly ubiquitinates TBK1-IKKi or functions as an adaptor for another E3 ligase. In this context, a recent study has demonstrated that the ubiquitin ligase NRDP1 functions as an E3 ligase for TBK1 (34). It is possible that TRAF3 and NRDP1 function together to promote TBK1-IKKi ubiquitination. Nevertheless, we found that TAX1BP1 and A20 specifically disrupted the interactions between TRAF3 and IKKi (Fig. 5, C and D) but not IPS-1 and TRAF3 (data not shown) or IKKi and IRF3 (supplemental Fig. 5B). This further indicates that IRF3 interaction with IKKi does not require IKKi ubiquitination, which was blocked by TAX1BP1. Furthermore, A20 and TAX1BP1 inhibited the synergy between TRAF3, TBK1, and IKKi in activating IFN (Fig. 5, E and F). Therefore, it is plausible that loss of TRAF3 binding to IKKi prevents the Lys63-linked polyubiquitination of IKKi and perhaps further unmasks a potential mechanism by which TRAF3 synergizes with these kinases, ultimately underscoring the critical importance of TRAF3 ubiquitin ligase function in antiviral signaling. In summary, we have identified a novel DUB-independent function of A20 in antiviral signaling, where A20 disrupts the interactions between an E3 ligase and its cognate substrates.
Acknowledgments
We are grateful to Drs. John Bethea and Genhong Cheng for providing the TRAF3-deficient MEFs, Dr. Khaled Tolba for 293-TLR4 cells, Dr. Shao-Cong Sun for the HA-TRAF3 plasmid, Dr. Rudi Beyaert and Belgian Coordinated Collections of Microorganisms/Laboratory of Molecular Biology of Ghent University (BCCMTM/LMBP) for the FLAG-A20 1–367 plasmid, Joshua Heiber for purified VSV, and Dr. Hiroki Ishikawa for critical discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1GM083143 and P01CA128115.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- TLR
- Toll-like receptor
- DUB
- deubiquitination
- luc
- luciferase
- IKK
- IκB kinase(s)
- MEF
- mouse embryo fibroblast
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- Ub
- ubiquitin
- ELISA
- enzyme-linked immunosorbent assay
- IP
- immunoprecipitation
- IFN
- interferon
- siRNA
- small interfering RNA.
REFERENCES
- 1.Kawai T., Akira S. (2008) Ann. N.Y. Acad. Sci. 1143, 1–20 [DOI] [PubMed] [Google Scholar]
- 2.Hiscott J., Grandvaux N., Sharma S., Tenoever B. R., Servant M. J., Lin R. (2003) Ann. N.Y. Acad. Sci. 1010, 237–248 [DOI] [PubMed] [Google Scholar]
- 3.Sadler A. J., Williams B. R. (2008) Nat. Rev. Immunol. 8, 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medzhitov R. (2007) Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 5.Honda K., Taniguchi T. (2006) Nat. Rev. Immunol. 6, 644–658 [DOI] [PubMed] [Google Scholar]
- 6.Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F. J., Sjöstrand M., Eloranta M. L., Ní Gabhann J., Winqvist O., Sundelin B., Jefferies C. A., Rozell B., Kuchroo V. K., Wahren-Herlenius M. (2009) J. Exp. Med. 206, 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng L., Yu C., Zhang Z., Yang C., Cai X. (2009) Oral Surg. Oral Med. Oral Pathol. Oral. Radiol. Endod. 107, 661–668 [DOI] [PubMed] [Google Scholar]
- 8.Vereecke L., Beyaert R., van Loo G. (2009) Trends Immunol. 30, 383–391 [DOI] [PubMed] [Google Scholar]
- 9.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 10.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 11.Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 12.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 13.Wang Y. Y., Liu L. J., Zhong B., Liu T. T., Li Y., Yang Y., Ran Y., Li S., Tien P., Shu H. B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michallet M. C., Meylan E., Ermolaeva M. A., Vazquez J., Rebsamen M., Curran J., Poeck H., Bscheider M., Hartmann G., König M., Kalinke U., Pasparakis M., Tschopp J. (2008) Immunity 28, 651–661 [DOI] [PubMed] [Google Scholar]
- 15.Balachandran S., Thomas E., Barber G. N. (2004) Nature 432, 401–405 [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa H., Barber G. N. (2008) Nature 455, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong B., Yang Y., Li S., Wang Y. Y., Li Y., Diao F., Lei C., He X., Zhang L., Tien P., Shu H. B. (2008) Immunity 29, 538–550 [DOI] [PubMed] [Google Scholar]
- 18.Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., Fujita T. (1998) EMBO J. 17, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y. Y., Li L., Han K. J., Zhai Z., Shu H. B. (2004) FEBS Lett. 576, 86–90 [DOI] [PubMed] [Google Scholar]
- 21.Saitoh T., Yamamoto M., Miyagishi M., Taira K., Nakanishi M., Fujita T., Akira S., Yamamoto N., Yamaoka S. (2005) J. Immunol. 174, 1507–1512 [DOI] [PubMed] [Google Scholar]
- 22.Lin R., Yang L., Nakhaei P., Sun Q., Sharif-Askari E., Julkunen I., Hiscott J. (2006) J. Biol. Chem. 281, 2095–2103 [DOI] [PubMed] [Google Scholar]
- 23.Shembade N., Harhaj N. S., Liebl D. J., Harhaj E. W. (2007) EMBO J. 26, 3910–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balachandran S., Venkataraman T., Fisher P. B., Barber G. N. (2007) J. Immunol. 178, 2429–2439 [DOI] [PubMed] [Google Scholar]
- 25.Shembade N., Harhaj N. S., Parvatiyar K., Copeland N. G., Jenkins N. A., Matesic L. E., Harhaj E. W. (2008) Nat. Immunol. 9, 254–262 [DOI] [PubMed] [Google Scholar]
- 26.Shembade N., Parvatiyar K., Harhaj N. S., Harhaj E. W. (2009) EMBO J. 28, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faria P. A., Chakraborty P., Levay A., Barber G. N., Ezelle H. J., Enninga J., Arana C., van Deursen J., Fontoura B. M. (2005) Mol. Cell 17, 93–102 [DOI] [PubMed] [Google Scholar]
- 28.Iha H., Peloponese J. M., Verstrepen L., Zapart G., Ikeda F., Smith C. D., Starost M. F., Yedavalli V., Heyninck K., Dikic I., Beyaert R., Jeang K. T. (2008) EMBO J. 27, 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee E. G., Boone D. L., Chai S., Libby S. L., Chien M., Lodolce J. P., Ma A. (2000) Science 289, 2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M. (2004) Nature 430, 694–699 [DOI] [PubMed] [Google Scholar]
- 32.Bhoj V. G., Chen Z. J. (2009) Nature 458, 430–437 [DOI] [PubMed] [Google Scholar]
- 33.Friedman C. S., O'Donnell M. A., Legarda-Addison D., Ng A., Cárdenas W. B., Yount J. S., Moran T. M., Basler C. F., Komuro A., Horvath C. M., Xavier R., Ting A. T. (2008) EMBO Rep. 9, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C., Chen T., Zhang J., Yang M., Li N., Xu X., Cao X. (2009) Nat. Immunol. 10, 744–752 [DOI] [PubMed] [Google Scholar]
- 35.Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Häcker G., Mann M., Karin M. (2006) Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
- 36.Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006) Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 37.Saha S. K., Pietras E. M., He J. Q., Kang J. R., Liu S. Y., Oganesyan G., Shahangian A., Zarnegar B., Shiba T. L., Wang Y., Cheng G. (2006) EMBO J. 25, 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatot J. S., Gioia R., Chau T. L., Patrascu F., Warnier M., Close P., Chapelle J. P., Muraille E., Brown K., Siebenlist U., Piette J., Dejardin E., Chariot A. (2007) J. Biol. Chem. 282, 31131–31146 [DOI] [PubMed] [Google Scholar]
- 39.Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O'Rourke K. M., Eby M., Pietras E., Cheng G., Bazan J. F., Zhang Z., Arnott D., Dixit V. M. (2007) Science 318, 1628–1632 [DOI] [PubMed] [Google Scholar]
- 40.Li S., Zheng H., Mao A. P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., Shu H. B. (2010) J. Biol. Chem. 285, 4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komuro A., Bamming D., Horvath C. M. (2008) Cytokine 43, 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore C. B., Bergstralh D. T., Duncan J. A., Lei Y., Morrison T. E., Zimmermann A. G., Accavitti-Loper M. A., Madden V. J., Sun L., Ye Z., Lich J. D., Heise M. T., Chen Z., Ting J. P. (2008) Nature 451, 573–577 [DOI] [PubMed] [Google Scholar]
- 43.Huang J., Liu T., Xu L. G., Chen D., Zhai Z., Shu H. B. (2005) EMBO J. 24, 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K. Q., Ishii K. J., Kawai T., Akira S., Suzuki K., Okuda K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14050–14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diao F., Li S., Tian Y., Zhang M., Xu L. G., Zhang Y., Wang R. P., Chen D., Zhai Z., Zhong B., Tien P., Shu H. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11706–11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M., Wu X., Lee A. J., Jin W., Chang M., Wright A., Imaizumi T., Sun S. C. (2008) J. Biol. Chem. 283, 18621–18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M. J., Hwang S. Y., Imaizumi T., Yoo J. Y. (2008) J. Virol. 82, 1474–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arimoto K., Takahashi H., Hishiki T., Konishi H., Fujita T., Shimotohno K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7500–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong B., Zhang L., Lei C., Li Y., Mao A. P., Yang Y., Wang Y. Y., Zhang X. L., Shu H. B. (2009) Immunity 30, 397–407 [DOI] [PubMed] [Google Scholar]
- 50.Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K. P., Yamaoka S. (2006) Nat. Immunol. 7, 598–605 [DOI] [PubMed] [Google Scholar]
- 51.Li Y., Li C., Xue P., Zhong B., Mao A. P., Ran Y., Chen H., Wang Y. Y., Yang F., Shu H. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7945–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M., Tian Y., Wang R. P., Gao D., Zhang Y., Diao F. C., Chen D. Y., Zhai Z. H., Shu H. B. (2008) Cell Res. 18, 1096–1104 [DOI] [PubMed] [Google Scholar]
- 53.Higgs R., Ní Gabhann J., Ben Larbi N., Breen E. P., Fitzgerald K. A., Jefferies C. A. (2008) J. Immunol. 181, 1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu L., Xiao N., Liu F., Ren H., Gu J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mankouri J., Fragkoudis R., Richards K. H., Wetherill L. F., Harris M., Kohl A., Elliott R. M., Macdonald A. (2010) PLoS Pathog. 6, e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutti J. E., Turk B. E., Asara J. M., Ma A., Cantley L. C., Abbott D. W. (2007) Mol. Cell. Biol. 27, 7451–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huye L. E., Ning S., Kelliher M., Pagano J. S. (2007) Mol. Cell. Biol. 27, 2910–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao A. P., Li S., Zhong B., Li Y., Yan J., Li Q., Teng C., Shu H. B. (2010) J. Biol. Chem. 285, 9470–9476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shembade N., Ma A., Harhaj E. W. (2010) Science 327, 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Valck D., Jin D. Y., Heyninck K., Van de Craen M., Contreras R., Fiers W., Jeang K. T., Beyaert R. (1999) Oncogene 18, 4182–4190 [DOI] [PubMed] [Google Scholar]